Exosomes genetic cargo in lung cancer: a truly Pandora’s box
Biomarkers of non-small cell lung cancer (NSCLC)
Lung cancer is the second most common cancer and the leading cause of cancer death for both men and women besides the important therapeutic achievements, the global disease control rate is still low. Other factors that can contribute to this unsuccessful scenario is the delay in the early detection, including also the absence of reliable biomarkers (1) and new drugs to fulfill an unmet medical need, specifically in NSCLC.
Biomarkers play a crucial role in medicine, as indicators of normal or pathological processes or as tools to assess pharmacological responses to therapeutic interventions or as a prognostic and predictive aid to elucidate the risk of recurrence and progression and the possibilities of treatments effectiveness (2). For this reason, biomarkers have become one of the most studies aspects in cancer (3).
The introduction of molecular biology in oncology, and mainly in NSCLC, made possible to identify drugable targets, for example EGFR mutation and ALK translocations among others. In this personalize treatment era, tissue became one of the most important treasure in the diagnosis and recurrence of the disease. Unfortunately, the amount of material is a big issue in lung cancer, due to the impossibility to reach the tumor. At this point, new tools have to be discovered in order to reveal the molecular information of the tumor. In this regards liquid biopsy has become a more and more important instrument in cancer research and in the clinic.
Liquid biopsy
Nowadays important efforts in molecular profiling of the tumors are allowing to develop new technology that permits to analyze the tumor characteristics in peripheral blood. Accordingly, the liquid biopsy refers to the analysis of components that can be isolated and analyzed from a blood sample as, circulating tumor cells (CTCs), cell free circulating DNA (circulating tumor DNA, ctDNA) and exosomes (a part of the secreted micro-vesicles) (4-6). In this regard, CTCs have been demonstrated to be good predictors for risk of metastatic progression, to monitor the response of an undergoing treatment, or to identify new targets and resistance mechanisms (7). On the other hand, ctDNA the most used component of the liquid biopsies is now being studied as possible biomarker and prognostic factor in cancer, although it is still difficult to standardize (8-11).
Nevertheless, the ultimate techniques developed in the field of sequencing and DNA amplification such as next generation sequencing (NGS) and digital droplet PCR (ddPCR) are transforming the molecular profiling of the tumor in a fast, easy, reliable and affordable manner (12-15).
Exosomes
Exosomes are the last frontier in liquid biopsies. Exosomes are spherical nano-size vesicles with a diameter between 30 to 100 nm and with a well delimited round morphology by electron microscopy that are released by exocytosis from multivesicular bodies of late endosome (16). They are exocytosed in a constitutive manner by any cell in both physiological and pathological condition and can be found in several body fluids like urine, saliva, amniotic liquid or blood (17,18). Exosome’s composition may reflect that of the parental cells, so the study of cancer derived exosomes (DEX) are an important non-invasive surrogate of the tumor. Additionally, as exosomes are linked to cell-secretion pathway, they carry some common proteins independent of the cells of origin that allow the development of robust exosomes-isolation techniques. These proteins are well known exosomes markers, such as ALIX, CD9, CD63, CD81, HSP70 and TSG-101, among others (19).
Several methods are used to extract exosomes with no clear winner. The principal methods to isolate the exosomes are ultracentrifugation, sucrose density-gradient ultracentrifugation, exosome immunoprecipitation and the isolation using commercial kits (20).
Inside the exosomes
Exosomes contain a wide variety of components such as single-stranded RNA, long non-coding RNA (lncRNA), microRNA (miRNA), proteins and lipids. Recently also double-stranded DNA has also been found inside the exosomes giving a new chance for exosomes to become the principal diagnostic tool (21) (Figure 1).

