
A pan-cancer analysis of the oncogenic role of Keratin 17 (KRT17) in human tumors
Introduction
Given the complication of tumorigenesis, analyzing the expression of any related gene and evaluating its correlation with potential molecular mechanisms and clinical prognosis is crucial (1). The Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) public databases contain numerous functional genomics datasets of different tumors, which can be used to carry out the relevant pan-cancer analysis (2,3).
With the increased research into tumor-targeted therapy, the effects of the keratin family in human tumors have been studied consistently, and have identified the involvement of Keratin 1 (KRT1) (4), KRT15 (5), and KRT16 (6) in tumor metastasis. KRT is a protein family that is critical for hair formation and is abundant in the outer layer of the skin, thus protecting epithelial cells from damage. Previous studies illustrated that KRT17 is overexpressed in many cancers, including cervical cancer (7), gastric cancer (8), and lung cancer (9). Li et al. found that KRT17 was overexpressed in pancreatic cancer, and the knockdown of KRT17 significantly inhibited the proliferation, migration and invasion of pancreatic cancer cells, and found that its specific mechanism was related to the activation of mTOR pathway (10). In our study, KRT17 was found to belong to type I keratin, which is abundantly found in epithelial basal cells (11). As a multifaceted cytoskeletal protein, KRT17 can regulate many biological processes (BP), including cell proliferation and growth, skin inflammation, and hair follicle circulation (12). The mechanism of abnormal overexpression and function of KRT17 has been observed in numerous diseases, including psoriasis, malignant tumors [such as breast cancer (13), cervical cancer (14), oral squamous cell carcinoma (15), and gastric cancer], etc. However, at present, there is no report on the pan-cancer analysis between KRT17 and various tumors. Herein, we discovered the association between KRT17 and different types of cancer based on current cell or animal experiments.
The most immediate value of pan-cancer analysis may be to drive a shift from cancer pathology to molecular diagnosis, making the classification of different types of cancer more relevant to therapeutic mechanisms. Although the cost of testing will fall further and the technology will spread, it will at least give cancer research a new direction. For example, whether clinical treatment or scientific research design in the future, it will not simply say lung cancer, liver cancer, etc., but a certain type of cancer with molecular mechanism. In our study, we performed a pan-cancer analysis of KRT17 using TCGA and GEO databases. At the same time, our study also included other information, such as gene expression and changes, survival status, immune infiltration, protein phosphorylation, and possible related cellular mechanisms, so as to further investigate the potential molecular mechanism of KRT17 in the clinical prognosis or pathogenesis of various types of cancer. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2118).
Methods
Gene expression analysis
We used the “Gene DE” module of the Tumor Immune Estimation Resource, version 2 (TIMER2) (http://timer.cistrome.org/) to search for differences in the level of KRT17 between tumors and nearby normal tissues in specific tumor subtypes or different tumors in TCGA database. Using the “Expression analysis-Box Plots” module of the Gene Expression Profiling Interactive Analysis, version 2 (GEPIA2) webserver (http://gepia2.cancer-pku.cn/#analysis) (16), box plots were obtained based on the different expression between tumors that avoided normal cells or had severely limited normal tissues [such as TCGA-glioblastoma multiforme (TCGA-GBM), TCGA-acutemyeloid (TCGA-LAML)] under the settings of log2 fold change (FC) cutoff =1, P value cutoff =0.01, and “Match TCGA normal and GTEx data”. We also used GEPIA2’s “Pathological Stage Plot” module to obtain violin plots for the expression of KRT17 in various pathological stages (stages IV, III, II, I, and 0) of all TCGA tumors. For the box and violin plots, we used transformed expression data of log2 [transcripts per million (TPM) +1].
We analyzed the Clinical Proteomic Tumor Analysis Consortium (CPTAC) protein expression dataset using the UALCAN portal (http://ualcan.path.uab.edu/analysis-prot.html), which is an interactive web resource to study cancer omics data (17). By typing in “KRT17”, we were compared the expression levels of total protein and phosphoprotein (with phosphorylation at the S13, S24, S32, and S39 sites) of KRT17 (NP 000413.1) in primary tumors and normal tissues, respectively. Four tumor datasets were selected: colon cancer, ovarian cancer, lung adenocarcinoma (LUAD), and uterine corpus endometrial carcinoma (UCEC). The Tumor Hospital Affiliated to Nantong University’s Ethics Committee (Nantong, China) approved the study. This research was carried out in accordance with the Helsinki Declaration (as revised in 2013).
