The Role of ARF Family Proteins and Their Regulators and Effectors in Cancer Progression: A Therapeutic Perspective
- CEDOC, NOVA Medical School, Faculdade de Ciências Médicas, Universidade NOVA de Lisboa, Lisbon, Portugal
The Adenosine diphosphate-Ribosylation Factor (ARF) family belongs to the RAS superfamily of small GTPases and is involved in a wide variety of physiological processes, such as cell proliferation, motility and differentiation by regulating membrane traffic and associating with the cytoskeleton. Like other members of the RAS superfamily, ARF family proteins are activated by Guanine nucleotide Exchange Factors (GEFs) and inactivated by GTPase-Activating Proteins (GAPs). When active, they bind effectors, which mediate downstream functions. Several studies have reported that cancer cells are able to subvert membrane traffic regulators to enhance migration and invasion. Indeed, members of the ARF family, including ARF-Like (ARL) proteins have been implicated in tumorigenesis and progression of several types of cancer. Here, we review the role of ARF family members, their GEFs/GAPs and effectors in tumorigenesis and cancer progression, highlighting the ones that can have a pro-oncogenic behavior or function as tumor suppressors. Moreover, we propose possible mechanisms and approaches to target these proteins, toward the development of novel therapeutic strategies to impair tumor progression.
Introduction
The Adenosine diphosphate-Ribosylation Factor (ARF) family of proteins belongs to the RAS superfamily of small GTPases and comprises around 30 members in mammals (Sztul et al., 2019). This family includes 6 ARFs (5 in humans since ARF2 is absent), 21 ARLs, 2 Secretion-Associated RAS-related (SARs) and the TRIpartite Motif-containing protein 23 (TRIM23). ARF1-5 regulate vesicle budding at the Golgi apparatus by recruiting coat complexes (Li et al., 2004; Kahn et al., 2006). ARF6 localizes to the plasma membrane, as well as endosomes and is involved in actin cytoskeleton dynamics and endocytic recycling (Donaldson, 2003). The functions of ARL proteins are more heterogeneous and currently unknown for several of them. ARL2 and ARL3 interact with microtubules and function in tubulin assembly and cytokinesis, respectively, while ARL4C and ARL4D are involved in actin remodeling and regulate cell migration (Li C.-C. et al., 2007; Chiang et al., 2017). Our laboratory has shown that ARL13B binds actin and regulates cell migration (Barral et al., 2012; Casalou et al., 2014). Interestingly, several ARLs, namely ARL3, ARL6 and ARL13B are associated with the cilium and play different roles in ciliary biology and signaling pathways associated with this organelle (Marwaha et al., 2019). ARL8B is well characterized and has been shown to localize to lysosomes and regulate several aspects of lysosome biology, such as positioning and motility (Khatter et al., 2015). Finally, SARs play a well-described role in the budding of COPII-coated vesicles from the ER, while TRIM23 was implicated in antiviral defense and adipocyte differentiation (Arimoto et al., 2010; Watanabe et al., 2015; Saito et al., 2017).
Like other GTPases, ARF family proteins switch between an active state, in which proteins are GTP-bound and an inactive state, in which proteins are GDP-bound. For this reason, they are referred to as “molecular switches.” Nucleotide exchange is catalyzed by GEFs and GTP hydrolysis is promoted by GAPs. When they are active, ARF proteins associate with membranes via lipid modifications, namely myristoylation, palmitoylation or acetylation and bind effectors. These are responsible for the downstream functions of ARF family proteins and are highly diverse. Among the effectors identified are coat complexes and adaptors, cytoskeleton-binding proteins and tethering factors (Donaldson and Jackson, 2012). The functions of ARF and ARL proteins, as well as their GEFs and GAPs are thoroughly reviewed in two excellent recent reviews (Marwaha et al., 2019; Sztul et al., 2019).
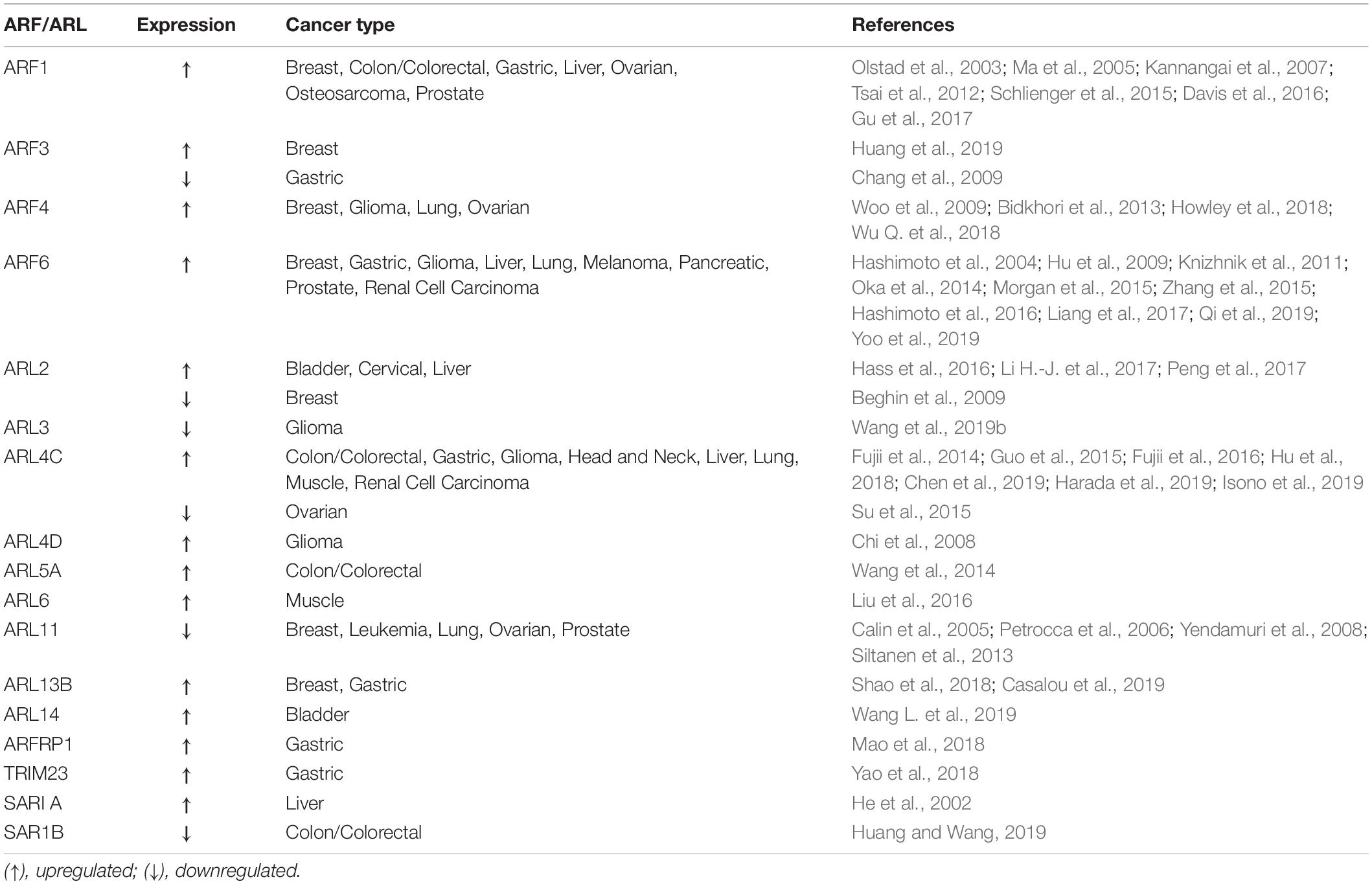
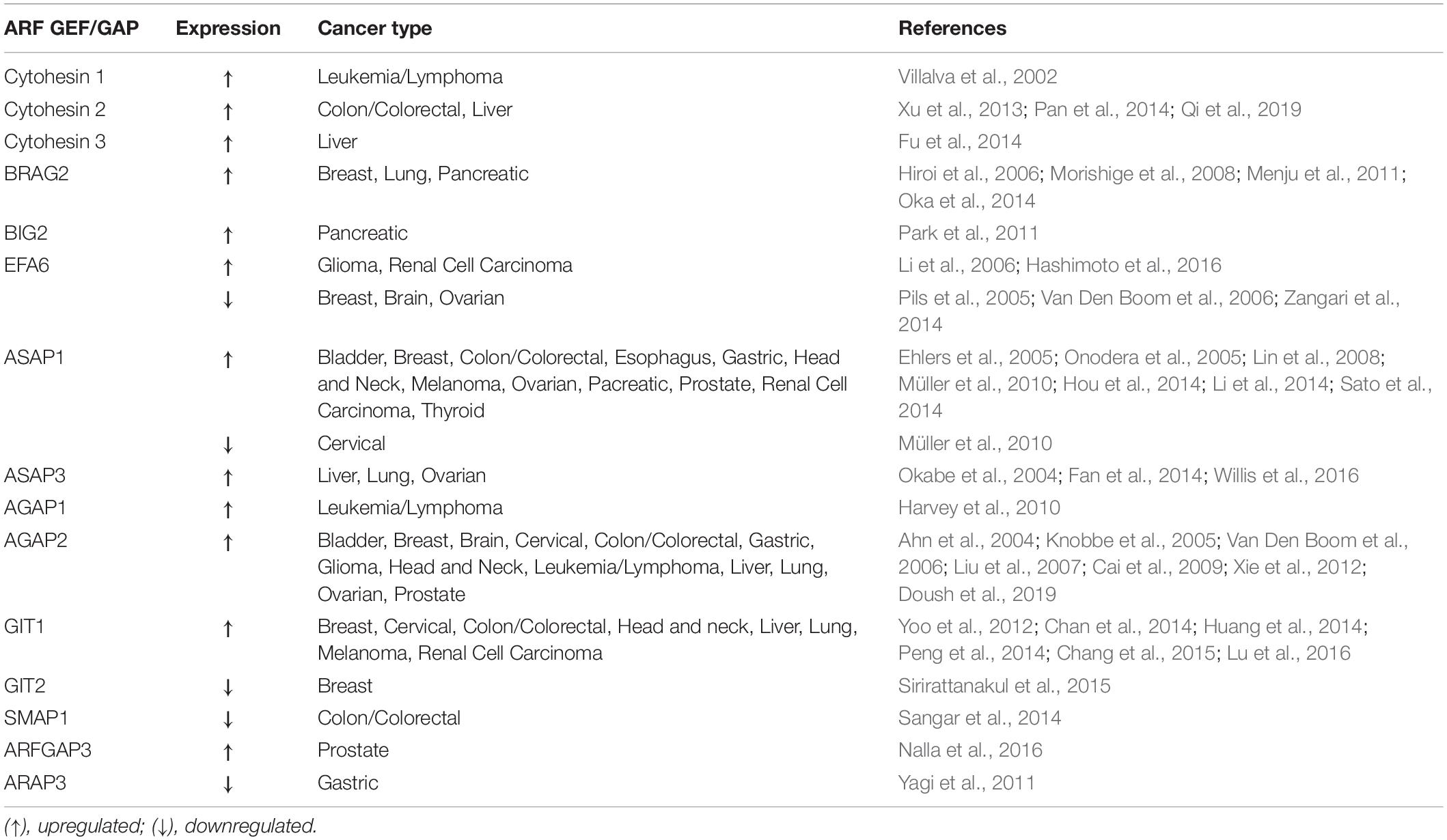
Since ARFs and their regulators play essential functions in cell cycle, cytoskeleton remodeling, cell migration and adhesion, it is not surprising that they can be subverted by cancer cells for proliferation, migration and invasion. Indeed, the expression and/or activity of several ARF family proteins and their GEFs and GAPs has been shown to be modulated in several types of cancer (Tables 1, 2). Moreover, the amplification and overexpression of ARF family genes, as well as the overexpression of their GEFs and GAPs, and variance in post-translational modifications are the most commonly detected alterations thought to be implicated in cancer. Here, we review the members of the ARF family and their activity regulators and effectors that have been implicated in cancer, and can either function as oncogenes or tumor suppressors and propose possible therapeutic approaches to target ARF family proteins or their effectors, GEFs and GAPs.
