
Modulation of the p53 family network by RNA-binding proteins
Introduction
It is fundamentally understood that the differential expression of a distinct assemblage of proteins is attributed to cancer cells’ ability to develop, proliferate, and metastasize (1). Indeed, increasing evidence has revealed that deregulated protein synthesis plays a critical role in cell transformation (2-4). Irregular protein expression may not only be due to gene mutations and altered transcription, but also through transformed posttranscriptional machineries such as mRNA storage, transport, splicing, translation, and degradation (5-8). These aforementioned mechanisms are directed by two distinct RNA-binding factors; microRNAs and RNA-binding proteins (RBPs) (9). While both microRNA and RBPs play a role in posttranscriptional regulation, the focus of this review is on the current understanding of RBP regulation of p53, p53 family members, and p53 downstream targets. For a detailed review on p53 regulation by microRNA please see (10). Multiple cancer related proteins, including tumor suppressors and oncoproteins, are tightly controlled through translational regulation and/or mRNA half-life, emphasizing the influence of RBPs on the expression of cancer associated proteins (5,11). Translation, as well as mRNA decay, is typically influenced by RBPs interacting with target gene transcripts. Characteristically, the sequences in the mRNA that modulate translation and mRNA stability usually reside in the 5'- and 3'-untranslated regions (UTRs).
Structurally, RBPs possess a high proportion of modularity, as seen by the majority of RBPs containing at least one or more RNA-binding and auxiliary domains (12). The differential combination and arrangements of these RNA-binding modules assist in the numerous interactions and regulatory abilities of these RBPs [reviewed here (13)]. To date, numerous classes of RNA-binding motifs have been identified including, RNA recognition motif (RRM), hnRNP K homology (KH) motif, RGG box, Pumilio, and double-stranded RNA-binding motif (13,14). Since many disease associated proteins are subjected to rigorous posttranscriptional regulation, it is without surprise that aberrant expression of RBPs have been tied to several human diseases including, neurological disorders, muscle atrophies, and cancer (11,15). Numerous RBPs have been linked with tumorigenesis (7). For example, aberrant eIF4E (eukaryotic translation initiation factor 4E) expression has been shown to lead to malignant transformation in mouse and rat fibroblasts (16), and further, targeting eIF4E with a cell-penetrating peptide leads to cell death in multiple cancer cell lines (17). Another important RBP in cancer, human antigen R (HuR), was one of the earliest RBPs identified to be aberrantly expressed in human malignances, including mammary and colon cancers (18,19). Extensive studies over the past decade revealed that HuR has the ability to induce the stability of many cancer-related transcripts including cytokines, growth factors, invasion factors, and other proto-oncogenes [for a review of HuR in cancer please see (20)]. Collectively, various lines of evidence have come to light establishing that posttranscriptional regulation by RBPs may play a role in multiple human diseases including cancer.
Regulation of p53
p53 is a transcription factor with an essential role in conserving the overall integrity of the genome and aiding in the prevention of cancer development. Highlighting the necessity for p53 in inhibiting tumorigenesis is the evidence that p53 inactivation occurs in more than 50% of human cancers (21). Under normal circumstances, p53 is highly regulated and protein expression is kept at low levels. However, in reaction to stress stimuli, p53 is activated and functions as a robust transcription factor inducing the activation of downstream targets that function in DNA repair, cell-cycle arrest and apoptosis (22-24). The importance of strict p53 protein regulation is underscored by the fact that too much p53 leads to premature ageing (25) and cell death due to excessive apoptosis (26), whereas too little p53 has been shown to be a key aspect of tumorigenesis (27).
Most work on the regulation of p53 has been demonstrated to be through posttranslational modifications, such as acetylation and phosphorylation. For example, phosphorylation of p53’s Ser15 (mouse Ser18) and Ser20 (mouse Ser23) at its N-terminus is believed to stabilize p53 by blocking its interactions with a key p53 inhibitor, MDM2 (28). Furthermore, in response to stress stimuli, p53 acquires increased acetylation correlating with increased p53 stabilization and activation (29-31). Subsequent studies revealed that not only were acetylation and phosphorylation important for regulating p53 stability and function, but other posttranslational modifications such as methylation, sumoylation, neddylation and ubiquitination were also able to modulate p53’s function and stability (32). Besides posttranslational modifications, it is now starting to be understood that p53 is also regulated through posttranscriptional mechanisms.