Survival prognosis analysis
Using GEPIA2’s “Survival Map” module, we obtained the disease-free survival (DFS) and overall survival (OS) significance map data of KRT17 across all TCGA tumors (16).
Low (50%) and high (50%) cutoff expression thresholds were used to divide the low- and high-expression cohorts. The hypothesis was tested using the log-rank test, while GEPIA2’s “Survival Analysis” module was used for the survival plots.
Genetic alteration analysis
Using the cBioPortal web (https://www.cbioportal.org/) (18,19), we selected the “TCGA Pan-cancer Atlas Studies” option in the “Quick select” section, and entered “KRT17” to search for KRT17 genetic alteration features. The “Cancer Types Summary” module displayed the results of the type of mutation, alteration frequency, and copy number alteration (CNA) among all TCGA tumors. Additionally, we utilized the “Comparison” module to obtain information about the OS, DFS, and progression-free survival (PFS) for the cases of TCGA cancer with and without KRT17 genetic mutations. Kaplan-Meier graphs with log-rank P values were also constructed.
Immune infiltration analysis
Using TIMER2’s “Immune-Gene” module, we explored the connection between immune infiltrates and KRT17 expression across all tumors of TCGA. The immune cells of cancer-associated fibroblasts were selected. CIBERSORT, TIMER, XCELL, CIBERSORT-ABS, MCPCOUNTER, QUANTISEQ, and EPIC algorithms were applied to estimate immune infiltration. The purity-adjusted Spearman’s rank correlation test was utilized to calculate the P values and partial correlation (cor) values. A scatter plot and heatmap were used to illustrate the data.
KRT17-related gene enrichment analysis
We used a single protein name (“KRT17”) and organism (“Homo sapiens”) to search the STRING database (https://string-db.org/). The following basic parameters were then set: (I) the lowest required interaction score [“low confidence (0.150)”]; (II) the maximum number of interactors to show (“no more than 50 interactors” in the first shell); (III) the meaning of network edges (“evidence”); and (IV) active interaction sources (“experiments”). Finally, the KRT17-binding proteins that could be found experimentally were obtained.
According to the normal tissues and all TCGA tumors datasets, GEPIA2’s module of “Similar Gene Detection” was employed to identify the top 100 KRT17-correlated targeting genes. A Pearson correlation study of selected genes and pairwise genes of KRT17 was also performed using GEPIA2’s “correlation analysis” module. For the dot plot, the log2 TPM was used. The correlation coefficient (R) and the P value were shown. We also used TIMER2’s “Gene_corr” module to provide heatmap data for the selected genes, including P value and partial correlation (cor) from the purity-adjusted Spearman’s rank correlation test.
An intersection study was conducted through Jvenn, an interactive Venn diagram viewer (20) to compare KRT17-binding and interacting genes. Furthermore, we joined the two datasets to conduct Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. We collected the data for the functional annotation chart by uploading the gene lists to the Database for Visualization, Integrated Discovery, and Annotation (DAVID) with the selected species (“Homo sapiens”) and identifier (“OFFICIAL GENE SYMBOL”). Finally, the enriched pathways were displayed using the R packages “tidyr (tidyr_1.1.3 version)” and “ggplot2 (3.3.5 version)”. We also used the R tool “clusterProfiler” to perform Gene Ontology (GO) enrichment analysis. Using the cnetplot function (colorEdge = T, circular = F, node label = T), the data for cellular component (CC), BP, and molecular function (MF) were shown as cnetplots. This research was conducted using the R programming language (R-3.6.3, 64-bit) (https://www.r-project.org/). P<0.05 with two-tailed significance was considered statistically significant.
Statistical analysis
Data analysis was conducted using SPSS 23.0. The data were expressed as mean ± standard deviation () and examined via t-test. Enumeration data were denoted by percentage (%), and the differences were compared by χ2 test. P<0.05 was considered statistically significant.
