
Implementing tumor mutational burden (TMB) analysis in routine diagnostics—a primer for molecular pathologists and clinicians
Introduction
Tumor mutational burden (TMB) is emerging as new predictive biomarker to select patients that benefit from immune checkpoint inhibitor therapy (1-7). It is commonly defined as the total number of somatic coding mutations and associated with the emergence of neoantigens that trigger anti-tumor immunity (8-10). As a defense mechanism, tumors acquire expression of checkpoint regulators, like programmed death-ligand 1 (PD-L1), the action of which can be overcome in clinical practice with therapeutic antibodies against PD-1 (programmed cell death protein 1) or PD-L1 alone or in combination with CTLA-4 (cytotoxic T-lymphocyte-associated protein 4) inhibitors or chemotherapy or more recently small-molecule kinase inhibitors (11-13). In numerous clinical trials over the past few years (14-21), these therapies have demonstrated impressive anti-tumor activity and are already approved for a large number of tumor entities in various indications (22) including non-small cell lung cancer (NSCLC) (23,24). Up to now, patient selection for immuno-oncologic (IO) treatment has been mainly based on expression of PD-L1 as assessed by immunohistochemistry (25,26). However, accumulating data suggest a much greater utility of TMB for this purpose.
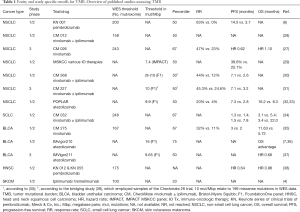
A retrospective analysis of the CheckMate (CM) 026 trial investigating first-line nivolumab vs. platinum-based chemotherapy in stage IV or recurrent NSCLC (27) uncovered that patients with high TMB showed higher response rates and longer progression-free survival (PFS) with PD-1 blockade (Table 1). Moreover, among TMB-high patients, a high (≥50%) PD-L1 expression distinguished an even more favorable subgroup with presumably “hot”, immunologically reactive tumors, but it had no predictive value among TMB-low cases. In keeping with these data, a post-hoc analysis in the nivolumab-ipilimumab arm of the CM 012 trial (28) also showed longer PFS of TMB-high cases under IO therapy, especially if PD-L1 was additionally present, but limited benefit in the TMB-low subgroup regardless of PD-L1 expression. Conversely, by focusing on PD-L1 negative patients, the CM 227 trial demonstrated a significant and incremental benefit from administration of nivolumab alone or in combination with ipilimumab for TMB-high cases, while additional presence of low TMB defined a truly refractory subset, for which novel IO approaches or non-IO treatment will be necessary (31,40). Notably, in all aforementioned analyses, PD-L1 expression was not associated with TMB levels, indicating that the two biomarkers are largely independent in NSCLC and probably also in other cancers (41). In addition, their results collectively may suggest a more fundamental role of a higher TMB for efficacy of currently available IO treatments, which therefore emerges as a potentially more suitable basis for the selection of NSCLC patients likely to benefit from them. At present, caution is definitely warranted, because overall survival (OS) data in conjunction with TMB are still pending and TMB measurement has not been used yet as an upfront stratification parameter in a prospective trial design. However, recent preliminary data suggest that higher TMB, but not higher PD-L1 expression levels, can reliably distinguish NSCLC patients predestined to experience long-term (>18 months) remissions with IO treatments, which is of utmost clinical importance, because it represents a proxy for “functional cure” (42). Since TMB appears to be predictive for efficacy of checkpoint blockade across diverse cancers (2,3), it is as an additional biomarker next to PD-L1 expected to improve care of many patients, which currently drives intense efforts for seamless integration of TMB assays in routine diagnostics.