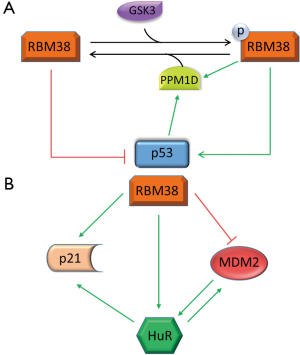
To date, multiple RBPs have been found to regulate p53 translation by interacting with the 5' or 3' UTR of p53 mRNA. An RBP extensively studied in our laboratory, RBM38, also known as RNPC1, was determined to be a target of p53, and a key regulator of p53 expression. Over-expression of RBM38 inhibits, while knockdown of RBM38 increases, p53 translation in both basal and stress conditions (33). The inhibition of p53 translation was attributed to RBM38 interacting with p53 5' and 3' UTRs. Upon binding to p53 mRNA, RBM38 is able to interact with eIF4E. As an essential component of mammalian translation, eIF4E binds to the 5' mRNA cap leading to the start of translation. The physical interaction between RBM38 and eIF4E causes the dissociation of eIF4E from p53 5' UTR, subsequently leading to decreased p53 translation. Furthermore, we demonstrated that phosphorylation of RBM38 at Ser195 modulates its ability to regulate p53 translation (Figure 1A). RBM38 phosphorylation by glycogen synthase kinase 3 (GSK3) enhances p53 translation by removing the RBM38 interaction with eIF4E, inducing p53 translation (34). Additionally, PPM1D phosphatase, a target of both p53 and RBM38, causes decreased phosphorylation of RBM38 at Ser195 leading to decreased p53 translation, as represented in Figure 1A (35). Physiologically, loss of RBM38 in mouse embryonic fibroblasts increased p53 protein levels triggering elevated premature senescence (36). RBM38 was additionally shown to be frequently elevated in dog lymphomas, often correlating with decreased expression of wild-type p53, emphasizing a novel auto-regulatory loop between p53 and a target RBP. The RBM38-p53 axis is not the only p53 target RBP autoregulation discovered thus far. For example, wild-type p53 induced gene 1 (WIG-1), a known p53 target and double-stranded-RNA-binding zinc finger protein, was shown to stabilize p53 mRNA by interacting with an AU-rich element (ARE) in p53 3' UTR leading to increased p53 expression in both normal and stressed cells (37).
Besides RBM38, various other RBPs have been revealed to regulate p53 translation. For example, ribosomal protein L26 (RPL26) augments p53 translation after DNA damage by interacting with a stem loop formed by p53 5' and 3' UTRs. RPL26 binding to p53 mRNA leads to an enhanced association with heavier polysomes, ultimately increasing p53 translation. The increased p53 translation results in G1 cell-cycle arrest and heightened irradiation-induced apoptosis (38). Another RBP, polypyrimidine tract-binding protein (PTB), a p53 internal ribosome entry site (IRES) interacting trans-acting factor, differentially controls the expression of p53 isoforms by preferentially binding to both p53 IRES elements (39). Moreover, HuR binds to target mRNAs with AREs and has been shown to modulate p53 translation via multiple approaches. HuR enhanced p53 translation in RKO cells treated with UVC by binding to the 3' UTR of p53 mRNA, whereas decreased HuR protein levels reduced p53 translation (40). Alternatively, HuR was demonstrated to induce p53 translation via von Hippel-Lindau (VHL)-dependent binding to p53 3' UTR (41), and further, in polyamine-depleted intestinal epithelial cells, HuR enhanced p53 mRNA stability (42). The above mentioned studies highlight the intricacy of HuR-mediated p53 expression. Like RBM38, other RBPs have been shown to inhibit p53 translation. For example, nucleolin competes with RPL26 to bind to p53 mRNA leading to decreased p53 translation. Over-expression of nucleolin subdued, whereas reduced endogenous nucleolin levels enhanced, IR-mediated p53 translation (38). Furthermore, thymidylate synthase binds to the C-terminal coding region of p53 mRNA leading to suppressed p53 translation (43). Interestingly, our laboratory recently demonstrated that PCBP4, a KH domain containing RBP and target of p53 (44), indirectly regulates p53 protein levels by modulating the mRNA stability of ZNF709. Knockout of PCBP4 led to increased ZNF709 protein expression ultimately leading to decreased p53 via a proteasome-dependent degradation pathway (45). These lines of evidence further solidify the overwhelming complexity of p53 regulation and open up the possibility for potential therapeutic intervention by modulating the regulation of p53 by RBP’s.