Results
Gene expression analysis
The TIMER2 method was used to examine the expression status of KRT17 among TCGA cancer types. According to Figure 1A, the level of KRT17 expression in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (P<0.01), cholangiocarcinoma (CHOL) (P<0.001), esophageal carcinoma (ESCA) (P<0.001), colon adenocarcinoma (COAD) (P<0.001), head and neck squamous cell carcinoma (HNSC) (P<0.001), LUAD (P<0.001), liver hepatocellular carcinoma (LIHC) (P<0.001), lung squamous cell carcinoma (LUSC) (P<0.001), pheochromocytoma and paraganglioma (PCPG) (P<0.05), rectum adenocarcinoma (READ) (P<0.001), stomach adenocarcinoma (STAD) (P<0.01), thyroid carcinoma (THCA) (P<0.01), and UCEC (P<0.01) was higher than their corresponding control tissues.
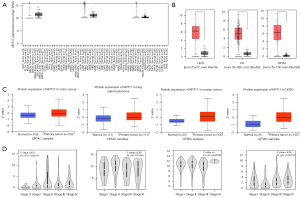
We also investigated variations in the expression of KRT17 between normal and tumor tissues of uterine carcinosarcoma (UCS), ovarian serous cystadenocarcinoma (OV), and thymoma (THYM) after being included the normal tissue as a control obtained from the GTEx dataset (Figure 1B) (P<0.01).
The CPTAC dataset revealed that primary tissues of LUAD, colon and ovarian cancers, and UCEC had a higher level of KRT17 than normal tissues (Figure 1C) (P<0.001).
We also utilized GEPIA2’s “Pathological Stage Plot” module to search for a connection between KRT17 expression and cancer pathological stages, such as skin cutaneous melanoma (SKCM), ESCA, LUSC, and THCA (Figure 1D) (all P<0.05).
Survival analysis data
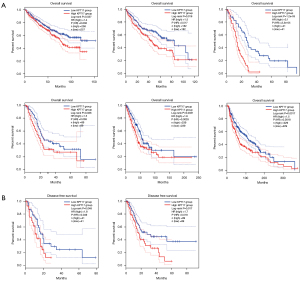
The cancer cases were separated into high- and low-expression groups based on the level of KRT17 expression, and the relationship between KRT17 expression and patient prognosis (in those suffering from various malignancies) was evaluated, primarily utilizing the TCGA and GEO databases, respectively. Within TCGA project, an elevated level of KRT17 was associated with poor OS in SKCM, pancreatic adenocarcinoma (PAAD), mesothelioma (MESO), LUAD, LIHC, and kidney renal clear cell carcinoma (KIRC) tumors (Figure 2A) (all P<0.05). The DFS analysis data (Figure 2B) exhibited an association between high KRT17 expression and a poor prognosis for TCGA cases of MESO (P=0.048) and PAAD (P=0.017).
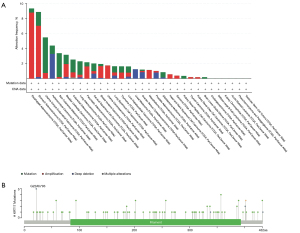
Genetic alteration data analysis
The genetic modification status of KRT17 was detected in several tumor samples from TCGA cohorts. KRT17 did not mutate in all cancer patients (<8%), as indicated in Figure 3A. In cases of esophageal adenocarcinoma and STAD (>5%), the “amplification” form of CNA was the most common. In Figure 3B, the types, locations, and case numbers of the KRT17 genetic mutations are further described. We observed that amplification of KRT17 was the main type of mutation, and G22Afs*93 is the main locus (Figure 3B).
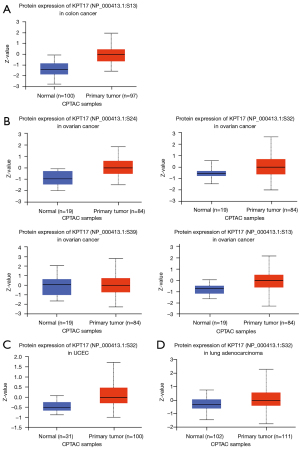
Protein phosphorylation data analysis
We also investigated how the levels of KRT17 phosphorylation differed between normal and primary tumor tissues. Four types of cancers (colon and ovarian cancers, UCEC, and LUAD) were studied using the CPTAC dataset. The KRT17 phosphorylation sites and substantial changes are summarized in Figure 4. We investigated the expression levels of KRT17 (NP 000413.1) total protein and phosphoprotein (with phosphorylation at the S13, S24, S32, and S39 sites) in primary tumor and normal tissues, respectively.