ARF Family Proteins and Their Activity Regulators and Effectors That Can Function as Oncogenes
Dysregulation of expression and/or activity of ARF family proteins and/or their effectors, GEFs and GAPs has been associated with enhanced cell migration, invasion and proliferation in several types of cancer. In this section, we review the ARF family members, as well as their activity regulators and effectors that have been found overexpressed in cancer and play essential roles in cancer progression (Tables 1, 2).
ARF1
ARF1 plays a central role in maintaining the structure and function of the Golgi apparatus and is highly expressed in breast, prostate and ovarian cancers (Schlienger et al., 2015; Davis et al., 2016; Gu et al., 2017). In the context of cancer, ARF1 has an important function in inter- and intracellular signaling, cell cycle regulation and DNA repair, as well as necrosis and apoptosis (D’Souza-Schorey and Chavrier, 2006; Gu et al., 2017). Moreover, ARF1 regulates breast cancer cell adhesion and proliferation, being essential for EGF-mediated phosphorylation of Focal Adhesion Kinase (FAK) and Src (Schlienger et al., 2015). Furthermore, ARF1 sensitizes MDA-MB-231 breast cancer cells to the anti-tumor drugs actinomycin D and vinblastine through ERK and Akt signaling (Luchsinger et al., 2018). In prostate cancer, ARF1 promotes tumorigenesis by controlling MAPK activation and cell growth (Davis et al., 2016). In myeloma cells, ARF1 expression promotes cell proliferation and inhibits cell adhesion, controlling proliferation- and cell adhesion-mediated drug resistance (Xu et al., 2017). Finally, ARF1 is upregulated in ovarian tumors, when compared with adjacent non-cancerous tissues and its overexpression is associated with ovarian cancer cell proliferation and migration through the PhosphoInositide 3-Kinase (PI3K) pathway (Gu et al., 2017).
ARF3
Like ARF1, ARF3 is involved in the recruitment of coat complexes to the Golgi apparatus, activation of PhosphoLipase D (PLD) and PI-kinases. Recently, ARF3 expression was positively correlated with breast cancer clinical stages, being upregulated in 92.8% of malignant cases, relative to benign ones (Huang et al., 2019). Indeed, ARF3 mRNA and protein expression levels are upregulated in human breast cancer cell lines and tissues (Huang et al., 2019). Moreover, ARF3 overexpression promotes breast cancer cell proliferation by regulating the cell-cycle G1/S transition, through inhibition of FOXO1 transcription factor activity (Huang et al., 2019). Additionally, ARF3 was found to be a candidate gene involved in the progression of pregnancy-associated breast cancer, based on integrated analysis of microarray profile datasets (Zhang et al., 2019).
ARF4
Together with the upregulation of COPB1 and USO1, which encode for the COPI subunit β1 and General vesicular transport factor p115, respectively and regulate ER-Golgi trafficking, ARF4 has been reported to be upregulated in breast cancer patient samples (Howley et al., 2018). This establishes a role for ARF4, COPB1, and USO1 in the regulation of breast cancer cell growth and invasion through the retrograde transport of proteins from the Golgi to ER via COPI-coated vesicles. ARF4 has also been associated with the regulation of breast cancer cell migration in response to Phorbol-12-Myristate 13-Acetate (PMA) (Jang et al., 2012). Finally, ARF4 has been found upregulated in other types of epithelial cancers, such as ovarian cancer (Wu Q. et al., 2018) and lung adenocarcinomas (Bidkhori et al., 2013). In U373MG human glioblastoma-derived cells, ARF4 has an anti-apoptotic function by reducing the generation of ROS in response to the expression of B-cell lymphoma 2 (Bcl-2)-Associated X protein (Bax) or the synthetic retinoid derivative N-(4-hydroxyphenyl) retinamide (Woo et al., 2009).
ARF6
ARF6 is well characterized in the context of cancer and known to regulate cancer cell invasion and metastasis, as well as tumor angiogenesis and growth (reviewed in Hongu et al., 2016; Li R. et al., 2017). Clinically, ARF6 expression and activation of its downstream signaling pathways was determined and associated with poor overall survival of breast, lung adenocarcinoma, pancreatic ductal adenocarcinoma and head and neck cancer patients (Li R. et al., 2017). Also, elevated ARF6 expression has been reported in prostate and non-small cell lung and squamous cell lung cancers (Knizhnik et al., 2011; Morgan et al., 2015). Moreover, a direct correlation between ARF6 protein expression levels and breast cancer cell invasiveness was shown in breast cancer cell lines with different invasive abilities (Hashimoto et al., 2004). Furthermore, ARF6 silencing impairs invasion of breast cancer, melanoma and glioma (Hashimoto et al., 2004; Hu et al., 2009; Grossmann et al., 2014), providing evidence that ARF6 is an important driver of cancer cell invasion and metastasis. In lung adenocarcinoma, the combined expression of ARF6, its GEF BRAG2/GEP100 and EGFR is associated with decreased patient survival (Oka et al., 2014). ARF6 is known to recruit actin binding proteins, adhesion molecules and proteases, which are essential for invadopodia formation and ExtraCellular Matrix (ECM) degradation (Schweitzer et al., 2011). Indeed, ARF6 activation was shown to promote invadopodia formation through activation of Rho- and Rac1-dependent pathways (Muralidharan-chari et al., 2009). ARF6 is also necessary for Human Growth Factor (HGF)-induced tumor angiogenesis and growth (Hongu et al., 2015). It has also been shown that ARF6 coordinates signaling and function of several oncogenes, like EGFR, ERBB2, and CTNNB1, which encode for EGFR, HER2, and β-catenin, respectively (Morishige et al., 2008; Menju et al., 2011; Pellon-Cardenas et al., 2013; Yoo et al., 2016). In agreement, it was recently observed that ARF6 is a downstream target of mutant KRAS and maintains KRAS-induced ERK activation, promoting pancreatic tumorigenesis (Liang et al., 2017). Also, ARF6 was linked to liver cancer through the regulation of the endocytic recycling of CD147, a tumor-related adhesive protein that promotes invasion of liver cancer cells (Qi et al., 2019). Moreover, the increased expression of ARF6-CD147 signaling components, like Cytohesin 2/ARNO, an ARF6 GEF and Rac1 were associated with poor overall survival of hepatocellular carcinoma patients (Qi et al., 2019).
ARF GEFs and GAPs
Amplification of ARF GAPs has been associated with several types of cancer. Indeed, AGAP2, which acts on ARF1 and ARF5, promotes cancer cell survival, migration and invasion in gliobastomas (Qi et al., 2017). Moreover, ASAP1 expression is correlated with the metastatic potential of melanoma, prostate cancer and colorectal cancer and increased invasiveness of breast cancer and melanoma cells (Ehlers et al., 2005; Onodera et al., 2005; Lin et al., 2008; Müller et al., 2010). Cancer cell migration requires coordinated assembly and disassembly of cell-ECM contacts, mediated by FAs. Indeed, several ARF GAPs, namely ASAP1, ASAP2, GIT1 and GIT2 have been found to be localized at FAs (Casalou et al., 2016). GIT1, which inactivates ARF6 specifically, is highly expressed in several types of cancers, including breast, cervical, colon and liver (Yoo et al., 2012; Chan et al., 2014; Huang et al., 2014; Peng et al., 2014). Moreover, GIT1 interacts with Paxillin and p21-activated kinase Interacting eXchange factor (PIX) at FAs, regulating cancer cell migration (Nayal et al., 2006). Furthermore, GIT1 silencing has been shown to inhibit cell migration and invasion in oral squamous cell carcinoma and breast cancer (Chan et al., 2014; Huang et al., 2014). Although GIT1 is associated with several types of cancer, it is not clear whether ARF6 inactivation by GIT1 is a requirement for cancer progression.
Regarding the ARF GEFs, BRAG2/GEP100 and EFA6, which activate ARF6 specifically, are known to be involved in cancer progression. BRAG2 induces breast cancer cell invasion and metastasis (Morishige et al., 2008). After BRAG2 binding to phosphorylated EGFR, ARF6 is activated in breast cancer cells, leading to the formation of invadopodia with recruitment of Cortactin, Paxillin and the ARF GAP ASAP1 (Onodera et al., 2005; Morishige et al., 2008). In lung adenocarcinoma, the pathway involving EGFR, ARF6 and ASAP1 was reported to be associated with reduced patient survival (Oka et al., 2014). In melanoma cells, the stimulation of WNT5A, a member of the Wnt signaling pathway, induces ARF6 activation mediated by BRAG2, which facilitates the release of β-catenin from cadherin and stimulates tumor cell invasion (Grossmann et al., 2014).
Concerning the EFA6 GEFs, they regulate tumor progression either positively or negatively, depending on the cancer types. In glioma and renal carcinoma, EFA6 GEFs are upregulated, controlling cancer cell invasion (Li et al., 2006; Hashimoto et al., 2016). The ARF GEFs Cytohesin 1-3 function as regulators of cytoskeleton reorganization and integrin signaling (Kolanus, 2007) and target ARF6, among other ARFs. In prostate cancer, inhibiton of Cytohesin 1 by siRNA, reduces the pro-tumorigenic role of Insulin Growth Factor Receptor (IGFR) signaling (Weizhong et al., 2011), suggesting that this ARF GEF could be targeted to impair prostate cancer progression. Additionally, the ectopic expression of the constitutively active form of ARF6 (ARF Q67L) enhances melanoma progression and metastasis in vivo (Muralidharan-chari et al., 2009).
ARL2
ARL2 was first reported to behave as a tumor suppressor in breast cancer. However, several publications thereafter suggest that this might not be the case for other types of cancers. Indeed, it was shown that BART binds to active ARL2, inhibiting the inactivation of RhoA and thus impairing the invasive potential of pancreatic cancer cells (Taniuchi et al., 2011). Other studies evaluated the effect of ARL2-targeting microRNAs (miRs). In particular, miR-214 was found to suppress growth and increased apoptosis in colon cancer (Long et al., 2015). Moreover, miR-214 was studied in the context of cervical cancer, in which its expression is able to suppress proliferation, migration and invasion of cancer cells (Peng et al., 2017). Two other miRs were found to be involved in cancer progression. miR-497-5p overexpression leads to a decrease in osteosarcoma cell proliferation and an increase in apoptosis (Sun et al., 2017). On the other hand, miR-195, which is regulated by Urothelial Cancer Associated 1 (UCA1) targets ARL2 in bladder cancer (Li H.-J. et al., 2017). Studies performed in mice showed that bladder tumor size is reduced upon UCA1 downregulation and the expression of miR-195 is increased, resulting in ARL2 downregulation. The authors concluded that the effects in bladder cancer cells mediated by UCA1/miR-195/ARL2 are a consequence of mitochondrial metabolism modulation, which regulates cancer cell survival (Li H.-J. et al., 2017). Finally, ARL2 was found to be overexpressed in human hepatocellular carcinoma samples by gene expression analysis (Hass et al., 2016).