Regulation of p53 family member’s p63 and p73
Years after the discovery of p53, two highly homologous p53 family members were discovered, p63 and p73. With the discovery of p63 in 1997, and p73 in 1998, initial thoughts were that these two family members may share similar tumor suppressor functions to p53Jeny (46-49). However, while p63 harbors many p53-like attributes, such as inducing cell-cycle arrest, senescence and apoptosis, p63 is not a classic tumor suppresser, but rather, has been shown to be critical for proper development (50,51). Of interest, p63 is expressed as two isoforms, TAp63 and ΔNp63. Subsequent studies have demonstrated that TAp63 may function as a tumor suppressor promoting cell cycle arrest, senescence, and apoptosis (52). However, ΔNp63 acts as an oncogene, with the ability to bind p53-responisve promoters, leading to repressed gene expression of p53 targets (53). Further, ΔNp63α is frequently over-expressed in low-grade squamous cell carcinomas mostly attributed to chromosomal amplification (54). Likewise, p73 is expressed as two isoforms, TAp73 and ΔNp73. TAp73 functions as a tumor suppressor capable of inducing apoptosis and cell cycle arrest. Similar to ΔNp63, ΔNp73 may act as an oncogene inhibiting both TAp73 and p53 functions (55).
Captivatingly, our group uncovered that RBM38 negatively regulates p63 mRNA stability by interacting with AU-/U-rich elements in p63 3' UTR (56). In addition, our laboratory recently discovered that RBM24, which has high sequence homology to RBM38 (57), was able to bind multiple regions in p63 3' UTR, subsequently destabilizing the p63 transcript, leading to decreased p63 protein expression (58). Contrastingly, it was revealed that PCBP1 positively regulates p63 transcript by interacting with a CU-rich element (CUE) in p63 3' UTR leading to increased p63 mRNA and protein levels (59). The p53 family member p73 is likewise regulated by multiple RBPs. For example, PCBP2 interacts with CUEs in p73 3' UTR causing increased p73 mRNA stability and increased protein expression (60). Of interest, RBM38 was also shown to be a p73 target, and capable of binding a CUE in p73 3' UTR leading to enhanced p73 mRNA stability, emphasizing a novel positive feedback regulation between the two genes (61). Underlining the complex regulation of p53 family members by RBPs is demonstrated by the ability for RBM38 to decrease p53 mRNA translation, inhibit p63 expression via destabilization of its transcript, and promote p73 expression by increasing its mRNA stability.
Regulation of MDM2
As a key regulator and downstream target of p53, MDM2 was first identified as having gene amplification on double-minute chromosomes in transformed mouse fibroblasts (62). Further studies soon discovered that MDM2 was overexpressed in multiple human cancers, such as soft tissue sarcomas and osteosarcomas (63-65). Interestingly, high expression levels of MDM2 has been correlated with increased genomic instability revealed by amplified chromosome breaks, aneuploidy or polyploidy (66). Importantly, MDM2 interacts with p53 forming a regulatory feedback loop, where p53 induces MDM2 expression, and MDM2 negatively regulates p53 function and protein levels (67). MDM2 has been demonstrated to repress p53 transcriptional activity and lead to p53 protein degradation through three modes of action: (I) conceal the p53 transactivation domain; (II) cause the shuttling of p53 out of the nucleus; (III) target p53 for degradation as an E3 ubiquitin ligase (68-71).