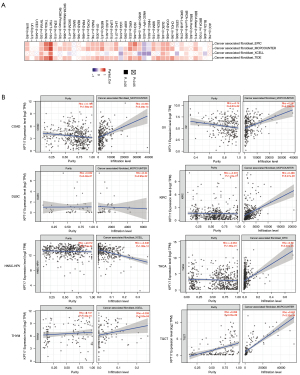
Immune infiltration data analysis
Cancer initiation, development, and metastasis are associated with tumor-infiltrating immune cells and are considered an important module of the tumor microenvironment (21,22). Cancer-associated fibroblasts have been shown to modulate the function of several tumor-infiltrating immune cells (23,24). Moreover, for the TCGA tumors of COAD, lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), KIRC, OV, testicular germ cell tumors (TGCT), THCA, and THYM, we found a statistically significant positive association between the expression of KRT17 and the estimated value of infiltration of cancer-associated fibroblasts, but a negative correlation for HNSC [human papillomavirus (HPV)−] (Figure 5).
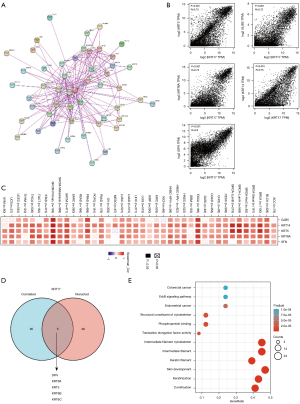
Enrichment analysis of KRT17-related partners
We attempted to screen out the genes that are correlated with KRT17 expression and targeting proteins (which are capable of binding with KRT17) for a series of pathway enrichment analyses to obtain a more precise understanding regarding the KRT17 molecular mechanism in tumorigenesis. Here, using the STRING program, we identified 50 KRT17-binding proteins, all of which were confirmed by scientific evidence, and their interaction networks are depicted in Figure 6A. We combined all the expression data of TCGA tumors with the GEPIA2 algorithm to identify the top 100 KRT17 expression-associated genes. According to Figure 6B, the expression level of KRT17 exhibited a positive correlation with gap junction protein beta 5 (GJB5) (R=0.72), KRT5 (R=0.73), KRT6A (R=0.75), KRT14 (R=0.75), and stratifin (SFN) (R=0.8) (all P<0.05). In the majority of described cancer types, the heatmap data revealed a positive connection between KRT17 and the top five genes (Figure 6C). SFN, KRT6A, KRT5, KRT6B, and KRT6C were identified as common members of the two groups after intersection analysis (Figure 6D). To conduct KEGG and GO enrichment analysis, we merged the two datasets. According to the KEGG data in Figure 6E, the “intermediate filament cytoskeleton” and “keratinization” may be involved along with KRT17 in tumor pathogenesis.
Discussion
Numerous studies have demonstrated the involvement of the KRT17 protein in many biological events, such as gene transcription regulation, subcellular localization (10), cell cycle (12), glycolysis (25), phosphorylation (13), ubiquitination (26), etc. A functional relationship between KRT17 and clinical disorders, particularly malignancies, has been also identified in recent publications (8,13-15). However, it remains unclear whether KRT17 plays a role in the development of many cancers via general processes. After conducting a literature search, we were unable to find any pan-cancer research on KRT17 from the standpoint of overall tumors. Thus, we investigated the KRT17 gene in 33 distinct tumors using the TCGA, CPTAC, and GEO databases, as well as molecular parameters such as gene expression, genetic alteration, protein phosphorylation, and DNA methylation.
KRT17 is highly expressed in numerous tumor tissues. In our study, we found that KRT17 is also highly expressed in UCS, OV, THYM, colon cancer, LUAD, ovarian cancer, and uterine corpus endocrinoma. However, the effect of KRT17 on the survival and prognosis of different tumors is not the same. In our study, we found positive correlations between the low expression of KRT17 and better OS in SKCM, PAAD, MESO, LUAD, LIHC and KIRC, as well as with the better DFS of MESO and PAAD. Moreover, the expression of KRT17 varied among the different tumor stages. For example, in SKCM and ESCA, it was mainly distributed in stage II tumors, while in LUSC and THCA, it was least distributed in stage III and stage II, respectively. This outcome could be influenced by different data processing or updated survival information. Thus, we further used, OncoLnc (http://www.oncolnc.org/) and the Kaplan Meier plotter method (27,28) to further update and analyze these data, the final results were consistent with the results of our study.