Since their discovery, the amount of information about his microvesicles has raised considerably, and a ‘microvesicle content database’ has been created to facilitate the access to all the newly discovered components (http://www.microvesicles.org). Currently (June 2016) this database contained 92,897 proteins, 27,642 mRNA, 4,936 miRNAs and 584 lipids based on a total of 538 studies. Although many of these proteins are common to all cell types, some others are privative for a certain cell alteration or tumorogenic process and can be useful for the identification of the origin of these vesicles and thus can be an adequate biomarker to get more information from the tumor of origin. In addition the study of the lipids is interesting due to their role in the internalization process of exosomes and to the bioactive function of some of them such as prostaglandins and leukotrienes (22).
Additionally, Valadi et al. described for the first time the presence of miRNAs inside (23) exosomes. miRNAs are short single-stranded and non-coding RNA molecules that act regulating gene expression as oncogenic or tumor suppressor function. They are involved in progression, cellular differentiation, apoptosis or cell signaling.
Exosomes biology
Although exosomes can be released directly from the plasma membrane (24), most of them are derived from the endosomal compartment as a part of the endocytic machinery (25). A very important part of the exosomes formation is controlled for the mechanism of endosomal sorting complex required for transport (ESCRT), which is the responsible for the accumulation and sorting of molecules in the endocytic machinery. In tumor cells, where ESCRT is significantly altered, the protein profile inside the exosomes and the amount of exosomes released can be modified (26,27). Once exosomes are released from the cells other cells can capture these vesicles as a physiological response to the endocytic machinery fusing them either with the plasma membrane or with lysosomes. If the exosomes fuse with lysosomes, their content will be degraded by proteolytic enzymes (28), but in contrast if the exosomes fuse with the plasma membrane, their content is released into the cytoplasm of the cell (29) being able to modify in a autocrine or paracrine manner the target cell behavior. This exosome’s uptake has reveal that they have a very important role in cell to cell communication or crosstalk (30,31) and three principal mechanisms governing this process have been described up to date (32). The first one, common to all type of vesicles is endocytosis, including pinocytosis, clathrin-mediated endocytosis, phagocytosis and others (30,33-35). A second mechanism is by direct binding of both cell and exosome membranes (36), leading to membranes fusion. Exosomes contain an enormous amount of surface proteins such as leptins, immunoglobulins, integrins and tetraspanins that are involved in their function, motility and internalization into the cells (34,37). The third mechanism is linked to ligand-receptor interaction that is the effect of the interplay of specific proteins in the exosome and the target cell surface (32). In this regard, tumor metastasis is not produced to random organs but to determine organs in each case and exosomes contribute to prepare the pre-metastatic niche. For example, integrin profiling in exosomes could be a feasible tool to identify possible organotropic metastasis before they have been produced (38).
Exosomes and cancer
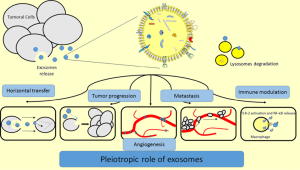
In recent years, more and more studies are ongoing with regard to the function of exosomes and their application for tumor detection and treatment and several reports have demonstrated that exosomes can play pleiotropic roles affecting pivotal aspects of tumor development and growth (Figure 2). For instance, exosomes derived from prostate tumor cells act in a paracrine fashion inducing a reprogramming of cell metabolism and enhancing of cell proliferation (39). The fusion gene TMPRSS2:ERG and EGFR, both related with advanced prostate have been described inside the exosomes (40,41). Cells from renal carcinoma are able to release exosomes containing some miRNAs and other RNAs that transform normal endothelial cells into an activated angiogenic phenotype, and directing towards lung cancer metastasis (42). In other studies it has been demonstrated that exosomes derived from highly metastatic melanoma cells promote the pre-metastatic niche formation through the education of the bone marrow and by reprogramming its progenitor cells to a pro-vasculogenic phenotype (43). On the other hand, another described role of exosomes in breast cancer and multiple myeloma is the ability to promote drug resistance through horizontal transfer (44,45). Exosomes are also involved in the creation of a pro-inflammatory microenvironment that promotes tumor growth by means of immunological proteins such as MHC-II, CD40 and CD40L. In breast cancer exosomes containing 27-Hydroxycholesterol (27-OHC), a lipid associated with proliferation and metastasis in estrogen receptor positive (ER+) tumor