- Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom
Human induced Pluripotent Stem Cell (hiPSC) models are a valuable new tool for research into neurodegenerative diseases. Neuroinflammation is now recognized as a key process in neurodegenerative disease and aging, and microglia are central players in this. A plethora of hiPSC-derived microglial models have been published recently to explore neuroinflammation, ranging from monoculture through to xenotransplantation. However, combining physiological relevance, reproducibility, and scalability into one model is still a challenge. We examine key features of the in vitro microglial environment, especially media composition, extracellular matrix, and co-culture, to identify areas for improvement in current hiPSC-microglia models.
Introduction
Microglia represent a branch of tissue-resident macrophages which originate primarily in the yolk sac (1) and finally mature in the central nervous system (CNS) (2, 3). Once within the CNS, microglia function as important homeostatic cells and innate immune sentinels, attracting the analogy of a double-edged sword through their contribution to both health and disease. They influence developing neural networks by regulating the rate of neurogenesis and pruning neuronal synapses [(4), reviewed by (5)]. As ramified cells, they aid brain homeostasis by surveying the brain parenchyma and clearing cellular debris. However, as amoeboid cells, they secrete cytokines in response to pathogens (e.g. viral infection), misfolded proteins (e.g. Alzheimer’s disease), or cellular damage (e.g. brain injury). The cytokine response can exacerbate neuronal damage if prolonged, contributing to chronic neurodegenerative disease [(6), reviewed by (7)]. This key pathological process is still inadequately understood, hindered by the lack of good quality in vitro models.
Why Use hiPSC-Microglia to Model Microglia?
Human induced Pluripotent Stem Cells (hiPSC) offer unprecedented advantages over other commonly used primary and immortalized cell cultures, notably human genetic background, normal karyotype, limitless self-renewal, and suitability for gene editing [reviewed by (8)]. The human genetic background solves the poor representation of disease-relevant gene orthologs in mouse models (9, 10) deemed partly responsible for the poor success-rate of translating therapeutic modalities from mouse studies into the clinic [reviewed by (11, 12)]. Many neurological diseases are multigenic, and therefore particularly difficult to model in mice, whereas hiPSC enable representation of extremes of polygenic risk score (13), and endogenous levels of protein expression.
Microglial identity is driven by both developmental origin and brain environment [reviewed by (14)]. They derive from early waves of primitive yolk sac macrophages, which migrate to the brain rudiment even before neural progenitors develop (1, 15), and are maintained by local self-renewal throughout life (16). Several strategies have been published recently to derive microglia from hiPSC, which generally mirror the main characteristics of primitive hematopoiesis, including dependence on PU.1 and IRF8 transcription factors and MYB independence [(15, 17) reviewed by (18)], though the in vitro developmental pathway does not necessarily conform exactly to in vivo primitive waves. While mesoderm/hemogenic endothelium/myeloid differentiation can be achieved using simple growth factor cocktails (minimally BMP4, VEGF, SCF, followed by IL-3, M-CSF), mature microglial identity subsequently relies on cues present in the CNS environment, and this is where protocols differ in terms of their final medium composition and use of monoculture versus co-culture to induce microglial maturation (19–28). Some protocols have been more widely adopted and implemented within independent labs and used at scale by companies, making them highly reproducible [see (19, 26, 29) and (25) adopted by (27, 30)].
Within hours of isolation from the brain, both human and rodent primary microglia downregulate several key mature microglial markers, particularly TMEM119, P2RY12, and SALL1 (2, 31), which can be restored upon re-transplantation into the brains of microglia-deficient mice (32). Likewise, hiPSC-microglia assume the closest identity to in vivo microglia when they are transplanted into rodent brains (9, 19, 33–35). However, this is clearly not practical for high-throughput experiments, and is still limited by the xenogenic environment. Therefore, further improving in vitro hiPSC-microglia models to better represent their in vivo counterparts remains paramount. The pros and cons of different hiPSC-microglia models, and their application for disease modeling, have been reviewed recently (18, 36–39), but how to improve the physiological relevance of these models is an under-covered area. In this mini review, we focus on how to better mimic the CNS habitat, with relevant media composition, extracellular matrix, and co-culture, which will further improve the physiological applications and reproducibility of existing in vitro hiPSC-microglia protocols.