Further analysis of gene mutation showed that KRT17 did not mutate in all tumors (<8%), so there was no corresponding mutant. Interestingly, we found that the “amplification” type of CNA was the most common in esophageal adenocarcinoma cases and STAD cases (>5%). We then further studied the case number sites and types of KRT17 genetic mutation and observed that amplification of KRT17 was the main type of mutation, and G22Afs*93 is the main locus.
In terms of phosphoprotein and total protein, we used the CPTAC dataset to explore the KRT17 molecular mechanism in colon cancer, ovarian cancer, UCEC, and LUAD. Here, we looked at the levels of expression of KRT17 (NP 000413.1) total protein and phosphoprotein (with phosphorylation at the S13, S24, S32, and S39 sites) in primary tumor and normal tissues. Interestingly, ovarian cancer had all four phosphorylation sites. It has been reported that KRT17 affects cell migration, proliferation, and invasion, which was associated with pancreatic cancer by affecting the level of phosphorylation of S6K1 (10). Therefore, in the future, we may be able to influence the prognosis of these tumors through appropriate intervention of phosphorylation sites. However, we still do not rule out that the changes of phosphorylation sites are by-products of dysfunctional signals in tumor cells, and thus, further experiments are needed to evaluate their potential effects.
Considering that many studies have shown that immune invasion has an impact on the prognosis of tumor, we performed the relevant analyses and found that tumor-associated fibroblasts are the main immune cells. Furthermore, we observed the statistically significant positive relationship between KRT17 expression and the estimated value of infiltration of cancer-associated fibroblasts in TCGA tumors (COAD, DLBC, KIRC, OV, TGCT, THCA, and THYM), but a statistically significant negative correlation in HNSC (HPV−). Through enrichment analysis, we constructed the network of interaction with the KRT17 protein, and screened the top five genes with the strongest correlation. The KRT17 expression level was found to be positively associated with GJB5 (R=0.72), KRT5 (R=0.73), KRT6A (R=0.75), KRT14 (R=0.75), and SFN (R=0.8) (all P<0.05). In the various cancer types described, the heatmap data revealed a positive connection between KRT17 and the top five genes. SFN, KRT6A, KRT5, KRT6B, and KRT6C were identified as common members of the above two groups after intersection analysis. To perform KEGG and GO enrichment analysis, the two datasets were combined. According to the KEGG data, “intermediate filament cytoskeleton” and “keratinization” may be involved in KRT17’s tumor pathogenesis effect.
Our study was the first pan-cancer of KRT17 to discover the statistical correlations of KRT17 expression with protein phosphorylation, clinical prognosis, amplification, and immune cell infiltration across multiple tumors, which helps us to better understand the role of KRT17 in tumorigenesis in terms of clinical tumor samples.