cells, open a new window to understand how exosomes genesis is achieved and its correlation with the content of the cells of origin (46). Other studies performed in exosomes derived from ovarian cancer cell lines show that exosomes can carry functional proteins that reprogram cell metabolism of the cell enhancing the pentose phosphate pathway that is crucial for resisting oxidant injury and favors tumor survival (47-49). Many miRNAs profiles have been identified in cancer-related exosomes. The oncogenic miRNA-21, upregulated in exosomes released by chronic myelogenous leukemia after Curcumin treatment, has been one of the most studied miRNA (50). Moreover, miRNA-21 has been found to be upregulated in different types of tumors such as esophageal squamous cell carcinoma (51) or glioblastoma (52), and its levels correlated with tumor progression and aggressiveness. miR-21 has also been described to be elevated in pancreatic carcinoma but no correlation was found with the stage of the disease (53). In ovarian cancer a profile of eight miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214) in both tumor cells and tumor exosomes has been already analyzed in a large cohort of 467 patients. The results revealed that levels of the miRNAs were different from those measured in benign disease, and the same miRNAs were undetectable in healthy donors (54).
Exosomes in NSCLC
As it was mentioned before, exosomes contain overexpressed or mutated proteins in the same proportion as the primary tumors. Therefore, in the specific case of exosomes obtained from NSCLC patients, EGFR (55), K-ras, claudins and other proteins can be analyzed. Recently, the translocation ALK-EML4 has also been identifying inside the exosomes (56). Al-Nedawi et al. demonstrated that exosomes carrying EGFR were able to interact with endothelial cells activating both MAPK and Akt pathways, resulting on an overexpression of VEGF and an augmentation of the vascularity of the tumor (57).
Many non-coding RNAs (ncRNAs) have been described in NSCLC, being involved in various processes of cancer formation and evolution (58,59) (Table 1). Rabinowits et al. carried one of the first studies concerning the possibility of using exosomes as a source of genetic material derived from the tumor comparing the miRNA content between the exosomes and the tumoral cells from patients with NSCLC and with healthy donors. Although the study revealed no differences between both compartments (72) it opened a window for tumor molecular identification. In a proof of concept, in a small cohort of patients with diagnosed NSCLC and a group of healthy donors, Rolfo et al. demonstrated the upregulation of two miRNA related with the EGFR pathway, 30b and 30c (60).

Full table
In a study that included 28 NSCLC patients, 365 miRNAs were analyzed and the level of let-7f and miR-30e-3p in were found to be associated with poor outcome. The authors also found that patients with more advanced stage of disease and lymph node metastases had higher exosome related miR-20b levels (62). miRNA-373 and miRNA-512 are tumor suppressor miRNA that are silenced in the tumor and whose reactivation produces a restriction of the growth and invasiveness of the tumor. Studying exosomes it has been demonstrated that the downregulation of these two miRNAs is associated with as a poor prognosis in NSCLC patients and that therapy that affects epigenetic status led to re-expression of both miRNAs which, on one side, augmented cisplatin-induced apoptosis while, on the other side inhibited cell migration (61). Other miRNAs present in the exosomes have been identified as possible biomarkers in response to certain treatments. miR-29a-3p and miR-150-5p were shown to decrease with increasing radiation dose so, and therefore identified as a reproducible circulating biomarkers that correlated with delivered radiotherapy dose (63). In another example, miR-208a and miR-1246 through the binding to p21 and DR5 mRNAs respectively, have been reported to promote tumor progression and resistance to radiotherapy (64,65). Also miR-302b has been described to block the cell proliferation and cell migration through interaction with its target, the mRNA of TGFβRII (66). Finally, our group recently described recently a correlation between the overexpression of miR-221-3p and 222-3p, both related with EGFR status, and a good clinical outcome in patients under osimertinib treatment (67). In addition to miRNAs, other ncRNAs have been identified inside the tumor-DEX, such as lncRNAs; non-coding RNA molecules with more than 200 bp with either tumor suppressor or oncogenic functions. However, the study of ncRNAs is less extended due to a lower general knowledge in the field and the lack of known function of many lncRNAs. In this regard, some lncRNAs have been described and studied in lung cancer but it has not been elucidated if are also present in exosomes. A good example of this is HOTAIR a largely studied onco-lncRNA related with resistance to cisplatin in NSCLC through p21 that has been also related with proliferation, migration and invasion (70,71).
