
Radiomics in head and neck cancer: from exploration to application
Introduction
Radiomics is concerned with the high-throughput extraction of large amounts of quantitative features whose subsequent analysis and selection can be incorporated in clinical decision-making. Radiomics complements, facilitates, and accelerates the advancement towards cancer precision medicine, as it is able to (I) non-invasively characterize the overall tumor accounting for heterogeneity; (II) produce prognostic and/or predictive biomarker value derived from routine, standard of care imaging data as-is; and (III) allow for a fast, low-cost, and repeatable means for longitudinal monitoring (1,2). Head and neck cancer presents a unique set of diagnostic and therapeutic challenges, including but not limited to the complex regional anatomy, the minute scale of critical structures, the variable appearance of primary and recurrent tumors, significant anatomic changes related to tumor response, and high intratumoral heterogeneity that varies depending on anatomic site.
In turn, radiomics holds the potential to address these barriers to personalized therapeutics. Contrast-enhanced computed tomography (CT), magnetic resonance (MR), and positron emission tomography (PET) imaging are routinely acquired during the diagnosis and staging process in head and neck cancer, and the immense data volume gathered from multiple imaging modalities in existing clinical datasets can greatly facilitate exploratory radiomic analysis. Also, the heterogeneous composition of head and neck cancers can be captured non-invasively, which can serve as an essential adjunct to clinical decision-making. In addition, radiomics can provide important day-to-day information regarding rapid anatomic change and tumor response during the course of treatment.
However, several key components are necessary to transition radiomics in head and neck cancers from exploratory studies to large-scale implementation as a clinical toolset. We first present an introductory overview of the radiomics workflow, texture analysis methods, and available software infrastructure. We then review key developments in head and neck cancer radiomics followed by a discussion of unmet challenges in its logistical and clinical application.
The radiomics workflow
To realize clinical application, an efficient series of iterative processes is required for reproducible and consistent extraction of radiomics data (Figure 1). This “radiomics workflow” begins with acquisition of high quality images with a standardized protocol. Segmentation of the tumor is then performed, followed by feature extraction from the defined tumor region. The extracted features that demonstrate the best performance, stability, or other defining metric are then selected for incorporation towards clinical applications (3).
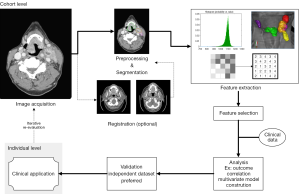
An overview of texture analysis methods and available software infrastructure for radiomics exploration
Methods for texture analysis in head and neck cancers are more or less the same as those used in other organ sites. These include first- and second-order texture methods as well as various transform-based methods. The most “direct” features are those based simply on intensity values within a region of interest (ROI). Similar but unique features may also be extracted from histograms of intensity values and Gaussian functions fitted to these histograms. Other “direct” features may be calculated from the shape of the ROI. Texture features in the head and neck are based on the same parent matrices that are utilized in other sites. Examples of these include the gray-level co-occurrence matrix (GLCM), the gray level run length matrix (GLRLM), the neighborhood intensity difference matrix (NIDM), neighborhood gray-level dependence matrix (NGLDM), and the intensity size-zone matrix (ISZM) (4-9). Other feature extraction methods are based on filters such as Fourier transform, Gabor transform, Laplacian of Gaussian filter (LoG), and multiscale wavelet decompositions (10-13). After processing the ROI according to the parent matrix or filter method, features such as coarseness, business, correlation, entropy, and energy are calculated. Details of these individual features are available in their respective references, and due to their multitude and complexity, will not be delineated in this article.
In addition to texture analysis methods, there exist multiple open-source, in-house developed, and commercial software solutions that facilitate the exploration and development of radiomics in head and neck cancer. A prime example of available open-source software is the Imaging Biomarker Explorer (IBEX) by Zhang et al., described as an “open infrastructure software platform that flexibly supports common radiomics workflow tasks such as multimodality image data import and review, development of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions.” (14). IBEX is compatible with CT, PET, and MR modalities, and is available at http://bit.ly/IBEX_MDAnderson. MazDa is another open-source solution for texture analysis that has been validated through multi-institutional studies (15). This software is built primarily for magnetic resonance imaging (MRI) texture analysis and supports various feature selection methods for model generation. MazDa is available at http://www.eletel.p.lodz.pl/programy/mazda/index.php?action=mazda. CGITA is yet another open-source texture analysis tool, built in the MATLAB environment. The software supports numerous heterogeneity indices, user-defined calculations, and batch processing with a focus on molecular imaging. CGITA supports CT, PET, and MR images and is available at http://code.google.com/p/cgita (16).