Improving hiPSC-Microglia Media Composition
To better understand, compare, and control hiPSC-microglia, we need fully defined, open-source media, which not only supply relevant survival/differentiation factors, but also reflect the composition of the brain interstitial fluid, mimicking ionic composition, energy-substrates, nutrients, minerals, vitamins, pH, and osmolarity. However, many conventional media compositions are not at all physiological, but rather a hangover from culturing cancer cell lines with high metabolic and proliferative rates. Firstly, media with high levels of glucose (up to 25 mM, versus physiological levels of 5 mM), can mask cellular phenotypes (40, 41), which is problematic, as microglial metabolic dysregulation is associated with disease phenotype (42). Secondly, while serum is a major source of nutrients, and its use made cell-culture of microglia possible in the first place (43), it is still often used for in vitro culture of glial cells, despite the fact that glia are not exposed to serum under normal conditions in vivo, as they are separated by the blood-brain barrier [reviewed by (44)]. Further, each serum batch is different, its composition is undefined, and it contains lipopolysaccharide (LPS)-binding proteins which aid binding to toll-like receptor 4 (TLR4), thereby enhancing microglial responses to LPS (45). Thirdly, a hangover from explant cultures are components which contain immunosuppressive molecules, notably B27 supplement, developed to prevent excessive cell death and reactivity, but which interfere with microglia responses (46).
Defined, more physiological media now exist [e.g. BrainPhys (47)], and serum-free conditions have been demonstrated to be viable for rodent microglia with the recently developed “TIC media.” TIC supplements DMEM/F12 with astrocyte-derived factors TGF-β, IL-34, and cholesterol (32). TGF-β promotes maturation and specialization of microglia within the developing brain, helps maintain the identity of cultured microglia [both when isolated as primary cells (2, 31) or derived from human iPSCs (19)], and encourages an anti-inflammatory quiescent microglia phenotype (48). IL-34 is the main ligand in the brain for Colony Stimulating Factor 1 receptor (CSF-1R), signaling through which is essential for microglial survival (1, 49). Cholesterol further improved the benefit conveyed by IL-34/TGF-β, despite myeloid cells being capable of de novo cholesterol synthesis [(50) reviewed by (51)]. However, despite TIC factors enabling survival, serum was still required to initiate microglial phagocytic activity (32). Overall, whilst the field is clearly moving towards defined, physiological media for hiPSC-microglia, serum confounds this, and use of immunomodulatory additives needs to be noted when comparing inflammatory responses across studies.
Improving hiPSC-Microglia Through Scaffolded 3D Extracellular Environment
Microglia in vivo are embedded in a three-dimensional network of macromolecules derived from both neuronal and glial cells, mainly glycosaminoglycans (e.g. hyaluronic acid), proteoglycans, glycoproteins, and low levels of fibrous proteins (including collagen, laminin, fibronectin, and vitronectin), creating a rather “soft,” viscoelastic brain environment of ~3 kPa (52, 53). Unfortunately, these basic features are mostly absent in conventional in vitro conditions. Mimicking this 3D extracellular environment should enable hiPSC-microglia to adopt a more authentic phenotype. This is particularly important as immune cells are especially prone to develop unwanted and inflammatory responses to a suboptimal environment, such as covalently cross-linked (purely elastic) synthetic matrices, which lack viscoelastic properties [(54), reviewed by (55)].