Exosomes have been considered also as possible vehicles for the selective and directed administration of drugs. In lung cancer two clinical trials were planned, a Phase I trial in which DEX immunotherapy were used, and a Phase II in which DEXs vaccination with exosomes carrying IL-15Ra and NKG2D in association with cyclophosphamide after platinum-based chemotherapy. The main objective of both studies was to measure the toxicity and they had positive results, concluding that DEXs are able to activate the adaptive and the innate immune system (68,73).
Discussion
Thanks to the lipid bilayer that makes the cargo very stable, exosomes allow the analysis of genetic material in order to identify the tumor of origin, mutations and possible resistances for the treatments, among others. However, some methodological issues are still focus of debate and need better implementations. For the moment, there are no standardized methods to extract exosomes. The most used protocol relies in ultracentrifugation in sucrose gradient, a step that is complicated to implement in the clinical setting due to the lack of expertise in the field of many laboratories. Some companies have started the development of user friendly and reproducible kits to improve the time, the quality and the yield of exosome extraction.
Some other controversial issues remain in exosome research. One of the key factors for exosome production and function is the cell or tissue microenvironment. For the in vitro analysis, most of the studies are performed in completely artificial monolayer of cultured tumor cells. To avoid this spatial problem Villasante et al. have recently published a technique based in a three dimensional tumor model to bypass the effect of culture microenvironment in tumor exosomes production (69).
In quantitative studies, another important problem is still the absence of endogenous control for exosomal miRNA titration. Some groups have described the miR-1228 as a good endogenous control in several investigations (67,74,75), but definitively, more studies are needed to validate this finding.
In conclusion, liquid biopsies have become a reality in lung cancer. The learning curve for oncologists includes CTCs and the current hot topic, circulating free DNA. Exosomes are still an area of research but they are likely to be implemented in the clinical practice in the near future, opening a new window in the cancer prediction, prognosis and treatment. Although more efforts are required in order standarize exosome isolation and analysis so that they can be implemented in the clinical setting, step by step we understand more of the exosome-related physiologic processes and their impact in carcinogenesis. The main point about exosomes is their pleiotropic role in cancer, not only as biomarkers but also as a sort of “Pandora box” that contains the "instructions" for migration and aggressiveness of the tumor and information about druggable targets. We are only in the beginning of the long, promising history of this new component of the liquid biopsy family.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Sturgeon CM, Duffy MJ, Stenman UH, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clin Chem 2008;54:e11-79. [Crossref] [PubMed]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001;69:89-95. [Crossref] [PubMed]
- Yap TA, Lorente D, Omlin A, et al. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res 2014;20:2553-68. [Crossref] [PubMed]
- Mäbert K, Cojoc M, Peitzsch C, et al. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol 2014;90:659-77. [Crossref] [PubMed]
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087-92. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. [Crossref] [PubMed]
- Yanagita M, Redig AJ, Paweletz CP, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Jansen MP, Martens JW, Helmijr JC, et al. Cell-free DNA mutations as biomarkers in breast cancer patients receiving tamoxifen. Oncotarget 2016. [Epub ahead of print]. [PubMed]
- Hadano N, Murakami Y, Uemura K, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer 2016;115:59-65. [Crossref] [PubMed]
- Camps C, Jantus-Lewintre E, Cabrera A, et al. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer 2011;72:365-9. [Crossref] [PubMed]
- Castéra L, Krieger S, Rousselin A, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 2014;22:1305-13. [Crossref] [PubMed]
- Hortobagyi GN, Piccart-Gebhart MJ, Rugo HS, et al. Correlation of molecular alterations with efficacy of everolimus in hormone receptor–positive, HER2-negative advanced breast cancer: Results from BOLERO-2. J Clin Oncol 2013;31:abstr LBA509.