ARL4
ARL4C was initially found to be upregulated at the mRNA level in both colorectal and lung cancers (Fujii et al., 2014). Moreover, the same authors found that ARL4C silencing leads to a decrease in cell migration and invasion in vitro, and proliferation both in vitro and in vivo, dependently on aberrant Wnt/β-catenin and EGF/RAS signaling. ARL4C was also found to be overexpressed in leiomyosarcoma type II (Guo et al., 2015). Furthermore, analysis of ARL4C expression in colorectal cancer samples revealed that this ARL is more expressed in tumor samples, comparing with adjacent normal tissue (Chen et al., 2016). The prognostic value of ARL4C in colorectal cancer was also evaluated and the same authors concluded that patients with higher expression of ARL4C have lower survival on average (Chen et al., 2016). In the case of lung and tongue squamous cell carcinoma, it was found that ARL4C promotes proliferation and migration of cells from these types of cancers (Fujii et al., 2016). Interestingly, ARL4C overexpression in lung tumors was shown to be due to hypomethylation of ARL4C in the 3′-UTR through Ten-Eleven Translocation methylcytosine dioxygenases (TETs) (Fujii et al., 2016). Recently, several groups investigated the role of ARL4C in different types of cancer. ARL4C was identified as a peritoneal dissemination-associated gene and found to be highly expressed in gastric cancer cells (Hu et al., 2018). Indeed, ARL4C silencing impairs migration and invasion of gastric cancer cells in vitro (Hu et al., 2018). Moreover, the reduced expression of ARL4C leads to a decrease of the epithelial-mesenchymal transition (EMT) marker SLUG, as well as a reduction in lamellipodia and filopodia formation in gastric cancer cells (Hu et al., 2018). ARL4C expression was also found to be increased in primary and metastatic hepatocellular carcinoma. Additionally, the decrease in ARL4C expression leads to the impairment of cancer cell proliferation and migration in vitro and in vivo, as well as a reduction in expression of PI3K Catalytic subunit Delta (PI3KCD) mRNA and activity of Akt (Harada et al., 2019). This suggests that the molecular mechanisms involved in the role of ARL4C in hepatocellular carcinoma are different from those in lung and colorectal cancers. Furthermore, upregulation of ARL4C was associated with a poor prognosis in endometriosis-associated ovarian cancer, Phosphatase and TENsin homolog deleted on chromosome 10 (PTEN)-deficient glioblastomas and renal cell carcinomas (Wakinoue et al., 2018; Chen et al., 2019; Isono et al., 2019). Another ARL4 isoform, ARL4D was first identified as a glioma-associated antigen (Nonaka et al., 2002). Later, a study revealed that ARL4D expression in gliomas is dependent on the loss of PTEN tumor suppressor and consequent activation of the Akt/mammalian Target of Rapamycin (mTOR) pathway (Chi et al., 2008).
ARL5, ARL6, ARL8, ARL14, and ARFRP1
ARL5A was found to be highly expressed in colorectal cancer and a target of miR-202-3P (Wang et al., 2014). Furthermore, the same study demonstrated that the downregulation of ARL5A and miR-202-3P expression leads to a similar reduction in colorectal cancer cell proliferation (Wang et al., 2014).
In rhabdomyosarcoma, ARL6 was demonstrated to be upregulated in cilia-dependent cancer cells and its silencing decreases cell proliferation (Liu et al., 2016). Additionally, ARL6 downregulation leads to an increase in apoptosis of rhabdomyosarcoma cancer cells due to defects in ciliogenesis and a reduction of Hedgehog (Hh) signaling (Liu et al., 2016).
In the case of ARL8, depletion of ARL8B leads to a reduction in invasion and protease secretion by prostate cancer cells (Dykes et al., 2016). Moreover, ARL8B silencing prevents the growth of prostate tumors in mice (Dykes et al., 2016). Furthermore, the same study revealed that the role of ARL8B in cancer progression is dependent on its function in regulation of lysosomal motility and fusion.
A recent study reported that ARL14 silencing in lung cancer cells blocks ERK1/2 and p28 signaling and upregulates the cell death inducing DFFA-like effector C (CIDEC), leading to cell cycle arrest (Guo et al., 2019). Also, hypermethylation of ARL14 was found to be correlated with a poor prognosis of bladder cancer patients (Wang L. et al., 2019).
Finally, in the case of ARFRP1, it was found upregulated in gastric cancer (Mao et al., 2018).
ARL13B
The role of ARL13B in medulloblastoma and gastric cancer progression, dependent on cilia and Hh signaling was described recently (Bay et al., 2018; Shao et al., 2018). Shao and co-authors showed that ARL13B promotes proliferation, migration and invasion of gastric cancer cells both in vitro and in vivo, through activation of Smoothened (Smo) and consequent activation of Hh signaling (Shao et al., 2018). In medulloblastoma, ARL13B depletion was reported to lead to a decrease in cilia-dependent oncogenic Hh signaling (Bay et al., 2018). Recently, our group found evidence that ARL13B plays a role in breast tumorigenesis and cancer progression, likely independently of cilia. Indeed, depletion of ARL13B in breast cancer cells leads to a reduction in cell migration and invasion in vitro and impaired tumor progression in vivo (Casalou et al., 2019). Moreover, our results revealed a new mechanism to explain the role of ARL13B in tumor progression, through the modulation of cell-ECM adhesion and integrin-mediated signaling.
Non-Muscle myosin heavy chain II A (NMIIA) was identified by us as an effector of ARL13B, since it binds to the active form of this protein (Casalou et al., 2014). In the same study, we found that NMIIA mediates ARL13B binding to actin and that both proteins are required for the formation of circular dorsal ruffles (CDRs), which are actin-rich structures required for cell migration (Casalou et al., 2014). Our group also found that GTP-bound ARL13B interacts with NMIIA in breast cancer cells (Casalou et al., 2019). Other studies reported the role of NMIIA in different types of cancers and indicate that NMIIA can function as a tumor suppressor or oncogene. For example, NMIIA was found to be overexpressed in gastric cancer (Liu et al., 2012) and promote tumor progression in different types of cancers (Derycke et al., 2011; Katono et al., 2015; Liao et al., 2017; Ye et al., 2017). On the other hand, NMIIA was described as a potential tumor suppressor gene in squamous cell carcinomas, since the downregulation of NMIIA associates with poor survival, increased cancer cell invasion and decreased p53 stabilization, in vitro and in vivo (Schramek et al., 2014). These studies are described in greater detail in two recent reviews (Pecci et al., 2019; Wang et al., 2019a). Other evidence suggests that NMIIA expression is increased in colorectal cancer and that NMIIA enhances tumor aggressiveness through activation of mitogen-activated protein kinase (MAPK) Akt signaling, which promotes EMT (Wang B. et al., 2019). NMIIA was also found to be a promoter of EMT in pancreatic cancer (Zhou et al., 2019). Moreover, in the same study it was observed that NMIIA downregulation results in decreased invasion and metastasis formation through the suppression of canonical Wnt/β-catenin signaling (Zhou et al., 2019).
TRIM23 and SAR1
In hepatocellular carcinomas, miR-194, which targets TRIM23 was found to be downregulated. Moreover, overexpression of miR-194 decreases cell migration, invasion and metastasis of hepatocellular carcinoma cells, through inhibition of Nuclear Factor (NF)-κb activity (Bao et al., 2015). Furthermore, TRIM23 was found to be overexpressed in gastric cancer, both in cell lines and tissues (Yao et al., 2018).
In the case of SAR1, it was found to be overexpressed in liver cancer (He et al., 2002). More recently, SAR1b was identified as a promoter of drug resistance, namely mTOR Complex (mTORC) inhibitors, in liver tumor initiating stem cells and hepatocellular carcinoma cells (Wu R. et al., 2018).
Arf Family Proteins and Their Activity Regulators and Effectors That Can Function as Tumor Suppressor Genes
In some cases, the expression of membrane traffic regulators, namely ARF family proteins or their effectors, GEFs or GAPs is found downregulated in tumor cells (Tables 1, 2).
ARF3
Besides its behavior as an oncogene in breast cancer, ARF3 has been found downregulated in gastric cancer (Chang et al., 2009). In fact, ARF3 expression is significantly decreased in gastric cancer stages I-III, when compared with paired normal gastric mucosa tissues, indicating that this protein could be a marker for gastric cancers without metastasis. The clinical significance of these results remains to be elucidated.
ARF GEFs and GAPs
SMAP1 or ARFGAP1 is a member of the ARF GAP family that is involved in clathrin-dependent endocytosis of the Transferrin receptor and E-cadherin (Kon et al., 2008; Kobayashi et al., 2014). In colorectal cancers with microsatellite instability, short deletions or insertions frequently occur in SMAP1, generating a premature termination codon. This results in reduced or abolished SMAP1 protein levels in colorectal tumors (Kon et al., 2014).
Unlike other phosphotyrosine proteins that are usually overexpressed or hyperphosphorylated in gastric tumor cells, the ARF GAP ARAP3 is downregulated in gastric cancer tissues (Yagi et al., 2011). Furthermore, GIT2 stabilizes FAs by reducing Rac1 activity in the breast cancer cell line MDA-MB-231 (Frank et al., 2017). Also, in a gene expression profile analysis of breast cancer patient samples, GIT2 was found downregulated in a group of lymph node-positive breast cancer patients (Sirirattanakul et al., 2015).
Finally, EFA6 GEFs are downregulated in breast, brain and ovarian cancers (Pils et al., 2005; Van Den Boom et al., 2006; Zangari et al., 2014).
ARL2
ARL2 has been shown to directly influence α/β-tubulin polymerization in the breast cancer cell line MCF-7 (Beghin et al., 2007). Moreover, MCF-7 cells expressing higher levels of ARL2 are more sensitive to cytotoxic agents, while cells with reduced expression of ARL2 show enhanced resistance to the same agents (Beghin et al., 2008). This resistance is mediated by Protein Phosphatase 2A (PP2A), whose activity is regulated by ARL2. When ARL2 is decreased, impaired dephosphorylation of p53 by PP2A occurs, leading to an increase of phosphorylated p53, which alters PP2A localization and causes a chemo-resistant phenotype (Beghin et al., 2008). Moreover, in vitro assays using breast cancer cells depleted for ARL2 show less contact inhibition, an enhanced clonogenic potential and increased proliferation than control cells (Beghin et al., 2009). Furthermore, using orthotopic mouse models, depletion of ARL2 was shown to impair cancer progression (Beghin et al., 2009). Additionally, ARL2 downregulation was recently correlated with more aggressive cases of glioma and a lower survival of the patients (Wang et al., 2018). Finally, ARL2 overexpression inhibits proliferation, as well as migration and tumorigenicity of glioma cells, through regulation of the receptor tyrosine kinase AXL, a known regulator of glioma tumorigenesis (Wang et al., 2018).
ARL3, ARL4, and SAR1
ARL3 mRNA and protein expression were shown to be downregulated in gliomas (Wang et al., 2019b). Furthermore, an extensive bioinformatics analysis suggested that ARL3 plays a role in angiogenesis and immune cell infiltration in the tumor microenvironment (Wang et al., 2019b).
ARL4C was associated with reduced metastatic potential of ovarian cancer cells, in which it inhibits cell motility but not cell proliferation (Su et al., 2015). Furthermore, ARL4C mRNA expression is lower in ovarian cancer samples of patients with a poor treatment response, while patients with higher ARL4C expression show increased overall survival (Su et al., 2015).
Finally, SAR1B was identified as a potential metastatic suppressor in colorectal cancer, through a targeted proteomic approach (Huang and Wang, 2019). Also, migration and invasion assays showed that SAR1B silencing leads to an increase in colorectal cancer cell motility and invasive capacity (Huang and Wang, 2019).