Beyond open-source software tools, a number of groups have also developed in-house tools for radiomic analyses, most often in the MATLAB environment, but these softwares are not publicly available to our knowledge (17-21). One such example is a modified version of Computational Environment for Radiotherapy Research (CERR) used for texture analysis.
Lastly, numerous commercial solutions for radiomic analyses are also emerging. For instance, TexRAD is a commercial software which uses a LoG special filter to delineate fine, intermediate, and coarse textures in a ROI for subsequent analysis. This software contains various decision support tools for thoracic and gastrointestinal imaging and has also demonstrated applicability in head and neck cancer textural analysis (22).
Novel applications of texture analysis methods and the emergence of new software tools have both spurred on developments in radiomics for head and neck cancer.
A review of recent developments in radiomics for head and neck cancer
Specific applications for texture analysis and radiomics in head and neck tumors have, to date, demonstrated exciting promise in several distinct arenas. We elaborate on the preliminary applications of these techniques in the following areas for head and neck cancer:
- Tumor segmentation and pathologic classification;
- Risk stratification, as prognostic and/or predictive biomarker(s);
- Monitoring of alteration in normal tissue as a sequelae of radiotherapy dose deposition.
A summary of the mentioned studies in this section can be found in the Table S1.
Radiomics for tumor segmentation and classification
Textural analysis has demonstrated preliminary evidence suggesting clinical utility in the classification of and segmentation process for head and neck cancers (Table 1).
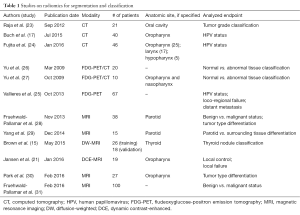
Full table
For instance, a number of studies have sought to classify head and neck cancers by human papillomavirus (HPV) status with textural analysis. Buch et al. investigated the use of texture analysis to distinguish between HPV(+) and HPV(−) status in 40 patients with primary oropharyngeal squamous cell carcinomas on contrast-enhanced CT (CE-CT) images. They identified three textural features (histogram median and entropy and GLCM entropy) that could make the distinction with statistical significance and concluded that textural analysis has the potential to be used as an adjunct to evaluate HPV status in squamous cell carcinomas (17). A follow-up study by Fujita et al. sought to distinguish the HPV status of 46 patients with non-oropharyngeal carcinoma using texture analysis extracted from CE-CT images. They identified three features demonstrating statistical significance after false discovery rate (FDR) correction (GLCM contrast, GLCM correlation and law L8). Consequently, they suggested that there are morphologic feature differences based on HPV status even in non-oropharyngeal cancer (OPC) patients (24). Exploring beyond CE-CT, Vallieres et al. aimed to evaluate whether features derived from fludeoxyglucose-positron emission tomography (FDG-PET) could be used as non-invasive biomarkers of HPV status. The study, including 67 patients with head and neck squamous cell carcinoma (HNSCC) and known HPV status, demonstrated that multivariate models built via logistic regression (LR) and support vector machine (SVM) using five features could reliably classify HPV status [area under curve (AUC) of 0.64 and 0.72 for LR and SVM, respectively] and had the potential to predict treatment failure (AUC 0.66) (25).
In addition to classification methodologies, segmentation (in particular, distinguishing between normal and abnormal tissue) has been another application of textural analysis for head and neck cancers. For instance, Yu et al. conducted a study examining 20 patients with head and neck cancer with 20 matched controls and found that neighborhood gray-tone difference matrix (NGTDM) features including PET coarseness, PET contrast, and CT coarseness extracted from co-registered FDG-PET/CT images yielded good discriminatory performance. Their multivariate model, constructed via a decision tree-based K-nearest neighbor (DT-KNN) classifier, was able to successfully discriminate between normal and abnormal ROIs [receiving operator characteristic area under the curve (ROC Az) of 0.95±0.007, Az90 of 0.087±0.003]. It was found that the combination of PET and CT features outperformed either PET or CT features individually in discriminatory ability. In addition, they found that features based on the NGTDM and SGLDM (spatial gray level dependence method) could classify ROIs with comparable accuracy to that of a human expert, suggesting that implementation of such analyses at the voxel level could lead to improvement in the accuracy of automated segmentation in head and neck cancer (26). To that end, Yu et al. published on such an implementation titled “co-registered multimodality pattern analysis segmentation system” (COMPASS) for ten head and neck cancer patients. They found that COMPASS outperformed other simpler PET-based thresholding methods for tumor segmentation and yielded contours that were quantitatively and qualitatively similar to those created manually by expert radiation oncologists (specificity 95%±2%, sensitivity 90%±12%) (27). In addition to studies that broadly examined segmentation of head and neck cancers, other studies have sought to segment tissue in specific anatomic regions, as Raja et al. aimed to do with regards to the buccal mucosa subsite of the oral cavity (23).