- Chen WW, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids 2013;2:e109. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012;10:e1001450. [Crossref] [PubMed]
- Conde-Vancells J, Rodriguez-Suarez E, Embade N, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res 2008;7:5157-66. [Crossref] [PubMed]
- Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009;9:4997-5000. [Crossref] [PubMed]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009;9:581-93. [Crossref] [PubMed]
- Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3.22.
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Record M, Carayon K, Poirot M, et al. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta 2014;1841:108-20.
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Booth AM, Fang Y, Fallon JK, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol 2006;172:923-35. [Crossref] [PubMed]
- Morvan J, Rinaldi B, Friant S. Pkh1/2-dependent phosphorylation of Vps27 regulates ESCRT-I recruitment to endosomes. Mol Biol Cell 2012;23:4054-64. [Crossref] [PubMed]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. [Crossref] [PubMed]
- Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 2013;126:5553-65. [Crossref] [PubMed]
- Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011;20:131-9. [Crossref] [PubMed]
- Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012;13:328-35. [PubMed]
- Fitzner D, Schnaars M, van Rossum D, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 2011;124:447-58. [Crossref] [PubMed]
- Atay S, Gercel-Taylor C, Taylor DD. Human trophoblast-derived exosomal fibronectin induces pro-inflammatory IL-1β production by macrophages. Am J Reprod Immunol 2011;66:259-69. [Crossref] [PubMed]
- Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014.3. [PubMed]
- Feng D, Zhao WL, Ye YY, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010;11:675-87. [Crossref] [PubMed]
- Escrevente C, Keller S, Altevogt P, et al. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011;11:108. [Crossref] [PubMed]
- Stephens L, Ellson C, Hawkins P. Roles of PI3Ks in leukocyte chemotaxis and phagocytosis. Curr Opin Cell Biol 2002;14:203-13. [Crossref] [PubMed]
- Parolini I, Federici C, Raggi C, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem 2009;284:34211-22. [Crossref] [PubMed]
- Christianson HC, Svensson KJ, van Kuppevelt TH, et al. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A 2013;110:17380-5. [Crossref] [PubMed]
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329-35. [Crossref] [PubMed]
- Zhao H, Yang L, Baddour J, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016;5:e10250. [Crossref] [PubMed]
- Kharmate G, Hosseini-Beheshti E, Caradec J, et al. Epidermal Growth Factor Receptor in Prostate Cancer Derived Exosomes. PLoS One 2016;11:e0154967. [Crossref] [PubMed]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7. [Crossref] [PubMed]
- Grange C, Tapparo M, Collino F, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 2011;71:5346-56. [Crossref] [PubMed]
- Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [Crossref] [PubMed]
- Ambudkar SV, Sauna ZE, Gottesman MM, et al. A novel way to spread drug resistance in tumor cells: functional intercellular transfer of P-glycoprotein (ABCB1). Trends Pharmacol Sci 2005;26:385-7. [Crossref] [PubMed]
- Wang J, Hendrix A, Hernot S, et al. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood 2014;124:555-66. [Crossref] [PubMed]
- Roberg-Larsen H, Lund K, Seterdal KE, et al. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J Steroid Biochem Mol Biol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Yi H, Zheng X, Song J, et al. Exosomes mediated pentose phosphate pathway in ovarian cancer metastasis: a proteomics analysis. Int J Clin Exp Pathol 2015;8:15719-28. [PubMed]
- D’Alessandro A, Amelio I, Berkers CR, et al. Metabolic effect of TAp63alpha: enhanced glycolysis and pentose phosphate pathway, resulting in increased antioxidant defense. Oncotarget 2014;5:7722-33. [Crossref] [PubMed]
- Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 2014;5:592-602. [Crossref] [PubMed]
- Taverna S, Giallombardo M, Pucci M, et al. Curcumin inhibits in vitro and in vivo chronic myelogenous leukemia cells growth: a possible role for exosomal disposal of miR-21. Oncotarget 2015;6:21918-33. [Crossref] [PubMed]
- Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159-67. [Crossref] [PubMed]
- Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Que R, Ding G, Chen J, et al. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol 2013;11:219. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Krug AK, Karlovich C, Koestler T, et al. Abstract B136: Plasma EGFR mutation detection using a combined exosomal RNA and circulating tumor DNA approach in patients with acquired resistance to first-generation EGFR-TKIs. Am Assoc Cancer Res 2016;14:B136.