A substantial amount of work has been done to unravel the regulatory mechanisms leading to increased MDM2 expression in cancer cells. In addition to the regulation by p53, multiple cancer cell lines exhibit enhanced MDM2 protein translation, such as cutaneous melanoma cells, breast cancer cells, and Burkitt’s lymphoma, underlining one potential mechanism for increased MDM2 protein levels (72-74). MDM2 transcription is under the control of two distinct promoters, P1 and P2 (75,76). Eloquent studies demonstrated that the P1 promoter, upstream of the first exon is responsible for the control of the basal expression of MDM2, whereas the P2 promoter located in the first intron is responsible for the inducible expression of MDM2. While both promoters encode identical transcripts, the translation efficiency due to differences in their 5' UTR is where they differ. The transcript from the P1 promoter contains two upstream open reading frames and was shown to have lower translation efficacy. Contrastingly, the 5' UTR from the P2 promoter was determined to be shorter allowing for efficient translation (75,76). Further, the Ras-driven Raf/MEK/MAP kinase pathway was discovered to induce MDM2 in a p53-independent fashion via activation of Ets and AP-1 sites in the P2 promoter (77).
MDM2 is also post-transcriptionally regulated by multiple RBPs. For example, our laboratory has reported that RBM38 influences MDM2 post-transcriptionally. Overexpression of RBM38 led to decreased MDM2 transcript and protein levels independent of p53 as represented in Figure 1B (78). This regulation was determined to be the result of RBM38 destabilizing MDM2 mRNA by binding to multiple AU-/U-rich elements in MDM2 3' UTR. With interest, HuR was discovered to be able to interact with, and stabilize, MDM2 mRNA (79). In addition, HuR is positively regulated by RBM38 via mRNA stability (80). Added, in a regulatory feedback loop, MDM2 interacts with, and stabilizes, HuR via MDM2-mediated NEDDylation, sequestering HuR in the nucleus protecting HuR from degradation (81). Both HuR and MDM2 contrastingly regulate p53, and stabilize each other, adding further complexity to the p53 regulation by RBPs. Further, BCR/ABL-expressing myeloid precursor cells showed enhanced MDM2 mRNA translation that required the interaction of the LA antigen with a 27-nucleotide segment in MDM2 5' UTR. Suppressing La by siRNA resulted in decreased MDM2 expression and heightened susceptibility to drug-induced apoptosis (82). Additionally, extensive work has revealed that numerous ribosomal proteins regulate MDM2, thus modulating p53 proteins levels (83). This intricate network of regulatory feedback between p53 and p53 targets with RBPs becomes more apparent as shown by the aforementioned regulations of the RBM38/MDM2/HuR/p53 axis.
Regulation of p21
A number of p53 targets are implicated in inhibiting tumorigenesis. For example, p21, a key regulator of p53 involved in cell cycle arrest, belongs to the Cip and Kip family of cyclin-dependent kinases (CDK) inhibitors. As a CDK inhibitor, p21 was discovered to be a major regulator in cell cycle transition from G1 to S by inhibiting the kinase activity of CDK2 and CDK1 (also known as CDC2) (84). Further, p21 directly interacts with proliferating cell nuclear antigen (PCNA), leading to decreased PCNA-dependent DNA polymerase activity, consequently inhibiting DNA replication and adversely affecting other PCNA-dependent DNA repair processes (85,86). Interestingly, p21 was shown to be a master regulator of multiple tumor suppressor pathways that were revealed to be independent of the p53 tumor suppressor pathway (87).