Much like the aforementioned studies performed in the CT and PET modalities, numerous investigations focused on the MR imaging modality have applied textural analysis towards classification and segmentation processes in head and neck cancer. Some of these MR imaging based studies (28-31) that differentiate between benign and malignant status and amongst different types of head and neck masses will not be elaborated upon in this text, but relevant findings and their statistical significance can be found in the Table S1.
MR imaging merits a distinct interest in radiomics. While CT Hounsfield units represent a standard physical phenomena, the dynamic range of information possible with distinct MRI sequences may allow greater imaging flexibility (in terms of voxel and subvoxel physiologic parameters) as well as the potential for multi-parametric texture/radiomics dataset acquisition in a single imaging series. Therefore, texture analysis of advanced MR sequences has generated interest for further investigation. For instance, in a multi-institutional study focused on preoperative stratification of thyroid tumors using diffusion-weighted (DW) MRI, Brown et al. reported that an algorithm constructed using linear discriminant analysis (LDA) with t21 features yielded an ROC AUC 0.97 (sensitivity 92%, specificity 96%) in a training dataset of 26 patients and correctly classified 89% of tumors in an 18 patient independent validation dataset (15). As expected, a significant difference in the ADC was observed between benign and malignant lesions, but ADC alone was not as effective in classification (AUC 0.73, sensitivity 70%, specificity 63%) as the model generated from radiomic analysis of the diffusion-weighted echo-planar imaging (DW-EPI) sequence. Furthermore, texture analysis of quantitative and semi-quantitative MRI data has also shown potential value. In a recent study of 19 HNSCC patients with pretreatment and intra-treatment dynamic contrast-enhanced (DCE), or DCE-MRI available, Jansen et al. analyzed the parametric maps of Ktrans and ve, which are measures of tumor vascularity. It was reported that the energy feature from the ve map was significantly higher on intra-treatment scans (0.41±0.22 vs. 0.30±0.11; P<0.04). The findings suggest that texture analysis may provide complementary information in addition to standard DCE-MRI measurements that have been shown to be predictive of treatment response in head and neck cancer patients (21,32,33).
Radiomics as prognostic and predictive biomarkers
Another application of radiomics is the development and refinement of radiomic signatures that can improve upon prognostic and/or predictive models for specific cancers, including those of the head and neck (Table 2). Heading the effort in this area, Aerts et al. conducted a radiomic analysis of 440 features extracted from a pre-treatment CT database of 1,019 patients with either lung or head and neck cancer. The features described tumor phenotype in four categories (tumor image intensity, shape, texture, and wavelet decomposition) and the strongest radiomic features from each of the four feature groups were identified to create a signature: statistics energy, shape compactness, gray level non-uniformity, and wavelet (HLH) gray level non-uniformity. A multitude of features were found to have association with oncologic outcomes in independent datasets of lung cancer and head and neck cancer patients. Of interest, a radiomic signature that was trained using a dataset of non-small cell lung cancer patients was found to have impressive translatability in two independent head and neck cancer validation datasets. They also suggested that the prognostic significance of the features can capture underlying intratumor heterogeneity and is associated with gene-expression patterns (34).

Full table
In a subsequent study, the aforementioned radiomic signature was externally validated on an independent cohort of oropharyngeal squamous cell carcinoma patients (542 patients) (37). The signature validated well, demonstrating good model fit and preservation of discrimination (Harrell’s c-index 0.628; P=2.72e-9). Interestingly, it was also found that the signature retained discriminatory ability in the presence of visible CT artifacts, which are often present in head and neck CT sequences due to dental hardware (Harrell’s c-index 0.647; P=5.35e-6).
In yet another application of radiomics as a predictive marker, Zhang et al. analyzed the predictive value of texture and histogram features in 72 HNSCC patients treated with induction chemotherapy. In multivariate Cox regression analysis incorporating both clinical and imaging variables, they found that in addition to expected factors such as tumor volume and N stage, primary mass entropy [hazard ratio (HR) =2.10 for each 0.5-unit increase; P=0.36] and histogram skewness (HR =3.67 for each 1.0-unit increase; P=0.009) were independent predictors of overall survival (OS) (22).
A common challenge in radiomics is to define a non-redundant set of imaging biomarkers from the vast amount of extracted features. An investigation by Parmar et al. examined the role of consensus clustering in reducing redundant features into a few robust and compact feature clusters. Upon analysis of features extracted from pre-treatment CT images from four independent lung and head and neck cancer cohorts (878 patients total), they showed that lung and head and neck radiomic clusters are significantly associated with patient survival and tumor stage. In addition to demonstrating prognostic value to clinical endpoints, their results revealed that clustering and the prognostic radiomic features are cancer specific (35).