Full table
TMB and cutoff values
Similar to the challenges of using PD-(L)1 expression as a biomarker for patient selection (43), there are many unsolved aspects of TMB assessment and interpretation which will need to be addressed before widespread adoption. Mutational burden is a continuous variable and the question arises how to define TMB-high tumors to enrich for patients likely to respond to IO therapy. Objective cut-points for TMB are not universally established. Clinical trials have mostly deferred study-specific cut-points using median TMB or dividing patients in tertiles or quartiles according to measured TMB (Table 1). Studies of NSCLC employing whole exome sequencing (WES) have for example set cut-points at 158 (median) (28), 200 (median) (6), and 243 (upper tertile) (27) mutations per exome. A study of SCLC determined 248 mutations per exome (34) to delineate the upper tertile, which is largely comparable to NSCLC. In a study of urothelial carcinoma, however, a lower number of 167 total mutations (35) was found to mark the upper tertile of patients. WES is considered the gold standard of TMB assessment but it bears noting that the size of the ‘exome’ depends on the enrichment method (exome capture kits) (44,45) used and that mutation types for TMB count vary between assays. Therefore, comparison of WES and panel sequencing in terms of TMB and cut-points not only requires detailed information of the panel including bioinformatics but also a clear definition of the technical reference standard (i.e., WES). For targeted gene panel sequencing in NSCLC cut-points were set at around 10 mutations per megabase (mut/Mbp) for Foundation Medicine panels (31,33) and 7.4 mut/Mpb for the MSK-IMPACT panel (29). A recent bridging study (39) using 44 samples from the Checkmate 26 trial demonstrated that the cut-point of 10 mut/Mbp (as determined by the FMI panel) relates to 199 missense mutations determined by WES. For urothelial carcinoma using the Foundation medicine (FMI) panel two different study-specific cut-points were applied with 9.65 mut/Mbp (median) (37) and 16 mut/Mbp (upper quartile) (36).
Factors influencing TMB detection
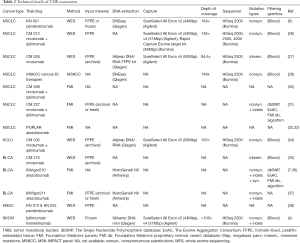
For accurate TMB assessment pre-analytical, analytical, and post-analytical variables have to be considered and results be carefully interpreted in the overall context. Based on the current literature, five major parameters influencing TMB measurement and related cutoff values can be delineated: (I) tumor type (46,47); (II) indication including drug types; (III) pre-analytics (including assessment of tumor cellularity); (IV) method (WES vs. type of panel sequencing including size and composition of panels, read depth and coverage); and (V) bioinformatics (including: limit of detection (LOD), threshold for allelic frequency [clonal vs. subclonal mutation], and filter settings for germline events and deamination artifacts, definition of mutation). Pre-analytical factors include input material, tumor cellularity, and DNA quality and quantity. Clinical trials have mostly employed formalin-fixed, paraffin-embedded (FFPE) tissue, either fresh (obtained for study purpose) or archived (routine clinical specimens) or occasionally obtained fresh frozen tissue plus blood for germline analysis. Table 2 gives an overview of the technical parameters of major TMB studies as provided in the respective papers. Assessment of tumor cellularity by a pathologist is warranted to ensure validity of input material (presence, percentage, and viability of tumor cells) and thereby aid subsequent interpretation of allelic frequencies. It should be noted, however, that tumor cellularity estimates itself influence allelic frequency calculations and related mutation counts (depending on LOD and filter settings). In daily clinical practice one most often has to resort to (archived) FFPE samples with usually suboptimal DNA quality (due to various degrees of DNA degradation and deamination artifacts) compared to fresh unfixed tissue. Quantities are also rather limited especially from routine clinical lung samples that are often small (transbronchial/endobronchial/image-guided) biopsies which are also used for standard pathology work-up including (limited) immunohistochemistry for tumor typing and PD-L1 staining. Most of the time this is done sequentially requiring repeated cutting of the paraffin block which causes additional tissue loss. A one-stop shop approach that yields all the required sections in one session is needed to optimize material usage (Figure 1). However, this will require upfront communication between clinicians, lab personnel, and pathologists, also to ensure that additional immunohistochemical and genetic studies are only performed when absolutely needed for accurate diagnosis. Looking at recent clinical trials, a rather high attrition rate of patients that could not be assessed for TMB is apparent, which might be due to sequential testing, enrichment method used (hybrid capture requires higher amount of input DNA), or unplanned addition of TMB testing. In the CM 026 trail (27), of 541 randomized patients only for 320 (59%) TMB could be assessed. Similar in the CM 227 trail (31) in which of 1,739 patients, only 1,004 (58%) had TMB-evaluable samples. Only 98 (34%) patients of 288 in the CM 568 trial had evaluable samples (30). Also in the IMvigor211 trial (37) only 544 (58%) of 931 patients had tumor samples that could be successfully tested.