In addition to the regulation by the p53 family, p21 is regulated by multiple p53-independent mechanisms (46,50,84,88) [for a comprehensive review please see (89)]. For example, the signaling through the Ras GTPase induces, whereas signaling through the Rho GTPase inhibits, p21 transcription (90). This transactivation of p21 through Ras signaling was later revealed to require the transcription factor E2F1 (91). Heterogeneous nuclear ribonucleoprotein K (hnRNP K) was also shown to specifically bind to CUEs in p21 3' UTR leading to p21 translational repression (92). Like p53, p21 is positivity regulated by HuR. HuR interacts with the 3' UTR of p21 mRNA causing increased inducibility and half-life of the p21 transcript (93). Further, RBM38 stabilizes both the basal and stress-induced p21 transcripts by directly binding to the 3' UTR of p21 mRNA (94). Increasing the complexity of p21 regulation by RBM38, it was later discovered that RBM38 and HuR physically interact and preferentially bind the upstream and downstream AREs in p21 3' UTR, respectively (95). Additionally, the RNA-binding activity of HuR to the p21 transcript was enhanced with increased RBM38 protein expression, suggestion a cooperative regulation of p21 by both RBPs as demonstrated in Figure 1B. Our group additionally established that RBM24 was able to interact with an AU/U rich region located in p21 3' UTR heightening p21 expression (57). PCBP1 and PCBP2 have also been revealed to cause decreased p21 mRNA stability via interaction with its 3' UTR (96). Moreover, PCBP4 was proven by our group to negatively regulate the p21 transcript. In PCBP4-deficient mice, it was discovered that p21 expression was noticeably enhanced, and this regulation by PCBP4 was due to its binding to p21 3' UTR, negatively regulating p21 mRNA stability (97). Collectively, the reciprocal regulation of p53, and p53 targets, by RBPs is indeed complex. Recapitulated by the p53/HuR/RBM38/p21 regulatory axis, this adds further credence for the additional study of the posttranscriptional regulation of p53 and downstream targets by RBPs.
Conclusions and perspectives
In light of recent works, the significance of gene regulation via posttranscriptional regulation has been revealed to not only regulate p53, but that of p53 family members and downstream targets. Until recently, most emphasis has been put towards understanding the transcriptional and posttranslational modifications dictating p53 function and downstream pathways. However, as highlighted in this review, an increased understanding of the posttranscriptional regulation of p53 and p53 targets will surely be needed to appreciate the role of p53 in tumor suppression, and subsequently, in the development of p53-based therapies. Herein, we reiterated the ability for multiple RBPs to regulate p53, and p53 pathways. While it is now understood that p53 and p53 targets are regulated by RBPs, the reciprocal regulation of these RBPs in a regulatory feed-back loop with p53 is just now starting to be revealed.
Even though the last 30 years of research have uncovered an enormous amount of detail about the biology of p53, nonetheless, numerous questions about p53 regulation continually reappear in a new context. With the exciting findings of the reciprocal regulation of p53 by RBPs, such as the RBM38/HuR/MDM2/p53 regulatory loop, our knowledge of the complex regulation of p53 and the p53 network allows for the potential intervention with therapeutic approaches to upregulate the expression of p53, or p53 targets like p21. For example, as depicted in Figure 1, the regulatory network by RBM38 is multifaceted. RBM38 is induced by p53 and negatively effects p53 translation, while also adversely affecting MDM2, a key regulator of p53. Further, HuR upregulates p53, but also increases MDM2 translation. In addition, RBM38 and HuR cooperatively influence p21 mRNA stability increasing its expression. One could imagine, with a therapeutic application, that by removing the inhibition of RBM38 on p53, this would lead to an increase in both p53 and p21, potentially inducing tumor suppression. Blocking RBM38 from binding to p53 mRNA using oligonucleotides, or inhibiting the interaction between RBM38 and eIF4E via small competing peptides are two possible approaches aimed at modulating RBM38 regulation of p53. Importantly, RBM38 deficiency was shown to decrease tumor penetrance in mice heterozygous for p53 by enhancing p53 expression (36). Accumulating research into RBPs is continually revealing new RBPs and targets, while many questions still remain unanswered. For example, with increasing evidence that multiple RBPs are capable of regulating a single target, how is this regulation synchronized? Further, is there a reciprocal coordination between p53 and RBPs to regulate mutual targets, such as p21 or MDM2? Nonetheless, additional work is needed to garnish the knowledge of the intricate regulation of p53 and p53 family members with the hope of devising potential therapeutic approaches targeting RBPs.