Machine-learning methods have also been investigated for prognostic value as biomarkers for head and neck cancers. From two independent head and neck cohorts totaling 196 patients, Parmar et al. investigated 13 feature selection methods and 11 machine-learning classification methods chosen for simplicity, efficiency, and popularity in the literature. Specifically, they identified three classifiers and feature selection methods that demonstrated high performance and stability for predicting 3-year OS in head and neck cancer, suggesting that these machine learning methods should be the starting point for radiomics-based prognostic analyses due to their consistency. Identifying optimal machine-learning methods for radiomic analysis is a prerequisite to the development of a robust, clinically-applicable radiomic workflow. These findings provide valuable information for methodology selection in future radiomics investigations. Such methods could allow for improvements in cancer biomarker identification and personalized medicine in oncology (36).
In addition to CT, applying radiomics for prognostic and/or predictive purposes for head and neck cancer has been investigated in the PET, MR, and histopathologic imaging modalities as well. For example, one study by El Naqa et al. examined shape and textural features as well as intensity volume histogram metrics extracted from pre-treatment PET images on nine head and neck cancer patients. Using the highest predictive features, they were able to construct a two-metric LR model predicting OS with an AUC of 1.0 (20). In addition, certain studies aim to predict p53 status, as a positive status is associated with poor prognosis in certain subsets of HNSCC patients (39-41). Dang et al. demonstrated that MRI texture analysis could predict p53 status in oropharyngeal squamous cell carcinoma with 81.3% accuracy (P<0.05). In a retrospective study of 16 patients, they identified and incorporated seven significant texture features into a predictive model. The variables featured significantly were those derived from post-gadolinium T1W1, T2W1, and ADC map, noted to be possibly due to differences in vascularity between p53(+) versus p53(−) status (38). In addition to PET and MR imaging, histopathological imaging coupled with textural analysis has demonstrated prognostic potential as well. For instance, a study of 53 cases of HNSCC was characterized by quantitative histologic texture analysis by generating a 2D planar tessellation of the tumor and then analyzing the reconstructed image using spatial statistics. Ultimately, mean nuclear area was found to be significant predictor of lymph node metastasis (42).
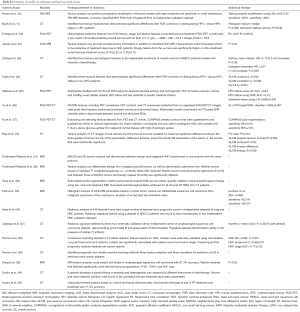
Radiomics for longitudinal monitoring of therapy response in non-tumor tissues
Several groups have compiled data indicative of the potential capacity for radiomics/texture techniques to afford longitudinal monitoring of tumor response. However, in addition to tumor imaging, the same approaches can readily be utilized to detect normal tissue physiologic alteration (Table 3). This is particularly beneficial for head and neck cancers, which exhibit significant anatomical changes with radiotherapy doses delivered to functional normal tissues. As an instance of this application, texture analysis has been applied to CT images to assess change in parotid gland structure during radiotherapy. In this study, a general decrease in parotid tissue complexity and heterogeneity was observed at different time points of radiotherapy. Also, volume and mean intensity variation were found to be correlated with pre-treatment dosimetric parameters, suggesting a relationship between dose plan and structural variation estimated after radiotherapy (44). The same group further investigated whether early variations of textural parameters [i.e., mean intensity and fractal dimension (FD)] could predict parotid shrinkage at the end of treatment. The study examined textural parameters extracted from CT images of 42 parotids of 21 nasopharyngeal cancer patients treated with IMRT. Using discriminant analysis based on volume and fractal dimensionality, they were able to predict final parotid shrinkage with 71.4% accuracy (43).

Full table
Unmet challenges for head and neck cancer radiomics
With respect to head and neck cancer radiomics, we first discuss the technical challenges inherent to the young field of big data, followed by the clinical challenges inherent to cancer medicine. The unmet technical challenges present today in the field of head and neck cancer radiomics are mostly similar to those in other disease sites. Technical challenges in radiomics for head and neck cancer include requirements for processing large amounts of high-quality imaging data, reproducibility in data processing, an assessment of radiogenomic associations, a suitable informatics infrastructure, and standardized reporting guidelines.
Radiomics inherently depends on large amounts of high quality imaging data. Results are highly dependent on segmentation, which can be a time-intensive process if carried out manually. However, manual segmentation adds a level of quality assurance and confidence in delineation that is not matched by auto-segmentation solutions, especially in the head and neck.