ARL11
ARL11, also known as ADP Ribosylation factor-Like Tumor Suppressor gene 1 (ARLTS1) was described as a potential low-penetrance tumor suppressor gene in different types of cancers, such as breast cancer, melanoma and chronic lymphocytic leukemia (Calin et al., 2005). Different variants of ARLTS1 have been associated with familial and sporadic cancers, where the mutations Trp149Stop and Cys148Arg are the most studied (Yendamuri et al., 2008). The nonsense mutation Trp149Stop leads to the production of a truncated protein unable to bind GTP, which results in decreased apoptotic potential of the cell (Petrocca et al., 2006). Both variants were found to be associated with predisposition to familial breast cancer and, more recently to familial hematological malignancies (Calin et al., 2005; Frank et al., 2006b; Hamadou et al., 2017). Additionally, the Cys148Arg variant was associated with melanoma and both familial and sporadic colorectal cancers (Frank et al., 2006a, c; Castellví-Bel et al., 2007). Furthermore, ARLTS1 expression was found to be decreased in different types of tumors, including ovarian, lung and prostate cancer, as well as chronic lymphocytic leukemia (Yendamuri et al., 2008; Siltanen et al., 2013). More recently, a study in ovarian cancer suggested that ARLST1 increases tumor cell sensitivity to chemotherapeutic agents by regulating apoptosis (Yang et al., 2011).
Therapeutic Strategies
As can be concluded from Tables 1, 2, several ARFs and ARF GEFs and GAPs are overexpressed in different types of cancers. Therefore, therapeutic strategies aiming to inhibit the expression of these proteins can be proposed. Other approaches like the use of small GTPase inhibitors that impair GTP binding or the binding to membranes, the blockade of GEF activity or ARF-GEF interaction, should also be considered. Furthermore, the stimulation of GAP activity/expression and the inhibition of the interaction with downstream effectors or the function/expression of these effectors can also be envisaged (Figure 1). For instance, the inhibitor LM11 can abolish specifically ARF1 activation through the blockade of the binding of the ARF GEF Cytohesin 2/ARNO (Flisiak et al., 2008; Xie et al., 2016). Indeed, it has been shown that the aggressiveness of breast tumors that overexpress ARF1 is reduced after treatment with this inhibitor through the decrease in cell invasion and proliferation and increased apoptosis (Schlienger et al., 2015; Xie et al., 2016). Also, the small inhibitor EXO2 reduces ARF1 activation and effectively impairs the proliferation of prostate cancer cells by blocking ERK1/2 activation (Lang et al., 2017). Moreover, EXO2 inhibits invasion of prostate cancer cells and induces their apoptosis. Furthermore, the same study shows that the simultaneous blockade of ARF1 and RAS activation in prostate cancer is a potential targeted strategy to prevent the development of this type of tumor (Lang et al., 2017).
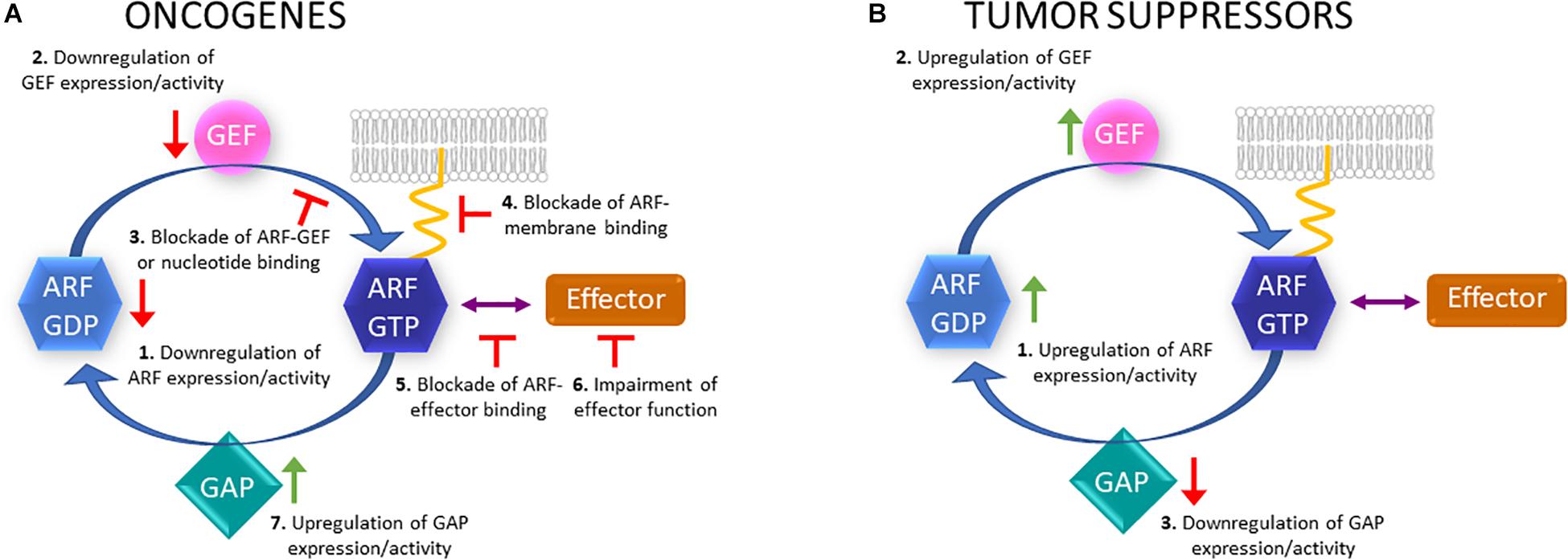
Figure 1. Putative therapeutic strategies to target ARF proteins, GEFs, GAPs and effectors. (A) In the case of ARF family members that can act as oncogenes, their expression or activity could be downregulated (1); GEF activity or expression downregulated (2); ARF-GEF binding blocked or nucleotide binding blocked (3); active ARF binding to membranes (4) or effectors blocked (5); effector function impaired (6); GAP expression or activity upregulated (7). (B) Regarding ARF proteins that can act as tumor suppressors, their expression or activity could be upregulated (1); GEF activity or expression upregulated (2); GAP activity or expression downregulated (3).
Since some ARF proteins like ARF6 and ARF1, are ubiquitously expressed and perform essential functions in all cell types (D’Souza-Schorey and Chavrier, 2006), targeting the proteins themselves could have dramatic and unwanted consequences. In alternative, targeting their regulators, such as ARF GEFs or GAPs, might represent a viable strategy for the development of specific anti-cancer therapies. Regarding the targeting of ARF GEFs, it has been shown that SecinH3, an ARF GEF inhibitor that impairs both ARF1 and ARF6-dependent signaling, is effective in decreasing the growth of breast cancer xenografts and reducing lung metastasis (Zhao et al., 2016), while suppressing angiogenesis of melanoma and lung carcinoma tumors (Grossmann et al., 2014; Hongu et al., 2015). Thus, inhibitors of the ARF6-dependent signaling pathway could be useful to control specifically tumor invasion and angiogenesis.
It has been observed that several ARF GAPs are overexpressed in cancer (Table 2), even though overexpression of ARF GAPs does not imply increased GAP activity. For instance, AGAP2 expression in chronic myeloid leukemia cells and prostate cancer is regulated by Specific Protein 1 (SP1) and ATRA (Doush et al., 2019). Additionally, the authors observed that the treatment of cells of these types of cancer with the polyphenol curcumin, leads to a decrease in ATRA-mediated AGAP2 expression (Doush et al., 2019; Giordano and Tommonaro, 2019). This data illustrates the relevance of regulating ARF GAP expression levels in cancer. On the other hand, it was observed that QS11, the only inhibitor of ARF GAPs known, binds to ARF GAP1 and inhibits the activity of this GAP on ARF1 and ARF6 (Zhang et al., 2007; Zhu et al., 2012). Interestingly, it was observed that QS11 blocks migration of metastatic breast cancer cells, in vitro (Zhang et al., 2007). Thus, inhibitors of ARF GAP activity could also be effective in controlling cancer cell migration and invasion.
Inhibition of the expression or function of downstream effectors of ARF family proteins is also a plausible strategy to impair the oncogenic potential of ARFs and ARLs. An interesting candidate is NMII. Indeed, several types of cancer exhibit differential expression and/or activation of NMII isoforms, leading to alterations in cell migration and invasion that are involved in tumorigenesis (Newell-Litwa et al., 2015). Moreover, we found that NMIIA is an effector of ARL13B (Casalou et al., 2014, 2019). Furthermore, blebbistatin inhibits the ATPase activity of NMIIA and has been found to block invasiveness of both breast cancer cells (Derycke et al., 2011) and pancreatic adenocarcinoma cells (Duxbury et al., 2004), and is phototoxic in human cancer cells under exposure to blue light (Mikulich et al., 2012).
Conclusion and Perspectives
Members of the RAS superfamily of small GTPases are master regulators of all the steps involved in membrane traffic. Thus, it is not surprising that many of them are hijacked by cancer cells to enhance their capacity to form a tumor and spread to other organs. In particular, ARF family proteins, their GEFs, GAPs and effectors are often upregulated in expression and/or activity in several types of cancer. Moreover, upregulated expression/activity can be linked to enhanced cancer progression and aggressiveness. Therefore, these proteins are good candidates to serve as therapeutic targets and, indeed several strategies have already been proposed and tested. These include the targeting of ARF proteins themselves or their GEFs, GAPs or effectors. While our knowledge of the GEFs, GAPs and effectors of ARFs is fairly complete, much less is known about the functions and identity of GEFs, GAPs and effectors of ARL subfamily members. Hence, the knowledge about these molecular players should be developed in order to find new therapeutic strategies for cancer types where ARLs or their regulators/effectors are subverted. Since most ARF family proteins are ubiquitous and required for essential cellular functions, the targeting of specific effectors and GEFs/GAPs could ensure tissue/function specificity. Nevertheless, specificity could also be achieved through targeted delivery of vectors/drugs.
In conclusion, the study of the mechanisms subverted by cancer cells involving ARF family proteins and their regulators of activity and effectors can shed light on the functions of these proteins and simultaneously provide clues about new therapeutic targets and strategies, which continue to be a pressing need in the cancer field.
Author Contributions
All authors conceived, wrote, reviewed, and edited the manuscript.
Funding
AF was funded by a Ph.D. fellowship (PD/BD/135506/2018) from the Fundação para a Ciência e a Tecnologia (FCT) and DB by the FCT Investigator Program (IF/00501/2014/CP1252/CT0001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those whose work we could not cite due to space limitations.
Abbreviations
4-HPR, N-(4-hydroxyphenil retinamide); Akt, Protein kinase B; ARF, ADP-Ribosylation Factor; ARL, ARF-Like; ARLTS1, ADP-Ribosylation factor-Like Tumor Suppressor gene 1; ATRA, All-Trans Retinoic Acid; BART, Binder of ARL Two; Bax, B-cell lymphoma 2-Associated X protein; CDR, Circular Dorsal Ruffle; CIDEC, Cell Death Inducing DFFA-like Effector C; ECM, ExtraCellular Matrix; EGF, Epidermal Growth Factor; EGFR, Epidermal Growth Factor Receptor; EMT, Epithelial-Mesenchymal Transition; ER, Endoplasmic Reticulum; ERK, Extracellular signal-Regulated Kinase; FA, Focal Adhesion; FAK, Focal Adhesion Kinase; FOXO1, Forkhead bOX O1; GAP, GTPase-Activating Protein; GDP, Guanosine DiPhosphate; GEF, Guanine nucleotide Exchange Factor; GTP, Guanosine TriPhosphate; HER2, Human Epidermal growth factor Receptor 2; HGF, Human Growth Factor; Hh, Hedgehog; IGFR, Insulin Growth Factor Receptor; MAPK, Mitogen-Activated Protein Kinase; miRs, microRNAs; mTOR, mammalian Target of Rapamycin; mTORC, mammalian Target of Rapamycin Complex; NF, Nuclear Factor; NMIIA, Non-Muscle Myosin heavy chain II A; PAK, p21-Activated Kinase; PI, PhosphoInositide; PI3K, PhosphoInositide 3-Kinase; PI3KCD, PhosphoInositide 3-Kinase Catalytic subunit Delta; PIX, PAK- Interacting eXchange factor; PLD, PhosphoLipase D; PMA, Phorbol-12-Myristate 13-Acetate; PP2A, Protein Phosphatase 2A; PTEN, Phosphatase and TENsin homolog deleted on chromosome 10; Rho, RAS homolog gene family member; Rac, RAS-related C3 botulinum toxin substrate; ROS, Reactive Oxygen Species; SAR, Secretion-Associated RAS-related; Smo, Smoothened; SP1, Specificity Protein 1; Src, Rous Sarcoma oncogene cellular homolog; TET, Ten-Eleven Translocation methylcytosine dioxygenase; TRIM23, TRIpartite Motif-containing protein 23; UCA1, Urothelial Cancer Associated 1.