Full table
In the future, blood based TMB assays might be an alternative in those clinical scenarios where tissue cannot be obtained and even more would allow for non-invasive monitoring of TMB over time. Assays are currently in development and first reports have shown correlation to TMB measured from tissue samples (48-50). However, current data are still preliminary and limited. One of the main factors influencing such approaches is the DNA yield obtained from a blood-draw: sequencing with sufficient sensitivity is well feasible but (tumor-related) DNA molecules that are not present cannot be sequenced (51).
Bioinformatics
Clinical interpretation of conventional sequencing approaches—though based on quantitative data like allele frequencies and base changes—mostly results in a qualitative result, e.g., presence or absence of a BRAF V600E mutation. In contrast, TMB as a continuous numerical value is a quantitative measurement. As such its measurement is more complex with the associated risk of capturing significant background noise. In daily clinical practice, germline samples are not routinely available for comparative sequencing due to regulatory constraints not permitting germline sequencing, logistic challenges, or financial considerations. Therefore, filtering of germline variants is paramount as not to count them as tumor specific mutations. Known polymorphisms with minor allele frequencies (MAF) <0.0001 annotated in the dbSNP and ExAC databases (52) should be filtered. Some groups have also introduced algorithms to predict germline mutations (53). Clonal hematopoiesis of indeterminate potential (Chip) might also contribute to few falsely attributed mutations in tumor-only sequencing (54) but is likely to be of only minor relevance in TMB assessment. Importantly, low allelic frequency C>T transitions that are introduced by fixation in formalin (i.e., deamination artifacts) need to be filtered and not mistaken as true mutational events.
FMI excludes recurrent mutations occurring in tumors and truncating mutations of tumor suppressor genes from their TMB calculation to avoid overestimation of TMB since gene panels are usually heavily targeted at recurrently mutated genomic regions (55). Our own data indicate that this is a valid approach though the influence of hot spot mutations on TMB is not prominent (Buchhalter I, 2018, submitted). Studies have used 5% allelic frequency as a cutoff to count mutations (LOD) to distinguish clonal vs. subclonal mutations which are entity specific and also influenced by therapy (56). In addition, patients with elevated clonal neo-antigen load and little subclonal heterogeneity were overrepresented in the group of patients benefitting from IO therapy (57). Whereas most WES studies included only non-synonymous mutations (mostly restricted to missense mutations) primarily for technical reasons (i.e., clear and reliable identification of this mutation type), panel sequencing approaches add insertions and deletions (indels) (Table 2). Some like FMI also include synonymous mutations, which even though they are not themselves giving rise to neoantigens, can be viewed as a surrogate marker for non-synonymous mutations elsewhere in the exome (55). Our own in-silico simulations indicate that inclusion of all mutations, even if they may not directly contribute to immunogenicity, increases precision of TMB estimation when using targeted gene panels (Buchhalter I, 2018, submitted).
Another aspect to consider is that sequencing of larger portions of the exome will result in many more detected genetic variants. How should this be dealt with in inherently resource-limited health care settings? Should all of these variants be clinically evaluated and reported? They probably will have to, which will bind additional manpower and strain IT resources. What about a potentially increased discovery rate of incidental genetic findings that might warrant genetic counselling? Refinancing of extensive manpower, reagent, and infrastructure associated costs will be challenging.
Composition and size of panel
Though one would wish to gather as much information from a patient’s tumor by conducting comprehensive WES, this is currently not feasible in clinical practice due to significant associated costs, long turn-around time, and suboptimal, limited tissue samples.
Targeted panel sequencing to identify therapeutically actionable oncogenic driver genes has emerged as a routine sequencing assay in many academic centers. Given its comparatively low cost, rather quick turn-around time, and more and more widespread availability, it would be desirable to leverage it to also assess TMB. After deciding on enrichment method (amplification-based or hybrid capture) and desired read depth, the most important question arising is one of sequencing panel size: How much of the exome needs to be sequenced to make an accurate prediction of a tumor’s total mutational burden (Figure 2)? The precision of TMB estimation is combinatorially limited by the number of bases that are sequenced. An upper limit of precision can be derived based on a model where each base in the genome is mutated at with the same probability. We calculated combinatorial confidence intervals of TMB for sequencing panels of sizes between 0.1 and 10 Mbp (Figure 3). Good separation of a tumor with 20 mut/Mbp from a tumor with 5 mut/Mbp was possible for panel sizes of 1 Mbp or larger.