Reproducibility of results is another unmet need in this field. Not only do radiomic features need to demonstrate reproducibility in the same patient through test-retest studies, but these features need to be evaluated across different device manufacturers, imaging acquisition parameters, and institutions. Several studies have been done on test-retest variability of radiomic features in other disease sites (19,45-48). Recently, Mackin et al. published a study on feature variability utilizing a phantom imaged by 16 CT scanners from four manufacturers at four separate facilities (48). They found that feature values tended to cluster by manufacturer and that the variability between manufacturers was not insignificant and even of comparable size to the variability among different non-small cell lung cancer tumors themselves. The findings highlight the need for collaboration among institutions to study the plethora of variables that may contribute to radiomic values and to develop a framework to minimize the variability.
Another major unmet goal specific to head and neck radiomics is in the radiogenomic assessment of head and neck cancers. In other disease sites including brain, breast, lung, and liver, studies have shown various associations between genomic expression patterns and radiographic features for a number of cancers (49-54). Such findings are supportive of the central hypothesis of radiomics that the genotype of a tumor is associated with the radiographic phenotype of a tumor. Researchers in other sites have leveraged genomic information from The Cancer Genome Atlas (TCGA) in combination with imaging data from The Cancer Imaging Archive (TCIA) for such studies. In the head and neck space, a radiogenomic assessment of the TCGA head and neck cohort is underway by the authors of this review.
From a practical standpoint, integration of radiomic data into the clinical workflow will require facilitation by software. An electronic health record (EHR) software and picture archiving communication system (PACS) that integrate radiomic analysis with all pertinent imaging metadata and clinical information will greatly enhance both the feasibility of a radiomics workflow and the potential value of acquired images. With respect to creating a standard methodology for analyzing and reporting radiomic data, two national collaborative efforts have been created to address this issue: the Quantitative Imaging Network (QIN), sponsored by the National Cancer Institute (NCI), and the Quantitative Imaging Biomarker Alliance (QIBA), sponsored by the Radiological Society of North America (RSNA) (55,56). In addition to these efforts, the NIH has required a plan for data sharing in all major research funding applications since 2003; ideally these efforts will spur sharing of datasets and methodologic transparency, in addition to increasing access to direct software resources developed for radiomics applications.
There are also several challenges unique to cancer medicine that must be addressed in radiomics. In clinical oncology, the ultimate goal of radiomics is to apply standardized signatures towards specific oncologic functions and outcomes, thereby enabling personalized cancer care that can be directly actionable. However, the literature investigating actionable radiomic signatures have not yet developed to a level sufficient for broad implementation. For instance, studies have examined the role of texture analysis in differentiating HPV status and p53 status in subpopulations of head and neck cancer patients. While the ability to infer oncology-specific parameters from imaging is promising, further investigations and collaborations are needed to incorporate these findings into a management scheme that can directly impact decision-making.
There is a need for radiomic signatures with specific oncologic function (i.e., defining oncologic pathophysiology such as metastasis). The complex management of each and every cancer patient also mandates radiomic signatures specific for clinical parameters tell-tale for different phases of treatment (i.e., pre- vs. post-procedure, during and after chemotherapy, and at different timepoints during radiation therapy). In addition, we must also refine clinical endpoints specific to oncology. For instance, in addition to investigating general outcomes like OS, radiomic signatures specific to the nodal metastasis probability would be clinically useful towards clinical management. In order to realize cancer-specific radiomics, we need extensive prospective multicenter trials and external validation to begin standardizing and refining radiomic signatures.
To that end, one fruitful and practical approach may be to leverage imaging data from ongoing and proposed randomized clinical trials which contain a well-defined imaging component. Validations should be done against completely independent large datasets, preferably from other institutions. A prime example of this process gaining ground is demonstrated in the efforts of Leijenaar et al. (37), whose group externally validated a prognostic radiomic signature on an independent cohort of oropharyngeal squamous cell patients. In addition, recent investigations revealing that radiomic signatures have translational capacity between cancer types, yet retain cancer-specific cluster features, are highly promising towards the effort to standardize and refine radiomic signatures (34,35). Finally, another means to accelerate developments in head and neck cancer radiomics takes the form of formal challenges posed to the research community to solve defined issues. The Medical Image Computing and Computer Assisted Interventions (MICCAI) challenge is one such challenge that bridges international solutions to user feedback. Their challenges have sought solutions applicable to obstacles in cancer imaging, including those of head and neck cancer. The potential for radiomics to realize personalized cancer care has been demonstrated by numerous investigations. The field and its applicability to oncology promise to develop with time, but only if we direct our efforts to specific oncologic function and oncologic outcomes, with external validation through multi-institutional collaborative efforts.
Moving the field forward
The data summarized herein suggest cumulatively that there is great potential for radiomics and texture analysis techniques to improve upon multiple aspects of the tumor assessment, risk stratification, and outcome evaluation aspects in head and neck cancer therapy. However, at present the vast majorities of these studies are primarily exploratory, or at best, seek to perform model refinement and validation methods. At present, there is a fundamental need for several key efforts as an oncologic community which will solidify the role of radiomics in texture analysis techniques in a manner that can realize clinical utilization.