References
Ahn, J. Y., Rong, R., Kroll, T. G., Van Meir, E. G., Snyder, S. H., and Ye, K. (2004). PIKE (Phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates akt activity and mediates cellular invasion. J. Biol. Chem. 279, 16441–16451. doi: 10.1074/jbc.M312175200
Arimoto, K. I., Funami, K., Saeki, Y., Tanaka, K., Okawa, K., Takeuchi, O., et al. (2010). Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 107, 15856–15861. doi: 10.1073/pnas.1004621107
Bao, C., Li, Y., Huan, L., Zhang, Y., Zhao, F., Wang, Q., et al. (2015). NF-κB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1α. Sci. Signal. 8, 1–9.
Barral, D. C., Garg, S., Casalou, C., Watts, G. F. M., Sandoval, J. L., Ramalho, J. S., et al. (2012). Arl13b regulates endocytic recycling traffic. Proc. Natl. Acad. Sci. U.S.A. 109, 1–6. doi: 10.1073/pnas.1218272110
Bay, S. N., Long, A. B., and Caspary, T. (2018). Disruption of the ciliary GTPase Arl13b suppresses Sonic hedgehog overactivation and inhibits medulloblastoma formation. Proc. Natl. Acad. Sci. U.S.A. 115, 1–6. doi: 10.1073/pnas.1706977115
Beghin, A., Belin, S., Sleiman, R. H., Manquat, S. B., Goddard, S., Tabone, E., et al. (2009). ADP ribosylation factor like 2 (Arl2) regulates breast tumor aggressivity in immunodeficient mice. PLoS One 4:e007478. doi: 10.1371/journal.pone.0007478
Beghin, A., Honore, S., Messana, C., Matera, E., Aim, J., Burlinchon, S., et al. (2007). ADP ribosylation factor like 2 (Arl2) protein influences microtubule dynamics in breast cancer cells. Exp. Cell Res. 13, 473–485. doi: 10.1016/j.yexcr.2006.10.024
Beghin, A., Matera, E., and Brunet-manquat, S. (2008). Expression of Arl2 is associated with p53 localization and chemosensitivity in a breast cancer cell line. Cell Cycle 7, 3074–3082. doi: 10.4161/cc.7.19.6777
Bidkhori, G., Narimani, Z., Ashtiani, S. H., Moeini, A., Nowzari-, A., and Masoudi-nejad, A. (2013). Reconstruction of an integrated genome-scale co- expression network reveals key modules involved in lung adenocarcinoma. PLoS One 8:e0067552. doi: 10.1371/journal.pone.0067552
Cai, Y., Wang, J., Li, R., Ayala, G., Ittmann, M., and Liu, M. (2009). GGAP2/PIKE-A directly activates both the akt and nuclear factor- B pathways and promotes prostate cancer progression. Cancer Res. 69, 819–827. doi: 10.1158/0008-5472.CAN-08-2537
Calin, G. A., Trapasso, F., Shimizu, M., Dumitru, C. D., Yendamuri, S., Godwin, A. K., et al. (2005). Familial cancer associated with a polymorphism in ARLTS1. N. Engl. J. Med. 352, 1667–1676.
Casalou, C., Faustino, A., and Barral, D. C. (2016). Arf proteins in cancer cell migration. Small GTPases 7, 270–282. doi: 10.1080/21541248.2016.1228792
Casalou, C., Faustino, A., Silva, F., Ferreira, I. C., Vaqueirinha, D., Ferreira, A., et al. (2019). Arl13b regulates breast cancer cell migration and invasion by controlling integrin-mediated signaling. Cancers 11:1461. doi: 10.3390/cancers11101461
Casalou, C., Seixas, C., Portelinha, A., Pintado, P., Barros, M., Ramalho, J. S., et al. (2014). Arl13b and the non-muscle myosin heavy chain IIA are required for circular dorsal ruffle formation and cell migration. J. Cell Sci. 127, 2709–2722. doi: 10.1242/jcs.143446
Castellví-Bel, S., Castells, A., de Cid, R., Muñoz, J., Balaguer, F., Gonzalo, V., et al. (2007). Association of the ARLTS1 Cys148Arg variant with sporadic and familial colorectal cancer. Carcinogenesis 28, 1687–1691. doi: 10.1093/carcin/bgm098
Chan, S., Huang, W., Chang, J., Chang, K., Kuo, W., Wang, M., et al. (2014). MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 33, 4496–4507. doi: 10.1038/onc.2014.10
Chang, J. S., Su, C. Y., Yu, W. H., Lee, W. J., Liu, Y. P., Lai, T. C., et al. (2015). GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget 6, 36278–36291. doi: 10.18632/oncotarget.5531
Chang, W., Ma, L., Lin, L., Gu, L., Liu, X., Cai, H., et al. (2009). Identification of novel hub genes associated with liver metastasis of gastric cancer. Int. J. Cancer 125, 2844–2853. doi: 10.1002/ijc.24699
Chen, Q., Weng, H., Tang, X., Lin, Y., Yuan, Y., Li, Q., et al. (2019). ARL4C stabilized by AKT/mTOR pathway promotes the invasion of PTEN-deficient primary human glioblastoma. J. Pathol. 247, 266–278. doi: 10.1002/path.5189
Chen, X., Su, Z., Wang, S., and Xu, H. (2016). Clinical and prognostic significance of Arl4c expression in colorectal cancer. Cancer Biomarkers 16, 253–257. doi: 10.3233/CBM-150562
Chi, J. H., Panner, A., Cachola, K., Crane, C. A., Murray, J., Pieper, R. O., et al. (2008). Increased expression of the glioma-associated antigen ARF4L after loss of the tumor suppressor PTEN. J. Neurosurg. 108, 299–303. doi: 10.3171/JNS/2008/108/2/0299
Chiang, T. S., Wu, H. F., and Lee, F. J. S. (2017). ADP-ribosylation factor-like 4C binding to filamin-A modulates filopodium formation and cell migration. Mol. Biol. Cell 28, 3013–3028. doi: 10.1091/mbc.E17-01-0059
Davis, J. E., Xie, X., Guo, J., Huang, W., Chu, W. M., Huang, S., et al. (2016). ARF1 promotes prostate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget 7, 39834–39845. doi: 10.18632/oncotarget.9405
Derycke, L., Stove, C., Wever, O. D. E., Dollé, L., Colpaert, N., Depypere, H., et al. (2011). The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int. J. Dev. Biol. 55, 835–840. doi: 10.1387/ijdb
Donaldson, J. G. (2003). Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278, 41573–41576. doi: 10.1074/jbc.R300026200
Donaldson, J. G., and Jackson, C. L. (2012). Arf Family G Proteins and their regulators: roles in membrane. Nat. Rev. Mol. Cell Biol. 12, 362–375. doi: 10.1038/nrm3117.Arf
Doush, Y., Surani, A. A., Navarro-Corcuera, A., McArdle, S., Billett, E. E., and Montiel-Duarte, C. (2019). SP1 and RARα regulate AGAP2 expression in cancer. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-018-36888-x
D’Souza-Schorey, C., and Chavrier, P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358. doi: 10.1038/nrm1910
Duxbury, M. S., Ashley, S. W., and Whang, E. E. (2004). Inhibition of pancreatic adenocarcinoma cellular invasiveness by blebbistatin: a novel myosin II inhibitor. Biochem. Biophys. Res. Commun. 313, 992–997. doi: 10.1016/j.bbrc.2003.12.031
Dykes, S. S., Gray, A. L., Coleman, D. T., Saxena, M., Stephens, C. A., Carroll, J. L., et al. (2016). The Arf-like GTPase Arl8b is essential for three-dimensional invasive growth of prostate cancer in vitro and xenograft formation and growth in vivo. Oncotarget 7, 31037–31052.
Ehlers, J. P., Worley, L., Onken, M. D., and Harbour, J. W. (2005). DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 11, 3609–3613. doi: 10.1158/1078-0432.CCR-04-1941
Fan, C., Tian, Y., Miao, Y., Lin, X., Zhang, X., Jiang, G., et al. (2014). ASAP3 expression in non-small cell lung cancer: association with cancer development and patients’ clinical outcome. Tumor Biol. 35, 1489–1494. doi: 10.1007/s13277-013-1205-1201
Flisiak, S., Zeeh, J. C., Guibert, B., Cherfils, J., and Zeghouf, M. (2008). An Arf1 GTPase mutant with different responses to GEF inhibitors. Biochem. Biophys. Res. Commun. 377, 156–160. doi: 10.1016/j.bbrc.2008.09.107
Frank, B., Hemminki, K., Brenner, H., Hoffmeister, M., Chang-Claude, J., and Burwinkel, B. (2006a). ARLTS1 variants and risk of colorectal cancer. Cancer Lett. 244, 172–175. doi: 10.1016/j.canlet.2005.12.006
Frank, B., Hemminki, K., Meindl, A., Wappenschmidt, B., Klaes, R., Schmutzler, R. K., et al. (2006b). Association of the ARLTS1 Cys148Arg variant with familial breast cancer risk. Int. J. Cancer 118, 2505–2508. doi: 10.1002/ijc.21687
Frank, B., Meyer, P., Boettger, M. B., Hemminki, K., Stapelmann, H., Gast, A., et al. (2006c). ARLTS1 variants and melanoma risk. Int. J. Cancer 119, 1736–1737. doi: 10.1002/ijc.22008
Frank, S. R., Köllmann, C. P., van Lidth de Jeude, J. F., Thiagarajah, J. R., Engelholm, L. H., Frödin, M., et al. (2017). The focal adhesion-associated proteins DOCK5 and GIT2 comprise a rheostat in control of epithelial invasion. Oncogene 36, 1816–1828. doi: 10.1038/onc.2016.345
Fu, Y., Li, J., Feng, M. X., Yang, X. M., Wang, Y. H., Zhang, Y. L., et al. (2014). Cytohesin-3 is upregulated in hepatocellular carcinoma and contributes to tumor growth and vascular invasion. Int. J. Clin. Exp. Pathol. 7, 2123–2132.
Fujii, S., Matsumoto, S., Nojima, S., Morii, E., and Kikuchi, A. (2014). Arl4c expression in colorectal and lung cancers promotes tumorigenesis and may represent a novel therapeutic target. Oncogene 34, 1–11. doi: 10.1038/onc.2014.402
Fujii, S., Shinjo, K., Matsumoto, S., and Harada, T. (2016). Epigenetic upregulation of ARL4C, due to DNA hypomethylation in the 3’-untranslated region, promotes tumorigenesis of lung squamous cell carcinoma. Oncotarget 7, 81571–81587.