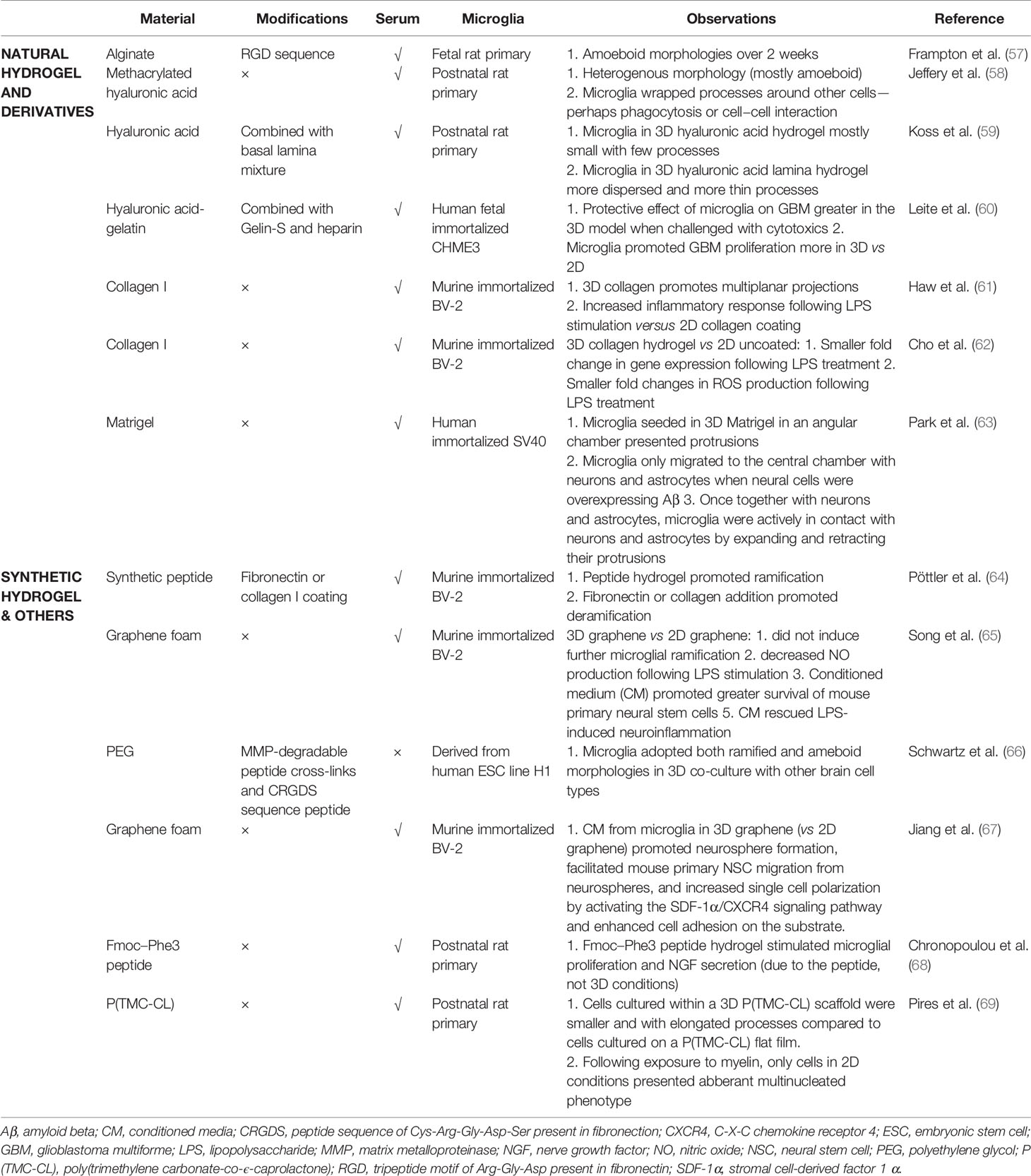
3D cell culture, both scaffolded and scaffold-free, is becoming more widely used, as it preserves natural cell shape, supports cell-to-cell and cell-to-matrix communication, enhances cell differentiation, and pushes gene and protein expression towards that found in vivo [reviewed by (56)]. Surprisingly, only a few groups have cultured microglial cells in a 3D scaffolded environment (Table 1), and several of them primarily focus on other cell types co-cultured with microglia, including neurons (70) and glioblastoma cells (71, 72). To properly distinguish the effects attributable to 3D conditions, systematic studies using relevant 2D controls are required for appropriate comparisons to be made.