- Brinkmann K, Enderle D, Koestler T, et al. Abstract 545: Plasma-based diagnostics for detection of EML4-ALK fusion transcripts in NSCLC patients. Cancer Res 2015;75:545. [Crossref]
- Al-Nedawi K, Meehan B, Kerbel RS, et al. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A 2009;106:3794-9. [Crossref] [PubMed]
- Usó M, Jantus-Lewintre E, Sirera R, et al. miRNA detection methods and clinical implications in lung cancer. Future Oncol 2014;10:2279-92. [Crossref] [PubMed]
- Giallombardo M, Chacártegui Borrás J, Castiglia M, et al. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients' Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J Vis Exp 2016;(111).
- Rolfo C, Chacartegui J, Giallombardo M, et al. 71P Exosomes isolated in plasma of non-small cell lung cancer patients contain microRNA related to the EGFR pathway: Proof of concept. J Thorac Oncol 2016;11:S85. [Crossref] [PubMed]
- Adi Harel S, Bossel Ben-Moshe N, Aylon Y, et al. Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth. Cell Death Differ 2015;22:1328-40. [Crossref] [PubMed]
- Silva J, García V, Zaballos Á, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 2011;37:617-23. [Crossref] [PubMed]
- Dinh T-KT, Fendler W, Chalubinska-Fendler J, et al. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat Oncol 2016;11:61. [Crossref] [PubMed]
- Yuan D, Xu J, Wang J, et al. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016;7:32707-22. [PubMed]
- Tang Y, Cui Y, Li Z, et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res 2016;35:7. [Crossref] [PubMed]
- Li J, Yu J, Zhang H, et al. Exosomes-Derived MiR-302b Suppresses Lung Cancer Cell Proliferation and Migration via TGFβRII Inhibition. Cell Physiol Biochem 2016;38:1715-26. [Crossref] [PubMed]
- Giallombardo M, Jorge Chacartegui J, Reclusa P, et al. Follow up analysis by exosomal miRNAs in EGFR mutated non-small cell lung cancer (NSCLC) patients during osimertinib (AZD9291) treatment: A potential prognostic biomarker tool. J Clin Oncol 2016;34:abstr e23035.
- Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med 2005;3:9. [Crossref] [PubMed]
- Villasante A, Marturano-Kruik A, Ambati SR, et al. Recapitulating the Size and Cargo of Tumor Exosomes in a Tissue-Engineered Model. Theranostics 2016;6:1119-30. [Crossref] [PubMed]
- Ono H, Motoi N, Nagano H, et al. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med 2014;3:632-42. [Crossref] [PubMed]
- Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One 2013;8:e77293. [Crossref] [PubMed]
- Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Viaud S, Thery C, Ploix S, et al. Dendritic cell-derived exosomes for cancer immunotherapy: what’s next? Cancer Res 2010;70:1281-5. [Crossref] [PubMed]
- Rolfo CD, Giallombardo M, Castiglia M, et al. Exosomes isolation and characterization in non small cell lung carcinoma patients: Proof of concept study. J Clin Oncol 2015;33:abstr 11101.
- Hu J, Wang Z, Liao BY, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J cancer 2014;135:1187-94. [Crossref] [PubMed]