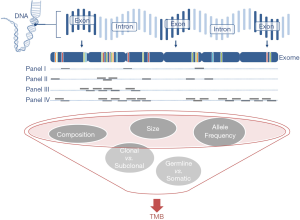
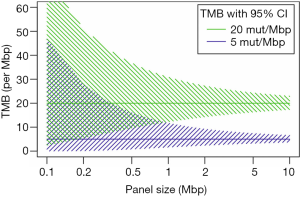
In NSCLC and melanoma, both tumors with high mutational load mainly caused by exogenous agents tobacco and UV irradiation, respectively (58,59), cancer-type specific algorithms were reported to allow for accurate prediction of TMB with a small panel of 170 genes (60). However, in clinical practice the challenge is posed by patients with intermediate TMB, accurate identification of which is difficult with small gene panels, because TMB values represent a continuum rather than distinct clusters. The 468-gene MSK-IMPACT panel (29,61) and 315-gene FoundationOne panel (62,63) have demonstrated diagnostic utility in predicting TMB comparably to WES (Table 3). In our experience with in-silico modeling (Buchhalter I, 2018, submitted) and resequencing of samples that previously underwent WES (Endris et al., in preparation), panels covering less than 1 Mbp are not suitable for a reliable prediction of TMB, but larger panels, ideally 1.5–3 Mbp can provide a good approximation of mutational load, as shown before (68). Sequencing of larger panels will also require capable sequencing machines, like the S5XL (Thermo Fisher) or NextSeq and beyond (Illumina).

Full table
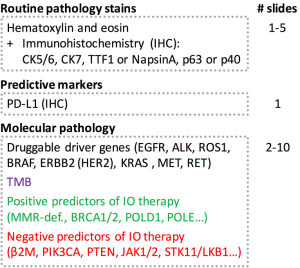
But of course, panel size is not the only factor, as composition is equally important. An ideal panel must ensure one-stop shop analysis with a combination of driver gene mutation analysis and TMB assessment to ensure maximum yield of clinically relevant information with limited DNA, technical resources, and economic constrains. In the case of lung cancer, this includes clinically actionable mutations in EGFR exons 19–21 including T790M alterations, as well as genetic aberrations including gene fusions that affect ALK, ROS1, RET, NTRK, BRAF, ERBB2 (HER2), KRAS, and MET (69). Furthermore, panels should ideally incorporate additional positive and negative predictors for checkpoint inhibitor therapy in order to provide a better grasp of the complex interaction between tumor and the immune system. It is of particular interest to detect tumors with defects in DNA repair genes since these tumors are prone to an increased number of somatic mutations, potentially leading to formation of tumor specific antigens, and recognition by tumor infiltrating lymphocytes. Mismatch repair-deficient tumors that are microsatellite instable (MSI-H) respond better to anti PD-1 therapy (70-72), and MSI-H status represents the first tissue-agnostic biomarker for checkpoint blockade with pembrolizumab (73,74). However, immune evasion mechanisms might still render an otherwise MSI-H tumor unresponsive to IO therapy (75,76). Defects in homologous recombination as caused by BRCA2 mutations confer a better response to checkpoint inhibitor therapy in malignant melanoma (77) and BRCA1/2-mutated high grade serous ovarian cancer exhibit a high mutational load (78). Mutations in POLD1 were noted in the lung cancer of a never-smoker with high TMB (6). POLE and POLD1 mutations occur in hypermutated colorectal and endometrial cancers (79,80). Loss-of-function mutations in chromatin remodeling gene PBRM1 sensitize tumors to IO therapy (22,81). On the flip side, loss of PTEN and subsequent activation of the PI3K-AKT pathway induced resistance to the antitumor T cell immune response in preclinical models of melanoma (82). In human lung adeno- and squamous cell carcinomas, expression of PD-L1 was found to be closely linked to mTOR activation (83). Amplification of MDM2 family proteins or aberrations in EGFR were found in patients experiencing hyperprogressive disease under treatment with checkpoint inhibitors (84) though this association was not seen in a subsequent study (29). Collectively, these data suggest that hyperprogression is still poorly understood and requires further investigation. JAK1/2 mutations as well as STAT family members were shown to confer primary resistance to PD-1 blockade (85-88). Defects in the IFN-γ pathway (89) and antigen-presenting protein beta-2-microglobulin (85,88,90,91) were shown to interfere with successful IO therapy. Patients with mutations in STK11 did not respond to treatment (28,29,92). While these data strongly suggest that the concept of negative IO-response predictors has great potential and will enter clinical stage soon, it should be noted that data of prospective clinical trials, which demonstrate the true clinical utility of these biomarkers have not been reported yet. Results from this comprehensive molecular tumor profiling should ideally be available within 10 working days from sample submission in order to ensure timely patient management.