First, there needs to be a global effort towards standardization. This effort requires standardization not only of individual radiomics algorithms, but also of specific acquisition parameters. Fundamentally, performance quality assurance and quality improvement must be utilized such that a specific imaging acquisition protocol matched with a specific texture and analytic protocol (see Figure 1) can be shown to perform within an estimated error range as a diagnostic, predictive, and evaluative tool. It is only when such data are well defined that we will truly be able to implement radiomics and texture analytic techniques in the clinic.
Individual work at the level of algorithm standardization is already underway, with the recent publication by Parmar et al. (57) serving as an excellent model. In this seminal head and neck radiomics manuscript investigating the quality assurance/quality improvement methodology, Parmar et al. carefully and rigorously evaluated the process variability in their radiomics development process, discovering that the majority of performance variation could be identified in the classification process. Only by similar efforts in investigating the relative performance characteristics and error estimators in each individual step of the radiomics process will we achieve reliable and reproducible tools which can scale across institutions and imaging data sets. Additionally, individual image acquisition protocols should be standardized in a similar manner. For example, it is unclear at present whether there is comparability between individual predictive/prognostic features in inter-/intra-sequence MRI data sets, and between similar textural features in CT or PET-CT datasets. Ideally, multimodality data sets should be interrogated to determine whether similar and/or related textual features can be representative across imaging modalities.
Finally, the bulk of our efforts should be directed towards determining the underlying mechanistic underpinnings of the observed clinical findings demonstrated by radiomics and texture analyses. For example, it is unclear specifically which features are individually representative of what underlying physiological processes drive tumor response or normal tissue injury changes observed in the affirmation data sets. While the leading candidates represent measures of tumor heterogeneity, or vascular perfusion differentials across tumor lines, it is imperative that future efforts derived at clinical, radiologic, histopathologic, or genomic characterization interrogate the underlying molecular physiologic processes which drive the meso/macro-scale features observed in clinical datasets. One of the most compelling examples of how to approach such studies is that of Panth et al. (58), who grew colon cancer-derived xenografts with doxycycline-inducible (GADD34) cells in the flanks of nude mice. As GADD34 overexpression decreases hypoxic fraction, changes in gene function and hypoxia could be observed through serial CT imaging over time. Radiomics analyses were performed at 40 kVp and again at 80 kVp for validation, before and after radiotherapy. These data showed not only reproducible, robust changes seen at multiple kVp levels, but that specific temporal kinetic differences could be observed with regard to genotypic and phenotypic radiomics signatures. Work of this quality and scope will need to be undertaken in head and neck specific models to interrogate the mechanistic underpinnings of radiomics in vivo.
Conclusions
In summary, we believe that the preponderance of evidence suggests that radiomics is in fact revealing real information regarding tumor and normal tissue information that is above and beyond visual analysis However, these quantitative methods require further hardening of the underlying methodologic processes, as well as a greater coupling of quantitative imaging phenomena to the underlying biologic processes. Our hope is that these needs can be met sooner than later.
Full table
Acknowledgments
Funding: Dr. Fuller received/receives grant and/or salary support from: the National Institutes of Health National Cancer Institute’s Paul Calabresi Clinical Oncology Award Program (K12 CA088084-06) and Clinician Scientist Loan Repayment Program (L30 CA136381-02) and Head and Neck SPORE Career Development Award (P50CA097007-10); the National Institutes of Health National Institute of Dental and Craniofacial Research research grant (R56DE025248-01,R01DE025248-01); a General Electric Healthcare/MD Anderson Center for Advanced Biomedical Imaging In-Kind Award; an Elekta AB/MD Anderson Department of Radiation Oncology Seed Grant; the Center for Radiation Oncology Research at MD Anderson Cancer Center seed grant program; and the MD Anderson Institutional Research Grant Program. This work was supported in part infrastructure support by National Institutes of Health Cancer Center Support (Core) Grant CA016672 to The University of Texas MD Anderson Cancer Center. Dr. Fuller has received travel funding from Elekta AB.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Laurence E. Court, Arvind Rao and Sunil Krishnan) for the series “Radiomics in Radiation Oncology” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.07.18). The series “Radiomics in Radiation Oncology” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Transactions on Systems, Man and Cybernetics 1973;SMC-3:610-21. [Crossref]
- Castellano G, Bonilha L, Li LM, et al. Texture analysis of medical images. Clin Radiol 2004;59:1061-9. [Crossref] [PubMed]
- Tang X. Texture information in run-length matrices. IEEE Trans Image Process 1998;7:1602-9. [Crossref] [PubMed]
- Soh LK. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans Geosci Remote Sens 1999;37:780-95. [Crossref]
- Galloway MM. Texture analysis using gray level run lengths. Computer Graphics and Image Processing 1975;4:172-9. [Crossref]
- Lam SW. Texture feature extraction using gray level gradient based co-occurence matrices. Systems, Man, and Cybernetics, 1996., IEEE International Conference on 1996;1:267-71.
- Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging 2013;13:400-6. [Crossref] [PubMed]
- Zhang X, Stockel J, Wolf M, et al. A new method for spherical object detection and its application to computer aided detection of pulmonary nodules in CT images. Med Image Comput Comput Assist Interv 2007;10:842-9. [PubMed]
- Guo X, Liu X, Wang H, et al. Enhanced CT images by the wavelet transform improving diagnostic accuracy of chest nodules. J Digit Imaging 2011;24:44-9. [Crossref] [PubMed]
- Bastawrous HA. Detection of Ground Glass Opacities in Lung CT Images Using Gabor Filters and Neural Networks. 2005 IEEE Instrumentationand Measurement Technology Conference Proceedings 2005;1:251-6.
- Zhang L, Fried DV, Fave XJ, et al. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42:1341-53. [Crossref] [PubMed]
- Brown AM, Nagala S, McLean MA, et al. Multi-institutional validation of a novel textural analysis tool for preoperative stratification of suspected thyroid tumors on diffusion-weighted MRI. Magn Reson Med 2016;75:1708-16. [Crossref] [PubMed]
- Fang YH, Lin CY, Shih MJ, et al. Development and evaluation of an open-source software package "CGITA" for quantifying tumor heterogeneity with molecular images. Biomed Res Int 2014;2014:248505.
- Buch K, Fujita A, Li B, et al. Using Texture Analysis to Determine Human Papillomavirus Status of Oropharyngeal Squamous Cell Carcinomas on CT. AJNR Am J Neuroradiol 2015;36:1343-8. [Crossref] [PubMed]
- Alobaidli S, McQuaid S, South C, et al. The role of texture analysis in imaging as an outcome predictor and potential tool in radiotherapy treatment planning. Br J Radiol 2014;87:20140369 [Crossref] [PubMed]
- Leijenaar RT, Carvalho S, Velazquez ER, et al. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 2013;52:1391-7. [Crossref] [PubMed]
- El Naqa I, Grigsby P, Apte A, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit 2009;42:1162-71. [Crossref] [PubMed]
- Jansen JF, Lu Y, Gupta G, et al. Texture analysis on parametric maps derived from dynamic contrast-enhanced magnetic resonance imaging in head and neck cancer. World J Radiol 2016;8:90-7. [Crossref] [PubMed]
- Zhang H, Graham CM, Elci O, et al. Locally advanced squamous cell carcinoma of the head and neck: CT texture and histogram analysis allow independent prediction of overall survival in patients treated with induction chemotherapy. Radiology 2013;269:801-9. [Crossref] [PubMed]
- Raja JV, Khan M, Ramachandra VK, et al. Texture analysis of CT images in the characterization of oral cancers involving buccal mucosa. Dentomaxillofac Radiol 2012;41:475-80. [Crossref] [PubMed]
- Fujita A, Buch K, Li B, et al. Difference Between HPV-Positive and HPV-Negative Non-Oropharyngeal Head and Neck Cancer: Texture Analysis Features on CT. J Comput Assist Tomogr 2016;40:43-7. [Crossref] [PubMed]
- Vallieres M, Kumar A, Sultanem K, et al. FDG-PET Image-Derived Features Can Determine HPV Status in Head-and-Neck Cancer. Int J Radiat Oncol Biol Phys 2013;87:S467. [Crossref]
- Yu H, Caldwell C, Mah K, et al. Coregistered FDG PET/CT-based textural characterization of head and neck cancer for radiation treatment planning. IEEE Trans Med Imaging 2009;28:374-83. [Crossref] [PubMed]
- Yu H, Caldwell C, Mah K, et al. Automated radiation targeting in head-and-neck cancer using region-based texture analysis of PET and CT images. Int J Radiat Oncol Biol Phys 2009;75:618-25. [Crossref] [PubMed]
- Fruehwald-Pallamar J, Czerny C, Holzer-Fruehwald L, et al. Texture-based and diffusion-weighted discrimination of parotid gland lesions on MR images at 3.0 Tesla. NMR Biomed 2013;26:1372-9. [Crossref] [PubMed]
- Yang X, Wu N, Cheng G, et al. Automated segmentation of the parotid gland based on atlas registration and machine learning: a longitudinal MRI study in head-and-neck radiation therapy. Int J Radiat Oncol Biol Phys 2014;90:1225-33. [Crossref] [PubMed]
- Park M, Kim J, Choi YS, et al. Application of Dynamic Contrast-Enhanced MRI Parameters for Differentiating Squamous Cell Carcinoma and Malignant Lymphoma of the Oropharynx. AJR Am J Roentgenol 2016;206:401-7. [Crossref] [PubMed]