Acknowledgments
Funding: This article is supported in part by NIH grants CA076069, CA081237, and CA121137.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.08.30). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Kosciuczuk EM, Saleiro D, Platanias LC. Dual targeting of eIF4E by blocking MNK and mTOR pathways in leukemia. Cytokine 2016. pii: S1043-4666(16)30024-2.
- Pandolfi PP. Aberrant mRNA translation in cancer pathogenesis: an old concept revisited comes finally of age. Oncogene 2004;23:3134-7. [Crossref] [PubMed]
- Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013;5:a012336 [Crossref] [PubMed]
- Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep 2009;42:125-30. [Crossref] [PubMed]
- López de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3'-untranslated region in human cancer. Cell Oncol 2007;29:1-17. [PubMed]
- Lukong KE, Chang KW, Khandjian EW, et al. RNA-binding proteins in human genetic disease. Trends Genet 2008;24:416-25. [Crossref] [PubMed]
- Sager R. Expression genetics in cancer: shifting the focus from DNA to RNA. Proc Natl Acad Sci U S A 1997;94:952-5. [Crossref] [PubMed]
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 2010;1:214-29. [Crossref] [PubMed]
- Hünten S, Siemens H, Kaller M, et al. The p53/microRNA network in cancer: experimental and bioinformatics approaches. Adv Exp Med Biol 2013;774:77-101. [Crossref] [PubMed]
- Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol 2014;15:R14. [Crossref] [PubMed]
- Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008;582:1977-86. [Crossref] [PubMed]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007;8:479-90. [Crossref] [PubMed]
- Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J 2013;280:3734-54. [Crossref] [PubMed]
- Castello A, Fischer B, Hentze MW, et al. RNA-binding proteins in Mendelian disease. Trends Genet 2013;29:318-27. [Crossref] [PubMed]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature 1990;345:544-7. [Crossref] [PubMed]
- Masse M, Glippa V, Saad H, et al. An eIF4E-interacting peptide induces cell death in cancer cell lines. Cell Death Dis 2014;5:e1500 [Crossref] [PubMed]
- Denkert C, Koch I, von Keyserlingk N, et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol 2006;19:1261-9. [Crossref] [PubMed]
- Heinonen M, Bono P, Narko K, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res 2005;65:2157-61. [Crossref] [PubMed]
- Wang J, Guo Y, Chu H, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci 2013;14:10015-41. [Crossref] [PubMed]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307-10. [Crossref] [PubMed]
- Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci 2004;61:822-42. [Crossref] [PubMed]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer 2002;2:594-604. [Crossref] [PubMed]
- Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049-56. [Crossref] [PubMed]
- Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002;415:45-53. [Crossref] [PubMed]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [Crossref] [PubMed]
- Donehower LA. The p53-deficient mouse: a model for basic and applied cancer studies. Semin Cancer Biol 1996;7:269-78. [Crossref] [PubMed]
- Chao C, Hergenhahn M, Kaeser MD, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 2003;278:41028-33. [Crossref] [PubMed]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997;90:595-606. [Crossref] [PubMed]
- Ito A, Lai CH, Zhao X, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 2001;20:1331-40. [Crossref] [PubMed]
- Luo J, Su F, Chen D, Shiloh A, et al. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000;408:377-81. [Crossref] [PubMed]
- Kruse JP, Gu W. Modes of p53 regulation. Cell 2009;137:609-22. [Crossref] [PubMed]
- Zhang J, Cho SJ, Shu L, et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev 2011;25:1528-43. [Crossref] [PubMed]
- Zhang M, Zhang J, Chen X, et al. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev 2013;27:2246-58. [Crossref] [PubMed]