Overall, in several retrospective and prospective clinical trials across multiple tumor entities, TMB was shown to identify patients that will or will not benefit from IO therapy more accurately than PD-L1 expression. It is therefore a promising marker that in conjunction with other parameters, like PD-(L)1 immunohistochemistry, positive and negative molecular predictors, immune cell infiltrates and inflammatory signatures can facilitate a more accurate guidance of IO interventions. However, for widespread clinical use, harmonization of assays and reporting is crucial in order to ensure validity and comparability of results with these of clinical trials. Two major efforts are currently underway to address the issue of standardized TMB estimation:
In the United States, Friends of Cancer Research (FoCR) gathered stakeholders primarily of industry and to a lesser degree academia to conduct a three-step harmonization project (93). First, in silico TMB analysis of TCGA datasets was performed to uncover factors of variability between different assays used. Next, a reference standard employing WES will be created to enable comparison of different gene panels. Lastly, clinically meaningful cutoff values will be determined in a retrospective analysis of samples with patient outcome data.
In Germany, an academia-driven round robin test carried out by Qualitätssicherungs-Initiative Pathologie QuIP GmbH (QuIP) (94), a joint venture of the two major German pathology societies Deutsche Gesellschaft für Pathologie (DGP) and Bundesverband Deutscher Pathologen e.V. (BDP), is in progress. In contrast to the FoCR initiative this will not only involve diagnostic and pharmaceutical companies but also eleven pathology institutes across Germany and Switzerland. Testing will be carried out on human tissue samples with validation against WES data.
Since these two initiatives approach the process of harmonization from different angles, a collaboration was initiated with the intention of joining forces and laying the ground for international and cross-sector standardization of TMB measurement and reporting. Considering the importance of TMB for the complex interaction between tumors and the immune system (95), these efforts are expected to also boost our understanding of microenvironment-related biomarkers, including abundance and clonal composition of immune cell infiltrates, thus paving the way for even more individualized and effective IO therapeutic strategies.
Acknowledgements
This work was supported by the German Cancer Consortium (DKTK, to P Schirmacher, J Budczies, A Stenzinger).
Footnote
Conflicts of Interest: V Endris: advisory board and lecture fees from AstraZeneca and ThermoFisher. J Leichsenring: consultancy contract with AstraZeneca. S Fröhling: speaker’s honoraria from Amgen, Lilly, PharmaMar and Roche; research funding from AstraZeneca, Pfizer and PharmaMar. M Thomas: advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, Boehringer, speaker’s honoraria from Lilly, MSD, Takeda, research funding from AstraZeneca, BMS, Celgene, Novartis, Roche and travel grants from BMS, MSD, Novartis, Boehringer. P Schirmacher: advisory board honoraria from Pfizer, Roche, Novartis, AstraZeneca as well as speaker’s honoraria and research funding from Roche, AstraZeneca and Novartis. A Stenzinger: advisory board honoraria from BMS, AstraZeneca, Novartis, ThermoFisher, speaker’s honoraria from BMS, Illumina, AstraZeneca, MSD, Novartis, Roche, ThermoFisher, and research funding from Chugai. The other authors have no conflicts of interest to declare.