- Fruehwald-Pallamar J, Hesselink JR, Mafee MF, et al. Texture-Based Analysis of 100 MR Examinations of Head and Neck Tumors - Is It Possible to Discriminate Between Benign and Malignant Masses in a Multicenter Trial? Rofo 2016;188:195-202. [PubMed]
- Kim S, Loevner LA, Quon H, et al. Prediction of response to chemoradiation therapy in squamous cell carcinomas of the head and neck using dynamic contrast-enhanced MR imaging. AJNR Am J Neuroradiol 2010;31:262-8. [Crossref] [PubMed]
- Shukla-Dave A, Lee NY, Jansen JF, et al. Dynamic contrast-enhanced magnetic resonance imaging as a predictor of outcome in head-and-neck squamous cell carcinoma patients with nodal metastases. Int J Radiat Oncol Biol Phys 2012;82:1837-44. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [PubMed]
- Parmar C, Leijenaar RT, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci Rep 2015;5:11044. [Crossref] [PubMed]
- Parmar C, Grossmann P, Rietveld D, et al. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front Oncol 2015;5:272. [Crossref] [PubMed]
- Leijenaar RT, Carvalho S, Hoebers FJ, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 2015;54:1423-9. [Crossref] [PubMed]
- Dang M, Lysack JT, Wu T, et al. MRI texture analysis predicts p53 status in head and neck squamous cell carcinoma. AJNR Am J Neuroradiol 2015;36:166-70. [Crossref] [PubMed]
- Gross AM, Orosco RK, Shen JP, et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat Genet 2014;46:939-43. [Crossref] [PubMed]
- Maruyama H, Yasui T, Ishikawa-Fujiwara T, et al. Human papillomavirus and p53 mutations in head and neck squamous cell carcinoma among Japanese population. Cancer Sci 2014;105:409-17. [Crossref] [PubMed]
- Wichmann G, Rosolowski M, Krohn K, et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer 2015;137:2846-57. [Crossref] [PubMed]
- Mattfeldt T, Fleischer F. Characterization of squamous cell carcinomas of the head and neck using methods of spatial statistics. J Microsc 2014;256:46-60. [Crossref] [PubMed]
- Scalco E, Fiorino C, Cattaneo GM, et al. Texture analysis for the assessment of structural changes in parotid glands induced by radiotherapy. Radiother Oncol 2013;109:384-7. [Crossref] [PubMed]
- Scalco E, Moriconi S, Rizzo G. Texture analysis to assess structural modifications induced by radiotherapy. Conf Proc IEEE Eng Med Biol Soc 2015;2015:5219-22.
- Galavis PE, Hollensen C, Jallow N, et al. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010;49:1012-6. [Crossref] [PubMed]
- Fave X, Mackin D, Yang J, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys 2015;42:6784-97. [Crossref] [PubMed]
- Yang J, Zhang L, Fave XJ, et al. Uncertainty analysis of quantitative imaging features extracted from contrast-enhanced CT in lung tumors. Comput Med Imaging Graph 2016;48:1-8. [Crossref] [PubMed]
- Mackin D, Fave X, Zhang L, et al. Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol 2015;50:757-65. [Crossref] [PubMed]
- Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology 2015;276:313. [Crossref] [PubMed]
- Hong SJ, Kim TJ, Choi YW, et al. Radiogenomic correlation in lung adenocarcinoma with epidermal growth factor receptor mutations: Imaging features and histological subtypes. Eur Radiol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Rizzo S, Petrella F, Buscarino V, et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol 2016;26:32-42. [Crossref] [PubMed]
- Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham) 2015;2:041007 [Crossref] [PubMed]
- Zinn PO, Mahajan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One 2011;6:e25451 [Crossref] [PubMed]
- Jamshidi N, Jonasch E, Zapala M, et al. The Radiogenomic Risk Score: Construction of a Prognostic Quantitative, Noninvasive Image-based Molecular Assay for Renal Cell Carcinoma. Radiology 2015;277:114-23. [Crossref] [PubMed]
- Buckler AJ, Bresolin L, Dunnick NR, et al. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology 2011;258:906-14. [Crossref] [PubMed]
- Kalpathy-Cramer J, Freymann JB, Kirby JS, et al. Quantitative Imaging Network: Data Sharing and Competitive AlgorithmValidation Leveraging The Cancer Imaging Archive. Transl Oncol 2014;7:147-52. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Panth KM, Leijenaar RT, Carvalho S, et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol 2015;116:462-6. [Crossref] [PubMed]