Scaffolded 3D culture influences microglial morphology and attachment, but the outcome depends on the scaffold composition and structure. Choosing a material that optimizes microglial phenotype still remains a challenge. Hydrogels of synthetic peptide (68), collagen (61, 64), or PEG (66) enhanced ramification in both postnatal rat microglia and immortalized murine BV-2 cells. Likewise, primary rat microglia cultured within a 3D fibrous poly(trimethylene carbonate-co-ϵ-caprolactone) scaffold had smaller cells with elongated processes versus cells attached to flat solvent-cast films (69). However, this was not observed in 3D graphene scaffolds, where BV-2 cells remained mostly amoeboid (65). Some peptides, like neural cell adhesion molecule (NCAM)-derived KHIFSDDSSE, favor neuron and astrocyte attachment but not microglial attachment (73). Functionalizing hydrogels with extracellular matrix (ECM) components have provided mixed results; a basal lamina mixture prompted primary postnatal rat microglia within a glial co-culture to disperse throughout the hyaluronic acid-based scaffold (59), whereas coating a collagen hydrogel with fibronectin and laminin decreased ramification in BV-2 cells (64).
3D culture can also affect microglial behavior and interaction with other cell types. Neuronal survival in conditioned media from 3D-cultured BV-2 cells was higher than from 2D-cultured cells, and LPS-induced BV-2-mediated neuronal toxicity was also reduced as a result of 3D microglial culture (65). Meanwhile, more BV-2 cells upregulated CD40 (induced through NF-κB and STAT-1α) in response to LPS when in 3D collagen than in 2D (61), suggesting a more homogenous inflammatory response in 3D (74). Although BV-2 and other immortalized cell lines have been a valuable resource for studying microglia, it is likely that microglial responses to a 3D environment might be masked or even reversed by the aberrantly proliferative and primed state of such cells (75, 76). Nonetheless, 3D culture of microglia in specific scaffolds seems to promote a production of growth factors and an anti-inflammatory phenotype beneficial to other cell types.
Improving hiPSC-Microglia Through Co-Culture
The simplest approach to study cross-talk between microglia and neurons or astrocytes, is by transferring medium conditioned by these cells cultured separately. This provides a high level of experimental control, as the conditioned media can be analyzed independently to identify cytokines produced by stimulated microglia (77), assess the impact of individual factors on survival (32), or interrogate the process studied using drugs. However, conditioned media only allows study of uni-directional signaling. Transwells or Boyden chambers allow different cell types to exchange secreted factors bi-directionally [reviewed by (78)]. This improves physiological relevance but offers less insight into which cell type the secreted signals originated from, or control over which cells are targeted with any manipulation or drug given. Furthermore, these culture methods are also limited by the effective concentration of secreted signals, being much lower compared to local concentrations if cells were in physical contact.
To fully capture the physiology of cellular interactions, physical contact-mediated cues, through receptor-ligand interactions, are required. Microglial quiescence is maintained through neuronal CD200 glycoprotein interaction with the microglial CD200 receptor [(79) reviewed by (80)], as well as neuronal transmembrane (but possibly also secreted) CD22 interaction with microglial CD45 receptors (81). The mainly neuronal chemokine CX3CL1 (“fractalkine”) maintains microglia in a surveying state when the membrane-bound form makes contact with the microglial CX3CR1 receptors, whereas soluble CX3CL1 has a chemoattractant effect on microglia [(82), reviewed by (83)]. Furthermore, the idea that electrical activity from neurons might play a role in immune-response suppression has been around for two decades (84), but the nature of this interaction is not well understood. Generating physical-contact co-cultures would evidently enable microglia to experience all modes of interaction with other cell types, yet when increasing the physiological relevance of the culture model, the degree of control over the experimental system decreases, highlighting the importance of choosing the appropriate culture-model for each research question (Figure 1).