References
- Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275-87. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Gubin MM, Artyomov MN, Mardis ER, et al. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 2015;125:3413-21. [Crossref] [PubMed]
- Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer Cell 2015;27:12-4. [Crossref] [PubMed]
- Grizzi G, Caccese M, Gkountakos A, et al. Putative predictors of efficacy for immune checkpoint inhibitors in non-small-cell lung cancer: facing the complexity of the immune system. Expert Rev Mol Diagn 2017;17:1055-69. [Crossref] [PubMed]
- Ebert PJR, Cheung J, Yang Y, et al. MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity 2016;44:609-21. [Crossref] [PubMed]
- Liu L, Mayes PA, Eastman S, et al. The BRAF and MEK Inhibitors Dabrafenib and Trametinib: Effects on Immune Function and in Combination with Immunomodulatory Antibodies Targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res 2015;21:1639-51. [Crossref] [PubMed]
- Kirchberger MC, Ugurel S, Mangana J, et al. MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: Results of a retrospective multicentre analysis of 364 patients. Eur J Cancer 2018;98:10-6. [Crossref] [PubMed]
- Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711-23. [Crossref] [PubMed]
- Wolchok JD, Kluger H, Callahan MK, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 2013;369:122-33. [Crossref] [PubMed]
- Robert C, Ribas A, Wolchok JD, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet 2014;384:1109-17. [Crossref] [PubMed]
- Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 2015;373:1803-13. [Crossref] [PubMed]
- Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 2014;515:558-62. [Crossref] [PubMed]
- Bauml J, Seiwert TY, Pfister DG, et al. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results From a Single-Arm, Phase II Study. J Clin Oncol 2017;35:1542-9. [Crossref] [PubMed]
- Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 2015;372:311-9. [Crossref] [PubMed]
- Kaufman HL, Russell J, Hamid O, et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol 2016;17:1374-85. [Crossref] [PubMed]
- Pan D, Kobayashi A, Jiang P, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770-5. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064-74. [Crossref] [PubMed]
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-852.e4. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Ramalingam S, Hellmann M, Awad M, et al. Tumor Mutational Burden (TMB) as a Biomarker for Clinical Benefit From Dual Immune Checkpoint Blockade With Nivolumab + Ipilimumab in First-line Non-Small Cell Lung Cancer: Identification of TMB Cutoff From CheckMate 568. American Association for Cancer Research (AACR) Annual Meeting; 2018 Apr 14–18; Chicago, USA.
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Kowanetz M, Zou W, Shames D, et al. OA20.01 Tumor Mutation Burden (TMB) is Associated with Improved Efficacy of Atezolizumab in 1L and 2L+ NSCLC Patients. J Thorac Oncol 2017;12:S321-2. [Crossref]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853-861.e4. [Crossref] [PubMed]
- Galsky MD, Saci A, Szabo PM, et al. 848PD Impact of tumor mutation burden on nivolumab efficacy in second-line urothelial carcinoma patients: Exploratory analysis of the phase ii checkmate 275 study. Ann Oncol 2017;28:mdx371.003.
- Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67-76. [Crossref] [PubMed]
- Powles T, Duran I, van der Heijden MS, et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2018;391:748-57. [Crossref] [PubMed]
- Haddad RI, Seiwert TY, Chow LQ, et al. Genomic determinants of response to pembrolizumab in head and neck squamous cell carcinoma (HNSCC). J Clin Oncol 2017;35:abstr 6009.
- Szustakowski J, Green G, Geese W, et al. #5528 Evaluation of tumor mutation burden as a biomarker for immune checkpoint inhibitor effıcacy: A calibration study of whole exome sequencing with FoundationOne®. AACR Annual Meeting 2018. Chicago 2018.
- Borghaei H, Hellmann M, Paz-Ares L, et al. Nivolumab (Nivo) + platinum-doublet chemotherapy (Chemo) vs chemo as first-line (1L) treatment (Tx) for advanced non-small cell lung cancer (NSCLC) with <1% tumor PD-L1 expression: Results from CheckMate 227. 2018 ASCO Annual Meeting.
- Kazmi SM. 35PA retrospective analysis to evaluate prevalence and correlation between PD-L1 score and tumor mutational burden (TMB) levels in patients with solid tumor malignancies. Ann Oncol 2017;28:mdx711.016.
- Rizvi H, Plodkowski AJ, Tenet M, et al. Clinical and molecular features predicting long-term response (LTR) to anti-PD-(L)1 based therapy in patients with NSCLC. J Clin Oncol 2018;36:abstr 9022.
- Kerr KM, Tsao MS, Nicholson AG, et al. Programmed Death-Ligand 1 Immunohistochemistry in Lung Cancer: In what state is this art? J Thorac Oncol 2015;10:985-9. [Crossref] [PubMed]
- Chilamakuri CS, Lorenz S, Madoui MA, et al. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics 2014;15:449. [Crossref] [PubMed]
- Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011;12:745-55. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Gandara DR, Kowanetz M, Mok TSK, et al. 1295OBlood-based biomarkers for cancer immunotherapy: Tumor mutational burden in blood (bTMB) is associated with improved atezolizumab (atezo) efficacy in 2L+ NSCLC (POPLAR and OAK). Ann Oncol 2017;28:mdx380. [Crossref]
- Fabrizio D, Malboeuf C, Lieber D, et al. 102PAnalytic validation of a next generation sequencing assay to identify tumor mutational burden from blood (bTMB) to support investigation of an anti-PD-L1 agent, atezolizumab, in a first line non-small cell lung cancer trial (BFAST). Ann Oncol 2017;28:mdx363.018.