Giordano, A., and Tommonaro, G. (2019). Curcumin and cancer. Nutrients 11:2376. doi: 10.3390/nu11102376
Grossmann, A. H., Yoo, J. H., Clancy, J., Sorensen, L. K., Tong, Z., Ostanin, K., et al. (2014). The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci. Signal. 6, 1–29. doi: 10.1126/scisignal.2003398
Gu, G., Chen, Y., Duan, C., Zhou, L., and Chen, C. (2017). Overexpression of ARF1 is associated with cell proliferation and migration through PI3K signal pathway in ovarian cancer. Oncol. Rep. 37, 1511–1520. doi: 10.3892/or.2017.5388
Guo, F., Yuan, D., Zhang, J., Zhang, H., Wang, C., and Zhu, L. (2019). Silencing of ARL14 gene induces lung adenocarcinoma cells to a dormant state. Front. Cell Dev. Biol. 7:238. doi: 10.3389/fcell.2019.00238
Guo, X., Jo, V. Y., Mills, A. M., Zhu, S. X., Lee, C., Nucci, M. R., et al. (2015). Clinically relevant molecular subtypes in leiomyosarcoma. Clin. Cancer Res. 21, 3501–3511. doi: 10.1158/1078-0432.CCR-14-3141
Hamadou, W. S., Besbes, S., Mani, R., Bourdon, V., Ben Youssef, Y., Achour, B., et al. (2017). ARTS1: gène candidat potentiel dans les agrégations familiales des hémopathies malignes. Bull. Cancer 104, 123–127. doi: 10.1016/j.bulcan.2016.10.016
Harada, T., Matsumoto, S., Hirota, S., Kimura, H., and Fujii, S. (2019). Chemically modified antisense oligonucleotide against ARL4C inhibits primary and metastatic liver tumor growth. Mol. Cancer Ther. 18, 602–612. doi: 10.1158/1535-7163.MCT-18-0824
Harvey, R. C., Mullighan, C. G., Wang, X., Dobbin, K. K., Davidson, G. S., Bedrick, E. J., et al. (2010). Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116, 4874–4884. doi: 10.1182/blood-2009-08-239681
Hashimoto, S., Mikami, S., Sugino, H., Yoshikawa, A., Hashimoto, A., Onodera, Y., et al. (2016). Lysophosphatidic acid activates Arf6 to promote the mesenchymal malignancy of renal cancer. Nat. Commun. 6, 1–11. doi: 10.1038/ncomms10656
Hashimoto, S., Onodera, Y., Hashimoto, A., Tanaka, M., Hamaguchi, M., Yamada, A., et al. (2004). Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. U.S.A. 101, 6647–6652. doi: 10.1073/pnas.0401753101
Hass, H. G., Vogel, U., Scheurlen, M., and Jobst, J. (2016). Gene-expression analysis identifies specific patterns of dysregulated molecular pathways and genetic subgroups of human hepatocellular carcinoma. Anticancer Res. 36, 5087–5096. doi: 10.21873/anticanres.11078
He, H. U. A., Dai, F., Yu, L., She, X., Zhao, Y., and Jiang, J. (2002). Identification and characterization of nine novel human small GTPases showing variable expressions in liver cancer tissues. Gene Expr. 10, 231–242. doi: 10.3727/000000002783992406
Hiroi, T., Someya, A., Thompson, W., Moss, J., and Vaughan, M. (2006). GEP100/BRAG2: activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and alpha-catenin. PNAS 103, 10672–10677. doi: 10.1073/pnas.0604091103
Hongu, T., Funakoshi, Y., Fukuhara, S., Suzuki, T., Sakimoto, S., Takakura, N., et al. (2015). Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial β1 integrin recycling. Nat. Commun. 6:7925. doi: 10.1038/ncomms8925
Hongu, T., Yamauchi, Y., Funakoshi, Y., Katagiri, N., Ohbayashi, N., and Kanaho, Y. (2016). Pathological functions of the small GTPase Arf6 in cancer progression: Tumor angiogenesis and metastasis. Small GTPases 7, 47–53. doi: 10.1080/21541248.2016.1154640
Hou, T., Yang, C., Tong, C., Zhang, H., Xiao, J., and Li, J. (2014). Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 7, 280–287. doi: 10.1177/1933719115625845
Howley, B. V., Link, L. A., Grelet, S., El-sabban, M., and Howe, P. H. (2018). A CREB3-regulated ER-Golgi trafficking signature promotes metastatic progression in breast cancer. Oncogene 37, 1308–1325. doi: 10.1038/s41388-017-0023-0.A
Hu, B., Shi, B., Jarzynka, M. J., Yiin, J., Souza-schorey, C. D., and Cheng, S.-Y. (2009). ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1 – mediated pathway. Cancer Res. 69, 794–802. doi: 10.1158/0008-5472.CAN-08-2110
Hu, Q., Masuda, T., Sato, K., Tobo, T., and Nambara, S. (2018). Identification of ARL4C as a peritoneal dissemination-associated gene and its clinical significance in gastric cancer. Ann. Surg. Oncol. 25, 745–753. doi: 10.1245/s10434-017-6292-6296
Huang, D., Pei, Y., Dai, C., Huang, Y., Chen, H., Chen, X., et al. (2019). Up-regulated ADP-Ribosylation factor 3 promotes breast cancer cell proliferation through the participation of FOXO1. Exp. Cell Res. 384:111624. doi: 10.1016/j.yexcr.2019.111624
Huang, M., and Wang, Y. (2019). A targeted quantitative proteomic approach for probing altered protein expression of small GTPases associated with colorectal cancer metastasis a targeted quantitative proteomic approach for probing altered protein expression of small GTPases associated w. Anal. Chem. 91, 6233–6241. doi: 10.1021/acs.analchem.9b00938
Huang, W., Chan, S., Jang, T., Chang, J., Ko, Y., Yen, T., et al. (2014). miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 74, 751–764. doi: 10.1158/0008-5472.CAN-13-1297
Isono, T., Chano, T., Yoshida, T., Makino, A., Ishida, S., Suzaki, M., et al. (2019). ADP-ribosylation factor-like 4C is a predictive biomarker of poor prognosis in patients with renal cell carcinoma. Am. J. Cancer Res. 9, 415–423.
Jang, S. Y., Jang, S., and Ko, J. (2012). Regulation of ADP-ribosylation factor 4 expression by small leucine zipper protein and involvement in breast cancer cell migration. Cancer Lett. 314, 185–197. doi: 10.1016/j.canlet.2011.09.028
Kahn, R. A., Cherfils, J., Elias, M., Lovering, R. C., Munro, S., and Schurmann, A. (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172, 645–650. doi: 10.1083/jcb.200512057
Kannangai, R., Vivekanandan, P., Martinez-Murillo, F., Choti, M., and Torbenson, M. (2007). Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum. Pathol. 38, 639–644. doi: 10.1016/j.humpath.2006.07.019
Katono, K., Sato, Y., Jiang, S., Kobayashi, M., and Nagashio, R. (2015). Prognostic significance of MYH9 expression in resected non-small cell lung cancer. PLoS One 10:e0121460. doi: 10.1371/journal.pone.0121460
Khatter, D., Sindhwani, A., and Sharma, M. (2015). Arf-like GTPase Arl8: moving from the periphery to the center of lysosomal biology. Cell. Logist. 5, 1–8. doi: 10.1080/21592799.2015.1086501
Knizhnik, A. V., Kovaleva, O. V., Laktionov, K. K., Mochalnikova, V. V., Komelkov, A. V., Tchevkina, E. M., et al. (2011). Arf6, RalA, and BIRC5 protein expression in nonsmall cell lung cancer. Mol. Biol. 45, 275–282. doi: 10.1134/S0026893310061032
Knobbe, C. B., Trampe-Kieslich, A., and Reifenberger, G. (2005). Genetic alteration and expression of the phosphoinositol-3-kinase/Akt pathway genes PIK3CA and PIKE in human glioblastomas. Neuropathol. Appl. Neurobiol. 31, 486–490. doi: 10.1111/j.1365-2990.2005.00660.x
Kobayashi, N., Kon, S., Henmi, Y., Funaki, T., Satake, M., and Tanabe, K. (2014). The Arf GTPase-activating protein SMAP1 promotes transferrin receptor endocytosis and interacts with SMAP2. Biochem. Biophys. Res. Commun. 453, 473–479. doi: 10.1016/j.bbrc.2014.09.108
Kolanus, W. (2007). Guanine nucleotide exchange factors of the cytohesin family and their roles in signal transduction. Immunol. Rev. 218, 102–113. doi: 10.1111/j.1600-065X.2007.00542.x
Kon, S., Kobayashi, N., and Satake, M. (2014). Altered trafficking of mutated growth factor receptors and their associated molecules: implication for human cancers. Cell. Logist. 4:e28461. doi: 10.4161/cl.28461
Kon, S., Tanabe, K., Watanabe, T., Sabe, H., and Satake, M. (2008). Clathrin dependent endocytosis of E-cadherin is regulated by the Arf6GAP isoform SMAP1. Exp. Cell Res. 314, 1415–1428. doi: 10.1016/j.yexcr.2007.11.006
Lang, L., Shay, C., Zhao, X., and Teng, Y. (2017). Combined targeting of Arf1 and Ras potentiates anticancer activity for prostate cancer therapeutics. J. Exp. Clin. Cancer Res. 36, 1–10. doi: 10.1186/s13046-017-0583-584
Li, C.-C., Chiang, T.-C., Wu, T.-S., Pacheco-Rodriguez, G., Moss, J., and Lee, F.-J. S. (2007). ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol. Biol. Cell 18, 4420–4437. doi: 10.1091/mbc.e07-02-0149
Li, H.-J., Sun, X.-M., Li, Z.-K., Yin, Q.-W., Pang, H., Pan, J.-J., et al. (2017). LncRNA UCA1 promotes mitochondrial function of bladder cancer via the MiR- 195/ARL2 signaling pathway. Cell. Physiol. Biochem. 43, 2548–2561. doi: 10.1159/000484507
Li, R., Peng, C., Zhang, X., Wu, Y., Pan, S., and Xiao, Y. (2017). Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 182, 80–84. doi: 10.1016/j.lfs.2017.06.008
Li, M., Ng, S. S., Wang, J., Lai, L., Leung, S. Y., Franco, M., et al. (2006). EFA6A enhances glioma cell invasion through ADP ribosylation factor 6/extracellular signal – regulated kinase signaling. Cancer Res. 66, 1583–1591. doi: 10.1158/0008-5472.CAN-05-2424
Li, M., Tian, L., Yao, H., Lu, J., Ge, J., Guo, Y., et al. (2014). ASAP1 mediates the invasive phenotype of human laryngeal squamous cell carcinoma to affect survival prognosis. Oncol. Rep. 31, 2676–2682. doi: 10.3892/or.2014.3150
Li, Y., Kelly, W. G., Logsdon, J. M., Schurko, A. M., Harfe, B. D., Hill-Harfe, K. L., et al. (2004). Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 18, 1834–1850. doi: 10.1096/fj.04-2273com
Liang, C., Qin, Y., Zhang, B., Ji, S., and Shi, S. (2017). ARF6, induced by mutant Kras, promotes proliferation and Warburg effect in pancreatic cancer. Cancer Lett. 388, 303–311. doi: 10.1016/j.canlet.2016.12.014
Liao, Q., Li, R., Zhou, R., Pan, Z., Xu, L., Ding, Y., et al. (2017). LIM kinase 1 interacts with myosin-9 and alpha-actinin-4 and promotes colorectal cancer progression. Br. J. Cancer 117, 563–571. doi: 10.1038/bjc.2017.193
Lin, D., Watahiki, A., Bayani, J., Zhang, F., Liu, L., Ling, V., et al. (2008). ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 68, 4352–4359. doi: 10.1158/0008-5472.CAN-07-5237
Liu, D., Zhang, L., Shen, Z., Tan, F., Hu, Y., Yu, J., et al. (2012). Clinicopathological significance of NMIIA overexpression in human gastric cancer. Int. J. Mol. Sci. 13, 15291–15304. doi: 10.3390/ijms131115291
Liu, X., Hu, Y., Hao, C., Rempel, S. A., and Ye, K. (2007). PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene 26, 4918–4927. doi: 10.1038/sj.onc.1210290
Liu, X., Shen, Q., Yu, T., Huang, H., Zhang, Z., Ding, J., et al. (2016). Small GTPase Arl6 controls RH30 rhabdomyosarcoma cell growth through ciliogenesis and Hedgehog signaling. Cell Biosci. 6, 1–11. doi: 10.1186/s13578-016-0126-122
Long, L., He, B., Huang, G., Guo, Y., Liu, Y., and Huo, J. (2015). microRNA-214 functions as a tumor suppressor in human colon cancer via the suppression of ADP-ribosylation factor-like protein 2. Oncol. Lett. 9, 645–650. doi: 10.3892/ol.2014.2746
Lu, X., Wan, F., Zhang, H., Shi, G., and Ye, D. (2016). ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumor Biol. 37, 253–262. doi: 10.1007/s13277-015-3792-3795
Luchsinger, C., Aguilar, M., and Burgos, P. V. (2018). Functional disruption of the Golgi apparatus protein ARF1 sensitizes MDA-MB-231 breast cancer cells to the antitumor drugs Actinomycin D and Vinblastine through ERK and AKT signaling. PLoS One 13:e0195401. doi: 10.1371/journal.pone.0195401
Ma, W.-M., Jiang, B., Lai, Z.-S., Wang, X.-Y., Geng, Y., and Han, Y.-J. (2005). [Expression and significance of ADP-ribosylation factor 1 (ARF1) in colorectal laterally spreading tumor]. Ai Zheng 24, 690–694.