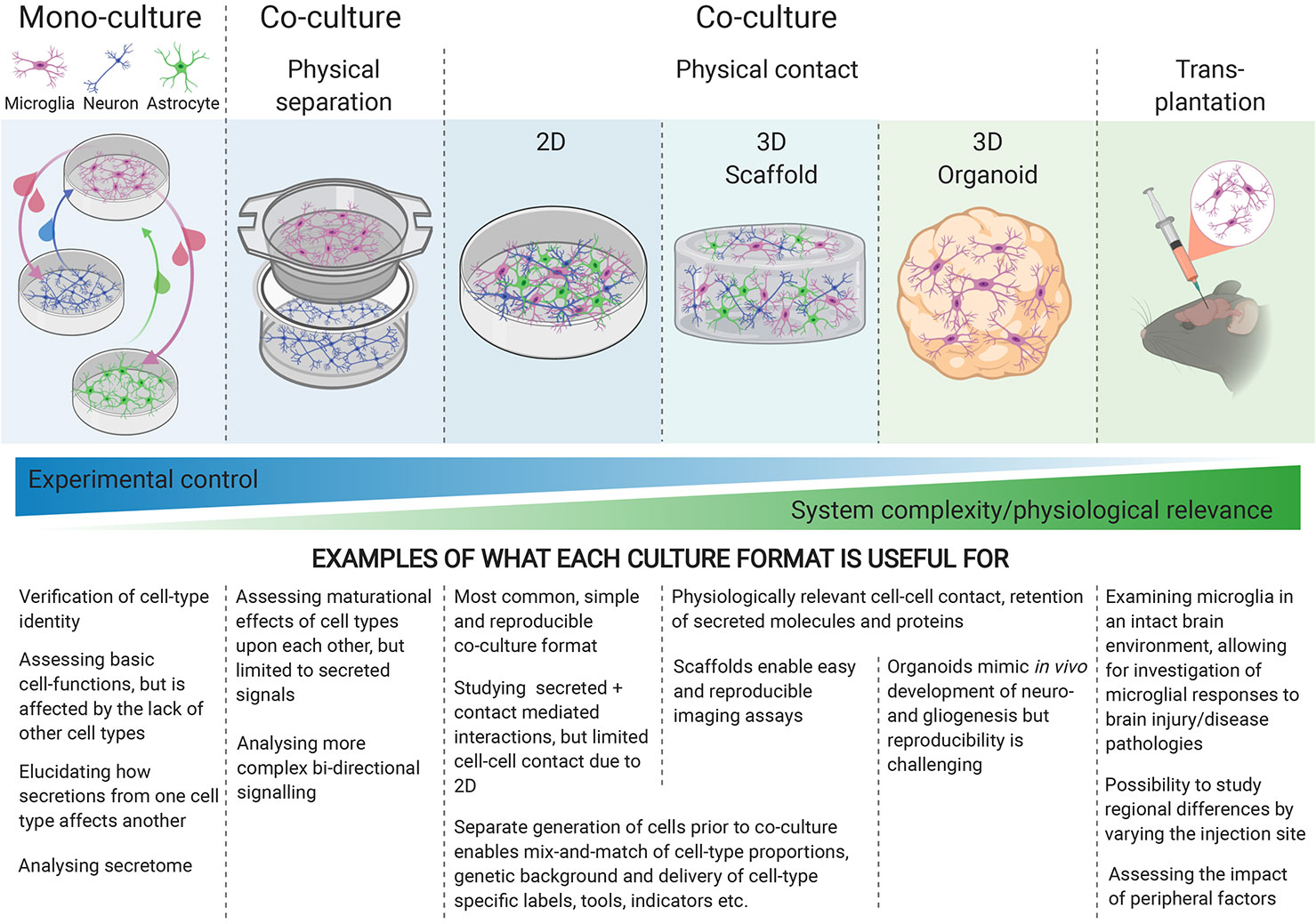
Figure 1 Different culture environments convey different levels of experimental control and physiological relevance, with mono-cultures at one end of the spectrum, and transplantation into intact brains at the other. The experimental question being investigated should guide the choice of culture-system.