- Koeppel F, Blanchard S, Jovelet C, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One 2017;12:e0188174. [Crossref] [PubMed]
- Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285-91. [Crossref] [PubMed]
- Sun JX, He Y, Sanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 2018;14:e1005965. [Crossref] [PubMed]
- Ptashkin RN, Mandelker DL, Coombs CC, et al. Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [Crossref] [PubMed]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res 2005;571:19-31. [Crossref] [PubMed]
- Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 2016;14:168. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Campesato LF, Barroso-Sousa R, Jimenez L, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015;6:34221-7. [Crossref] [PubMed]
- Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272-6. [Crossref] [PubMed]
- Memorial Sloan Kettering Cancer Center: MSK-IMPACT™ Is the First Tumor-Profiling Multiplex Panel Authorized by the FDA, Setting a New Pathway to Market for Future Oncopanels; New York; 2017 Nov 15 [accessed 2018 July 1]. Available online: https://www.mskcc.org/press-releases/msk-impact-first-tumor-profiling-multiplex-panel-authorized-fda-setting-new-pathway-market-future-oncopanels
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]
- Foundation Medicine, Inc.: FoundationOne CDx™ Technical Information; Cambridge, MA [accessed 2018 July 1]. Available online: https://assets.ctfassets.net/vhribv12lmne/6Rt6csmCPuaguuqmgi2iY8/e3a9b0456ed71a55d2e4480374695d95/FoundationOne_CDx.pdf
- Garofalo A, Sholl L, Reardon B, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med 2016;8:79. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. [Crossref] [PubMed]
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22:813-20. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Lemery S, Keegan P, Pazdur R. First FDA Approval Agnostic of Cancer Site - When a Biomarker Defines the Indication. N Engl J Med 2017;377:1409-12. [Crossref] [PubMed]
- Oncology Center of Excellence (OCE). FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. Silver Spring, MD: U.S. Food and Drug Administration 2017 May 23 [accessed 2018 Jun 19]. Available online: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm560040.htm
- Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer 2010;127:1001-10. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698-711. [Crossref] [PubMed]
- Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165:35-44. [Crossref] [PubMed]
- Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016;7:13587-98. [Crossref] [PubMed]
- Briggs S, Tomlinson I. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 2013;230:148-53. [Crossref] [PubMed]
- Howitt BE, Shukla SA, Sholl LM, et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol 2015;1:1319-23. [Crossref] [PubMed]
- Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801-6. [Crossref] [PubMed]
- Peng W, Chen JQ, Liu C, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016;6:202-16. [Crossref] [PubMed]
- Lastwika KJ, Wilson W 3rd, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res 2016;76:227-38. [Crossref] [PubMed]
- Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res 2017;23:4242-50. [Crossref] [PubMed]
- Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819-29. [Crossref] [PubMed]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov 2017;7:188-201. [Crossref] [PubMed]
- Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018;118:9-16. [Crossref] [PubMed]
- Budczies J, Bockmayr M, Klauschen F, et al. Mutation patterns in genes encoding interferon signaling and antigen presentation: A pan-cancer survey with implications for the use of immune checkpoint inhibitors. Genes Chromosomes Cancer 2017;56:651-9. [Crossref] [PubMed]
- Gao J, Shi LZ, Zhao H, et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016;167:397-404.e9. [Crossref] [PubMed]
- Grasso CS, Giannakis M, Wells DK, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 2018;8:730-49. [Crossref] [PubMed]
- Ozcan M, Janikovits J, von Knebel Doeberitz M, et al. Complex pattern of immune evasion in MSI colorectal cancer. Oncoimmunology 2018;7:e1445453. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Friends of Cancer Research (FoCR); Tumor Mutational Burden (TMB); Washington, DC; 2018 May 10 [accessed 2018 June 2]. Available online: https://www.focr.org/tmb
- Qualitätssicherungs-Initiative Pathologie QuIP GmbH; Tumor Mutational Burden (TMB): QuIP organises a study and liaises with FoCR; Berlin, Germany; 2018 May 14 [accessed 2018 June 9]. Available online: https://quip.eu/en_GB/2018/05/14/tumor-mutational-burden-tmb-quip-organisiert-studie-und-arbeitet-mit-focr-zusammen
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. [Crossref] [PubMed]