Mao, Y., Zhao, Q., Yin, S., Ding, X., and Wang, H. (2018). Genome-wide expression profiling and bioinformatics analysis of deregulated genes in human gastric cancer tissue after gastroscopy. Asia. Pac. J. Clin. Oncol. 14, 29–36. doi: 10.1111/ajco.12688
Marwaha, R., Dwivedi, D., and Sharma, M. (2019). Emerging roles of Arf-Like GTP-binding proteins: from membrane trafficking to cytoskeleton dynamics and beyond. Proc. Indian Natl. Sci. Acad. 85, 189–212. doi: 10.16943/ptinsa/2019/49574
Menju, T., Hashimoto, S., Hashimoto, A., Otsuka, Y., Handa, H., Ogawa, E., et al. (2011). Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PLoS One 6:e0025301. doi: 10.1371/journal.pone.0025301
Mikulich, A., Kavaliauskiene, S., and Juzenas, P. (2012). Blebbistatin, a myosin inhibitor, is phototoxic to human cancer cells under exposure to blue light. Biochim. Biophys. Acta Gen. Subj. 1820, 870–877. doi: 10.1016/j.bbagen.2012.04.003
Morgan, C., Lewis, P. D., Hopkins, L., Burnell, S., Kynaston, H., and Doak, S. H. (2015). Increased expression of ARF GTPases in prostate cancer tissue. Springerplus 4:342. doi: 10.1186/s40064-015-1136-y
Morishige, M., Hashimoto, S., Ogawa, E., Toda, Y., Kotani, H., Wei, S., et al. (2008). GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10, 85–92. doi: 10.1038/ncb1672
Müller, T., Stein, U., Poletti, A., Garzia, L., Rothley, M., Plaumann, D., et al. (2010). ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene 29, 2393–2403. doi: 10.1038/onc.2010.6
Muralidharan-chari, V., Hoover, H., Clancy, J., Schweitzer, J., Suckow, M. A., Schroeder, V., et al. (2009). ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res. 69, 2201–2210. doi: 10.1158/0008-5472.CAN-08-1301
Nalla, A. K., Williams, T. F., Collins, C. P., Rae, D. T., and Trobridge, G. D. (2016). Lentiviral vector-mediated insertional mutagenesis screen identifies genes that influence androgen independent prostate cancer progression and predict clinical outcome. Mol. Carcinog. 55, 1761–1771. doi: 10.1002/mc.22425
Nayal, A., Webb, D. J., Brown, C. M., Schaefer, E. M., Vicente-manzanares, M., and Horwitz, A. R. (2006). Paxillin phosphorylation at Ser273 localizes a GIT1–PIX–PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173, 587–599. doi: 10.1083/jcb.200509075
Newell-Litwa, K. A., Horwitz, R., and Lamers, M. L. (2015). Non-Muscle myosin II in disease: mechanisms and therapeutic opportunities. DMM Dis. Model. Mech. 8, 1495–1515. doi: 10.1242/dmm.022103
Nonaka, Y., Tsuda, N., Shichijo, S., Ito, M., Maeda, Y., Harada, M., et al. (2002). Recognition of ADP-ribosylation factor 4-like by HLA-A2-restricted and tumor-reactive cytotoxic T lymphocytes from patients with brain tumors. Tissue Antigens 60, 319–327. doi: 10.1034/j.1399-0039.2002.600406.x
Oka, S., Uramoto, H., Shimokawa, H., Yamada, S., and Tanaka, F. (2014). Receptor-GEP100-Arf6 axis affects the prognosis of lung adenocarcinoma. Clin. Transl. Res. 86, 263–270. doi: 10.1159/000360089
Okabe, H., Furukawa, Y., Kato, T., Hasegawa, S., Yamaoka, Y., and Nakamura, Y. (2004). Isolation of development and differentiation enhancing factor-like 1 (DDEFL1) as a drug target for hepatocellular carcinomas. Int. J. Oncol. 24, 43–48. doi: 10.3892/ijo.24.1.43
Olstad, O., Gautvik, V., Reppe, S., Rian, E., Jemtland, R., Ohlsson, C., et al. (2003). Molecular heterogeneity in human osteosarcoma demonstrated by enriched mRNAs isolated by directional tag PCR subtraction cloning. Anticancer. Res. 23, 2201–2216.
Onodera, Y., Hashimoto, S., Hashimoto, A., Morishige, M., Mazaki, Y., Yamada, A., et al. (2005). Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 24, 963–973. doi: 10.1038/sj.emboj.7600588
Pan, T., Sun, J., Hu, J., Hu, Y., Zhou, J., Chen, Z., et al. (2014). Cytohesins/ARNO: the function in colorectal cancer cells. PLoS One 9:e090997. doi: 10.1371/journal.pone.0090997
Park, J. Y., Kim, S. A., Chung, J. W., Bang, S., Park, S. W., Paik, Y. K., et al. (2011). Proteomic analysis of pancreatic juice for the identification of biomarkers of pancreatic cancer. J. Cancer Res. Clin. Oncol. 137, 1229–1238. doi: 10.1007/s00432-011-0992-992
Pecci, A., Ma, X., Savoia, A., and Adelstein, R. S. (2019). Myh9: structure, functions and role of non-muscle myosin IIA in human disease. Gene 664, 152–167. doi: 10.1016/j.gene.2018.04.048.MYH9
Pellon-Cardenas, O., Clancy, J., Uwimpuhwe, H., and Souza-schorey, C. D. (2013). ARF6-regulated endocytosis of growth factor receptors links cadherin-based adhesion to canonical Wnt signaling in epithelia. Mol. Cell. Biol. 33, 2963–2975. doi: 10.1128/MCB.01698-1612
Peng, H., Dara, L., Li, T. W. H., Zheng, Y., Yang, H., Maria, L., et al. (2014). Methionine adenosyltransferase 2B-GIT1 interplay activates MEK1-ERK1/2 to induce growth in human liver and colon cancer. Hepatology 57, 2299–2313. doi: 10.1002/hep.26258.Methionine
Peng, R., Men, J., Ma, R., Wang, Q., Wang, Y., Sun, Y., et al. (2017). miR-214 down-regulates ARL2 and suppresses growth and invasion of cervical cancer cells. Biochem. Biophys. Res. Commun. 484, 623–630. doi: 10.1016/j.bbrc.2017.01.152
Petrocca, F., Iliopoulos, D., Qin, H. R., Nicoloso, M. S., Yendamuri, S., Wojcik, S. E., et al. (2006). Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 66, 10287–10292. doi: 10.1158/0008-5472.CAN-06-2289
Pils, D., Horak, P., Gleiss, A., Sax, C., Fabjani, G., Moebus, V. J., et al. (2005). Five genes from chromosomal band 8p22 are significantly down-regulated in ovarian carcinoma: N33 and EFA6R have a potential impact on overall survival. Cancer 104, 2417–2429. doi: 10.1002/cncr.21538
Qi, Q., Kang, S. S., Zhang, S., Pham, C., Fu, H., Brat, D. J., et al. (2017). Co-amplification of phosphoinositide 3-kinase enhancer A and cyclin-dependent kinase 4 triggers glioblastoma progression. Oncogene 36, 4562–4572. doi: 10.1038/onc.2017.67
Qi, S., Su, L., Li, J., Zhang, C., Ma, Z., Liu, G., et al. (2019). Arf6-driven endocytic recycling of CD147 determines HCC malignant phenotypes. J. Exp. Clin. Cancer Res. 9, 1–17.
Saito, K., Maeda, M., and Katada, T. (2017). Regulation of the Sar1 GTPase cycle is necessary for large cargo secretion from the endoplasmic reticulum. Front. Cell Dev. Biol. 5:75. doi: 10.3389/fcell.2017.00075
Sangar, F., Schreurs, A. S., Umaña-Diaz, C., Clapéron, A., Desbois-Mouthon, C., Calmel, C., et al. (2014). Involvement of small ArfGAP1 (SMAP1), a novel Arf6-specific GTPase-activating protein, in microsatellite instability oncogenesis. Oncogene 33, 2758–2767. doi: 10.1038/onc.2013.211
Sato, H., Hatanaka, K. C., Hatanaka, Y., Hatakeyama, H., Hashimoto, A., Matsuno, Y., et al. (2014). High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients. Cell Commun. Signal. 12, 1–9. doi: 10.1186/1478-811X-12-17
Schlienger, S., Ramirez, R. A. M., and Claing, A. (2015). ARF1 regulates adhesion of MDA-MB-231 invasive breast cancer cells through formation of focal adhesions. Cell. Signal. 27, 403–415. doi: 10.1016/j.cellsig.2014.11.032
Schramek, D., Sendoel, A., Segal, J. P., Beronja, S., Heller, E., Oristian, D., et al. (2014). Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313. doi: 10.1126/science.1248627.Direct
Schweitzer, J. K., Sedgwick, A. E., and D’Souza-Schorey, C. (2011). ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin. Cell Dev. Biol. 22, 39–47. doi: 10.1016/j.semcdb.2010.09.002
Shao, J., Xu, L., Chen, L., Lu, Q., Xie, X., Shi, W., et al. (2018). Arl13b promotes gastric tumorigenesis by regulating Smo trafficking and activation of the Hedgehog signaling pathway. Cancer Res. 77, 4000–4013. doi: 10.1158/0008-5472.CAN-16-2461.Arl13b
Siltanen, S., Fischer, D., Rantapero, T., Laitinen, V., Mpindi, J. P., Kallioniemi, O., et al. (2013). ARLTS1 and prostate cancer risk - analysis of expression and regulation. PLoS One 8:e072040. doi: 10.1371/journal.pone.0072040
Sirirattanakul, S., Wannakrairot, P., Tencomnao, T., and Santiyanont, R. (2015). Gene expression profile in breast cancer comprising predictive markers for metastatic risk. Genet. Mol. Res. 14, 10929–10936. doi: 10.4238/2015.September.21.3
Su, D., Katsaros, D., Xu, S., Xu, H., Gao, Y., Biglia, N., et al. (2015). ADP-ribosylation factor-like 4C (ARL4C), a novel ovarian cancer metastasis suppressor, identified by integrated genomics. Am. J. Transl. Res. 7, 242–256.