- Zhang M, Xu E, Zhang J, et al. PPM1D phosphatase, a target of p53 and RBM38 RNA-binding protein, inhibits p53 mRNA translation via dephosphorylation of RBM38. Oncogene 2015;34:5900-11. [Crossref] [PubMed]
- Zhang J, Xu E, Ren C, et al. Mice deficient in Rbm38, a target of the p53 family, are susceptible to accelerated aging and spontaneous tumors. Proc Natl Acad Sci U S A 2014;111:18637-42. [Crossref] [PubMed]
- Bersani C, Xu LD, Vilborg A, et al. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene 2014;33:4407-17. [Crossref] [PubMed]
- Takagi M, Absalon MJ, McLure KG, et al. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005;123:49-63. [Crossref] [PubMed]
- Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 2008;7:2189-98. [Crossref] [PubMed]
- Mazan-Mamczarz K, Galbán S, López de Silanes I, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A 2003;100:8354-9. [Crossref] [PubMed]
- Galbán S, Martindale JL, Mazan-Mamczarz K, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol 2003;23:7083-95. [Crossref] [PubMed]
- Zou T, Rao JN, Liu L, et al. Polyamine depletion induces nucleophosmin modulating stability and transcriptional activity of p53 in intestinal epithelial cells. Am J Physiol Cell Physiol 2005;289:C686-96. [Crossref] [PubMed]
- Ju J, Pedersen-Lane J, Maley F, et al. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci U S A 1999;96:3769-74. [Crossref] [PubMed]
- Zhu J, Chen X. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol Cell Biol 2000;20:5602-18. [Crossref] [PubMed]
- Yan W, Scoumanne A, Jung YS, et al. Mice deficient in poly(C)-binding protein 4 are susceptible to spontaneous tumors through increased expression of ZFP871 that targets p53 for degradation. Genes Dev 2016;30:522-34. [Crossref] [PubMed]
- Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997;90:809-19. [Crossref] [PubMed]
- Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene 1997;15:1363-7. [Crossref] [PubMed]
- Trink B, Okami K, Wu L, et al. A new human p53 homologue. Nat Med 1998;4:747-8. [Crossref] [PubMed]
- Osada M, Ohba M, Kawahara C, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med 1998;4:839-43. [Crossref] [PubMed]
- Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998;2:305-16. [Crossref] [PubMed]
- Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999;398:714-8. [Crossref] [PubMed]
- Bergholz J, Xiao ZX. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenviron 2012;5:311-22. [Crossref] [PubMed]
- Westfall MD, Mays DJ, Sniezek JC, et al. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol 2003;23:2264-76. [Crossref] [PubMed]
- Massion PP, Taflan PM, Jamshedur Rahman SM, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003;63:7113-21. [PubMed]
- Rufini A, Agostini M, Grespi F, et al. p73 in Cancer. Genes Cancer 2011;2:491-502. [Crossref] [PubMed]
- Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci U S A 2010;107:9614-9. [Crossref] [PubMed]
- Jiang Y, Zhang M, Qian Y, et al. Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. J Biol Chem 2014;289:3164-75. [Crossref] [PubMed]
- Xu E, Zhang J, Zhang M, et al. RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol Cancer Res 2014;12:359-69. [Crossref] [PubMed]
- Cho SJ, Jung YS, Chen X. Poly (C)-binding protein 1 regulates p63 expression through mRNA stability. PLoS One 2013;8:e71724 [Crossref] [PubMed]
- Ren C, Zhang J, Yan W, et al. RNA-binding Protein PCBP2 Regulates p73 Expression and p73-dependent Antioxidant Defense. J Biol Chem 2016;291:9629-37. [Crossref] [PubMed]
- Yan W, Zhang J, Zhang Y, et al. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol Cell Biol 2012;32:2336-48. [Crossref] [PubMed]
- Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 1991;10:1565-9. [PubMed]
- Oliner JD, Kinzler KW, Meltzer PS, et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992;358:80-3. [Crossref] [PubMed]
- Leach FS, Tokino T, Meltzer P, et al. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 1993;53:2231-4. [PubMed]
- Cordon-Cardo C, Latres E, Drobnjak M, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 1994;54:794-9. [PubMed]