Acknowledgments
Funding: The research was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2118
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2118). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Tumor Hospital Affiliated to Nantong University’s Ethics Committee (Nantong, China) approved the study. This research was carried out in accordance with the Helsinki Declaration (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cui X, Zhang X, Liu M, et al. A pan-cancer analysis of the oncogenic role of staphylococcal nuclease domain-containing protein 1 (SND1) in human tumors. Genomics 2020;112:3958-67. [Crossref] [PubMed]
- Xiao H, Wang K, Li D, et al. Evaluation of FGFR1 as a diagnostic biomarker for ovarian cancer using TCGA and GEO datasets. PeerJ 2021;9:e10817. [Crossref] [PubMed]
- Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell 2018;173:530. [Crossref] [PubMed]
- Palko E, Poliska S, Sziklai I, et al. Analysis of KRT1, KRT10, KRT19, TP53 and MMP9 expression in pediatric and adult cholesteatoma. PLoS One 2018;13:e0200840. [Crossref] [PubMed]
- Rao X, Wang J, Song HM, et al. KRT15 overexpression predicts poor prognosis in colorectal cancer. Neoplasma 2020;67:410-4. [Crossref] [PubMed]
- Yuanhua L, Pudong Q, Wei Z, et al. TFAP2A Induced KRT16 as an Oncogene in Lung Adenocarcinoma via EMT. Int J Biol Sci 2019;15:1419-28. [Crossref] [PubMed]
- Li J, Chen Q, Deng Z, et al. KRT17 confers paclitaxel-induced resistance and migration to cervical cancer cells. Life Sci 2019;224:255-62. [Crossref] [PubMed]
- Chivu-Economescu M, Dragu DL, Necula LG, et al. Knockdown of KRT17 by siRNA induces antitumoral effects on gastric cancer cells. Gastric Cancer 2017;20:948-59. [Crossref] [PubMed]
- Liu J, Liu L, Cao L, et al. Keratin 17 promotes lung adenocarcinoma progression by enhancing cell proliferation and invasion. Med Sci Monit 2018;24:4782-90. [Crossref] [PubMed]
- Li D, Ni XF, Tang H, et al. KRT17 functions as a tumor promoter and regulates proliferation, migration and invasion in pancreatic cancer via mTOR/S6k1 pathway. Cancer Manag Res 2020;12:2087-95. [Crossref] [PubMed]
- Yang L, Zhang S, Wang G. Keratin 17 in disease pathogenesis: from cancer to dermatoses. J Pathol 2019;247:158-65. [PubMed]
- Depianto D, Kerns ML, Dlugosz AA, et al. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet 2010;42:910-4. [Crossref] [PubMed]
- Sizemore GM, Sizemore ST, Seachrist DD, et al. GABA(A) receptor pi (GABRP) stimulates basal-like breast cancer cell migration through activation of extracellular-regulated kinase 1/2 (ERK1/2). J Biol Chem 2014;289:24102-13. [Crossref] [PubMed]
- Dong M, Dong Z, Zhu X, et al. Long non-coding RNA MIR205HG regulates KRT17 and tumor processes in cervical cancer via interaction with SRSF1. Exp Mol Pathol 2019;111:104322. [Crossref] [PubMed]
- Mikami Y, Fujii S, Nagata K, et al. GLI-mediated Keratin 17 expression promotes tumor cell growth through the anti-apoptotic function in oral squamous cell carcinomas. J Cancer Res Clin Oncol 2017;143:1381-93. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Chen F, Chandrashekar DS, Varambally S, et al. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun 2019;10:5679. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Bardou P, Mariette J, Escudie F, et al. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 2014;15:293. [Crossref] [PubMed]
- Steven A, Seliger B. The role of immune escape and immune cell infiltration in breast cancer. Breast Care (Basel) 2018;13:16-21. [Crossref] [PubMed]
- Fridman WH, Galon J, Dieu-Nosjean MC, et al. Immune infiltration in human cancer: prognostic significance and disease control. Curr Top Microbiol Immunol 2011;344:1-24. [PubMed]
- Kwa MQ, Herum KM, Brakebusch C. Cancer-associated fibroblasts: how do they contribute to metastasis? Clin Exp Metastasis 2019;36:71-86. [Crossref] [PubMed]
- Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99-115. [Crossref] [PubMed]
- Yan X, Yang C, Hu W, et al. Knockdown of KRT17 decreases osteosarcoma cell proliferation and the Warburg effect via the AKT/mTOR/HIF1alpha pathway. Oncol Rep 2020;44:103-14. [Crossref] [PubMed]
- Yang L, Jin L, Ke Y, et al. E3 Ligase Trim21 Ubiquitylates and Stabilizes Keratin 17 to Induce STAT3 Activation in Psoriasis. J Invest Dermatol 2018;138:2568-77. [Crossref] [PubMed]
- Mo XC, Zhang ZT, Song MJ, et al. Screening and identification of hub genes in bladder cancer by bioinformatics analysis and KIF11 is a potential prognostic biomarker. Oncol Lett 2021;21:205. [Crossref] [PubMed]
- Shi C, Sun L, Liu S, et al. Overexpression of Karyopherin Subunit alpha 2 (KPNA2) Predicts Unfavorable Prognosis and Promotes Bladder Cancer Tumorigenicity via the P53 Pathway. Med Sci Monit 2020;26:e921087. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