Sun, Z., Li, A., Yu, Z., Li, X., Guo, X., and Chen, R. (2017). MicroRNA-497-5p suppresses tumor cell growth of osteosarcoma by targeting ADP ribosylation factor-like protein 2. Cancer Biother. Radiopharm. 32, 371–378. doi: 10.1089/cbr.2017.2268
Sztul, E., Chen, P., Casanova, J. E., Cherfils, J., Dacks, J. B., and Bement, W. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell 30, 1249–1271. doi: 10.1091/mbc.E18-12-0820
Taniuchi, K., Iwasaki, S., and Saibara, T. (2011). BART inhibits pancreatic cancer cell invasion by inhibiting ARL2-mediated RhoA inactivation. Int. J. Oncol. 39, 1243–1252. doi: 10.3892/ijo.2011.1156
Tsai, M. M., Lin, P. Y., Cheng, W. L., Tsai, C. Y., Chi, H. C., Chen, C. Y., et al. (2012). Overexpression of ADP-ribosylation factor 1 in human gastric carcinoma and its clinicopathological significance. Cancer Sci. 103, 1136–1144. doi: 10.1111/j.1349-7006.2012.02243.x
Van Den Boom, J., Wolter, M., Blaschke, B., Knobbe, C. B., and Reifenberger, G. (2006). Identification of novel genes associated with astrocytoma progression using suppression subtractive hybridization and real-time reverse transcription-polymerase chain reaction. Int. J. Cancer 119, 2330–2338. doi: 10.1002/ijc.22108
Villalva, C., Trempat, P., Greenland, C., Thomas, C., Girard, J. P., Moebius, F., et al. (2002). Isolation of differentially expressed genes in NPM-ALK-positive anaplastic large cell lymphoma. Br. J. Haematol. 118, 791–798. doi: 10.1046/j.1365-2141.2002.03671.x
Wakinoue, S., Chano, T., Amano, T., Isono, T., and Kimura, F. (2018). ADP-ribosylation factor-like 4C predicts worse prognosis in endometriosis-associated ovarian cancers. Cancer Biomarkers 1, 1–7. doi: 10.3233/CBM-181836
Wang, B., Qi, X., Liu, J., Zhou, R., Lin, C., Shangguan, J., et al. (2019). MYH9 promotes growth and metastasis via activation of MAPK/AKT signaling in colorectal cancer. J. Cancer 10, 874–884. doi: 10.7150/jca.27635
Wang, L., Shi, J., Huang, Y., Liu, S., Zhang, J., Ding, H., et al. (2019). A six - gene prognostic model predicts overall survival in bladder cancer patients. Cancer Cell Int. 19, 1–15. doi: 10.1186/s12935-019-0950-957
Wang, Y., Liu, S., Zhang, Y., and Yang, J. (2019a). Myosin heavy chain 9: oncogene or tumor suppressor gene? Med. Sci. Monit. 25, 888–892. doi: 10.12659/MSM.912320
Wang, Y., Zhao, W., Liu, X., Guan, G., and Zhuang, M. (2019b). ARL3 is downregulated and acts as a prognostic biomarker in glioma. J. Transl. Med. 17:210. doi: 10.1186/s12967-019-1914-3
Wang, Q., Huang, Z., Guo, W., Ni, S., Xiao, X., Wang, L., et al. (2014). MicroRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Hum. Cancer Biol. 20, 1146–1158. doi: 10.1158/1078-0432.CCR-13-1023
Wang, Y., Guan, G., Cheng, W., Jiang, Y., Shan, F., Wu, A., et al. (2018). ARL2 overexpression inhibits glioma proliferation and tumorigenicity via down-regulating AXL. BMC Cancer 18:599. doi: 10.1186/s12885-018-4517-0
Watanabe, M., Takahashi, H., Saeki, Y., Ozaki, T., Itoh, S., Suzuki, M., et al. (2015). The E3 ubiquitin ligase TRIM23 regulates adipocyte differentiation via stabilization of the adipogenic activator PPARγ. eLife 4:e05615. doi: 10.7554/eLife.05615
Weizhong, Z., Shuohui, G., Hanjiao, Q., Yuhong, M., Xiaohua, Y., Jian, C., et al. (2011). Inhibition of cytohesin-1 by siRNA leads to reduced IGFR signaling in prostate cancer. Braz. J. Med. Biol. Res. 44, 642–646. doi: 10.1590/s0100-879x2011007500072
Willis, S., Villalobos, V. M., Gevaert, O., Abramovitz, M., Williams, C., Sikic, B. I., et al. (2016). Single gene prognostic biomarkers in ovarian cancer: a meta-analysis. PLoS One 11:e0149183. doi: 10.1371/journal.pone.0149183
Woo, I. S., Eun, S. Y., Jang, H., Kang, E. S., Kim, G. H., Kim, H. J., et al. (2009). Identification of ADP-ribosylation factor 4 as a suppressor of N - (4-hydroxyphenyl) retinamide-induced cell death. Cancer Lett. 276, 53–60. doi: 10.1016/j.canlet.2008.10.031
Wu, Q., Ren, X., Zhang, Y., Fu, X., Li, Y., and Peng, Y. (2018). MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem. Biophys. Res. Commun. 497, 1162–1170. doi: 10.1016/j.bbrc.2017.01.002
Wu, R., Murali, R., Kabe, Y., French, S. W., Chiang, Y.-M., Liu, S., et al. (2018). Baicalein targets GTPase-mediated autophagy to eliminate liver tumor initiating stem cell-like cells resistant to mTORC1 inhibition. Hepatology 68, 1726–1740. doi: 10.1002/hep
Xie, X., Tang, S.-C., Cai, Y., Pi, W., Deng, L., Wu, G., et al. (2016). Suppression of breast cancer metastasis through the inactivation of ADP-ribosylation factor 1. Oncotarget 7, 58111–58120. doi: 10.18632/oncotarget.11185
Xie, Z., Jiang, Y., Liao, E. Y., Chen, Y., Pennypacker, S. D., Peng, J., et al. (2012). PIKE mediates EGFR proliferative signaling in squamous cell carcinoma cells. Oncogene 31, 5090–5098. doi: 10.1038/onc.2012.10
Xu, K., Gao, J., Yang, X., Yao, Y., and Liu, Q. (2013). Cytohesin-2 as a novel prognostic marker for hepatocellular carcinoma. Oncol. Rep. 29, 2211–2218. doi: 10.3892/or.2013.2366
Xu, X., Wang, Q., He, Y., Ding, L., Zhong, F., and Ou, Y. (2017). ADP-ribosylation factor 1 (ARF1) takes part in cell proliferation and cell adhesion-mediated drug resistance (CAM-DR). Ann. Hematol. 1, 847–858. doi: 10.1007/s00277-017-2949-2942
Yagi, R., Tanaka, M., Sasaki, K., Kamata, R., Nakanishi, Y., Kanai, Y., et al. (2011). ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene 30, 1413–1421. doi: 10.1038/onc.2010.522
Yang, X. Y., Yu, H., and Peng, Z. L. (2011). Involvement of ARLTS1 in chemotherapy and apoptosis in ovarian cancer cell line. Arch. Gynecol. Obstet. 284, 1241–1246. doi: 10.1007/s00404-010-1782-1789
Yao, Y., Liu, Z., Guo, H., Huang, S., Zhong, M., Deng, J., et al. (2018). Elevated TRIM23 expression predicts poor prognosis in Chinese gastric cancer. Pathol. Res. Pract. 214, 1–7. doi: 10.1016/j.prp.2018.10.010
Ye, G., Huang, K., Yu, J., Zhao, L., Zhu, X., Yang, Q., et al. (2017). MicroRNA-647 Targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer. Theranostics 7, 6350–6351. doi: 10.7150/thno.20512
Yendamuri, S., Trapasso, F., and Calin, G. A. (2008). ARLTS1 – a novel tumor suppressor gene. Cancer Lett. 264, 11–20. doi: 10.1016/j.canlet.2008.02.021
Yoo, J. H., Brady, S. W., Acosta-Alvarez, L., Rogers, A., Peng, J., Sorensen, L. K., et al. (2019). The small GTPase ARF6 activates PI3K in melanoma to induce a prometastatic State. Cancer Res. 79, 2892–2908. doi: 10.1158/0008-5472.CAN-18-3026
Yoo, J. H., Shi, D. S., Grossmann, A. H., Odelberg, S. J., Ostanin, K., and Li, D. Y. (2016). ARF6 is an actionable node that orchestrates article ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell 29, 889–904. doi: 10.1016/j.ccell.2016.04.015
Yoo, S. M., Antonyak, M. A., and Cerione, R. A. (2012). The adaptor protein and Arf GTPase-activating protein Cat-1/Git-1 is required for cellular transformation. J. Biol. Chem. 287, 31462–31470. doi: 10.1074/jbc.M112.353615
Zangari, J., Partisani, M., Cabaud, O., Chetaille, B., Berruyer-pouyet, C., Finetti, P., et al. (2014). EFA6B antagonizes breast cancer. Cancer Res. 74, 1–15. doi: 10.1158/0008-5472.CAN-14-0298
Zhang, J., Zhou, Y., Yu, Z., Chen, A., Yu, Y., and Wang, X. (2019). Identification of core genes and clinical roles in pregnancy-associated breast cancer based on integrated analysis of different microarray profile datasets. Biosci. Rep. 39:BSR20190019. doi: 10.1042/BSR20190019
Zhang, Q., Major, M. B., Takanashi, S., Camp, N. D., Nishiya, N., Peters, E. C., et al. (2007). Small-molecule synergist of the Wnt/β-catenin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 7444–7448. doi: 10.1073/pnas.0702136104
Zhang, Y., Du, J., Zheng, J., Liu, J., Xu, R., and Shen, T. (2015). EGF-reduced transcription induces mesenchymal transition via Arf6-ERK signaling in gastric cancer cells. Oncotarget 6, 7244–7261.
Zhao, H., Ahirwar, D. K., Oghumu, S., Wilkie, T., Powell, C. A., Nasser, M. W., et al. (2016). Endothelial Robo4 suppresses breast cancer growth and metastasis through regulation of tumor angiogenesis. Mol. Oncol. 10, 272–281. doi: 10.1016/j.molonc.2015.10.007
Zhou, P., Li, Y., Li, B., Zhang, M., Liu, Y., Yao, Y., et al. (2019). NMIIA promotes tumor growth and metastasis by activating the Wnt/β -catenin signaling pathway and EMT in pancreatic cancer. Oncogene 38, 5500–5515. doi: 10.1038/s41388-019-0806-806
Keywords: ARL, migration, invasion, tumorigenesis, guanine nucleotide exchange factor, GTPase-activating protein, membrane traffic
Citation: Casalou C, Ferreira A and Barral DC (2020) The Role of ARF Family Proteins and Their Regulators and Effectors in Cancer Progression: A Therapeutic Perspective. Front. Cell Dev. Biol. 8:217. doi: 10.3389/fcell.2020.00217
Received: 15 January 2020; Accepted: 12 March 2020;
Published: 21 April 2020.
Edited by:
Sunil Kumar Verma, Centre for Cellular & Molecular Biology (CCMB), IndiaReviewed by:
Wei-Hsiung Yang, Mercer University, United StatesIra Daar, National Cancer Institute (NCI), United States
Copyright © 2020 Casalou, Ferreira and Barral. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duarte C. Barral, duarte.barral@nms.unl.pt
†These authors have contributed equally to this work
 Cristina Casalou
Cristina Casalou Andreia Ferreira
Andreia Ferreira Duarte C. Barral
Duarte C. Barral