- Lushnikova T, Bouska A, Odvody J, et al. Aging mice have increased chromosome instability that is exacerbated by elevated Mdm2 expression. Oncogene 2011;30:4622-31. [Crossref] [PubMed]
- Wu X, Bayle JH, Olson D, et al. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993;7:1126-32. [Crossref] [PubMed]
- Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237-45. [Crossref] [PubMed]
- Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 1993;362:857-60. [Crossref] [PubMed]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299-303. [Crossref] [PubMed]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci U S A 1999;96:3077-80. [Crossref] [PubMed]
- Capoulade C, Bressac-de Paillerets B, Lefrère I, et al. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt's lymphoma cells. Oncogene 1998;16:1603-10. [Crossref] [PubMed]
- Polsky D, Bastian BC, Hazan C, et al. HDM2 protein overexpression, but not gene amplification, is related to tumorigenesis of cutaneous melanoma. Cancer Res 2001;61:7642-6. [PubMed]
- Okumura N, Saji S, Eguchi H, et al. Distinct promoter usage of mdm2 gene in human breast cancer. Oncol Rep 2002;9:557-63. [PubMed]
- Barak Y, Gottlieb E, Juven-Gershon T, et al. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev 1994;8:1739-49. [Crossref] [PubMed]
- Zauberman A, Flusberg D, Haupt Y, et al. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res 1995;23:2584-92. [Crossref] [PubMed]
- Ries S, Biederer C, Woods D, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 2000;103:321-30. [Crossref] [PubMed]
- Xu E, Zhang J, Chen X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene 2013;32:2169-78. [Crossref] [PubMed]
- Ghosh M, Aguila HL, Michaud J, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest 2009;119:3530-43. [Crossref] [PubMed]
- Cho SJ, Jung YS, Zhang J, et al. The RNA-binding protein RNPC1 stabilizes the mRNA encoding the RNA-binding protein HuR and cooperates with HuR to suppress cell proliferation. J Biol Chem 2012;287:14535-44. [Crossref] [PubMed]
- McLarnon A. Cancer: Mdm2-regulated stabilization of HuR by neddylation in HCC and colon cancer--a possible target for therapy. Nat Rev Gastroenterol Hepatol 2011;9:4. [PubMed]
- Trotta R, Vignudelli T, Candini O, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 2003;3:145-60. [Crossref] [PubMed]
- Kim TH, Leslie P, Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 2014;5:860-71. [Crossref] [PubMed]
- el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993;75:817-25. [Crossref] [PubMed]
- Waga S, Hannon GJ, Beach D, et al. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994;369:574-8. [Crossref] [PubMed]
- Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63-71. [Crossref] [PubMed]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400-14. [Crossref] [PubMed]
- Zhu J, Jiang J, Zhou W, et al. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res 1998;58:5061-5. [PubMed]
- Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res 1999;246:280-9. [Crossref] [PubMed]
- Olson MF, Paterson HF, Marshall CJ. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature 1998;394:295-9. [Crossref] [PubMed]
- Gartel AL, Najmabadi F, Goufman E, et al. A role for E2F1 in Ras activation of p21(WAF1/CIP1) transcription. Oncogene 2000;19:961-4. [Crossref] [PubMed]
- Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem 2005;280:12690-9. [Crossref] [PubMed]
- Wang W, Furneaux H, Cheng H, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 2000;20:760-9. [Crossref] [PubMed]
- Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev 2006;20:2961-72. [Crossref] [PubMed]
- Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res 2010;38:2256-67. [Crossref] [PubMed]
- Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem 2009;284:9039-49. [Crossref] [PubMed]
- Scoumanne A, Cho SJ, Zhang J, et al. The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res 2011;39:213-24. [Crossref] [PubMed]