Most hiPSC-microglia studies have focused on characterizing the gene expression and functionality of the resulting microglia in monoculture [reviewed by (8, 36)], but a few have included co-cultures with neurons (19, 24, 27) in 2D (3D co-cultures will be discussed later). Physical contact with neurons increases the expression of mature microglial genes, enhances ramified morphology compared to conditioned media, as well as eliciting a dampened response to LPS + IFNγ, high microglial motility and extension of processes to sites of injury (19, 24, 27), reflecting the homeostatic surveillance and response to injury associated with microglia in vivo (85). The exact cues responsible for these phenotypes are yet to be elucidated. Our understanding of interactions between microglia and astrocytes is limited, these cells typically being examined in isolation, but interest in their complex relationship, in terms of maintaining homeostasis in health and reactive cross-talk in disease, is growing [reviewed by (86)]. Pandya et al. used astrocytes as a feeder layer for maturing their hiPSC-microglia for 1–2 weeks before isolating the CD39+ microglia for monoculture (23), but interactions between mature hiPSC-microglia and astrocytes are otherwise currently under-explored.
The maturity and functionality of the non-microglial co-cultured cells is important to consider, as crucial microglial functions like synaptic pruning are likely only observable if the synaptic network is sufficiently mature, which may depend on more than bi-culture. Tri-culture of microglia with neurons and astrocytes will enable further maturation, more fully capture additional aspects of cross-talk and identify potential compensatory mechanisms between different brain cell types. Microglia and astrocytes operate in concert, both possessing that “double-edged sword” capacity to either support neuronal recovery (simplistically, “helpful” M2 and A2 phenotypes) or cause neuronal death (“harmful” M1 and A1 reactive phenotypes) [(77) reviewed by [87)]. A primary rodent tri-culture system of neurons, astrocytes and microglia in serum-free conditions (using TIC-supplemented media) shows promise in modeling different neuroinflammatory scenarios (88), in line with both in vivo and in vitro studies (77, 89, 90). However, TIC factors could not be omitted in the tri-cultures (88), indicating that endogenous secretion from the cultured astrocytes was not sufficient to retain a viable and physiologically active microglial phenotype. Interestingly, co-culture with hiPSC-neurons and astrocytes is capable of shifting the transcriptional profile of hiPSC-microglia further towards an ex vivo state than TIC media, although the gap between in vitro and ex vivo microglia is not fully closed (29).
Oligodendrocytes, endothelial cells, and/or vasculature are also likely to affect microglial function, yet they are rarely included in in vitro culture models, or assessment of their impact on microglia is lacking (66). Excitingly though, with the advent of single-cell RNAseq and single-cell proteomics, analyzing complex co-culture systems has become more straightforward and informative. These high-throughput methods allow for detection of overall cellular changes and activation/inactivation of biological pathways within each cell type, providing important clues to understand inter-cellular cross-talk. Using hiPSC, it is now possible to “mix and match” the genetic background of each cell type within the co-culture, such that the effect of a mutation can be studied in one cell type at a time. Strategies for co-culturing multiple hiPSC cell-types are increasingly popular and are beginning to move towards three dimensional environments as well.
Combining Co-Culture and 3D Environment for hiSC-Microglia
Providing microglia with a more physiologically relevant cellular environment relies on the principle of letting separately derived microglia populate pre-existing 3D neuronal cultures, brain organoids, or xenotransplantation into rodent brains [reviewed by (18)]. 3D scaffolded cultures confer a high degree of reproducibility and experimental flexibility, as individual cell types can be derived separately and added to the scaffold one at a time. This enables mix and match of co-cultures, i.e. cell types, ratios, genetic backgrounds, genetic tools, and fluorescent reporters. A 3D scaffold culture with microfluidic chambers has demonstrated that immortalized human SV40-derived microglia display ramified morphologies when seeded within Matrigel, migrate towards Amyloid-beta (Aβ)-expressing neurons, and cleave their axons while producing neurotoxic mediators (63). This encouraging study could be made more physiologically relevant by replacing the genetically aberrant SV40-microglial cells with hiPSC-derived microglia, and mouse sarcoma-derived Matrigel with a defined hydrogel more reminiscent of brain ECM.
3D organoids preserve the human model context, yet high variability between organoid batches remains a technical challenge. Remarkably, cerebral organoids spontaneously containing microglia have been obtained (91), but this is only apparent when mesoderm is not sufficiently suppressed. More conventionally, organoids and microglia are derived separately, due to the differing ontogeny between ectodermally derived CNS cells and mesodermally derived microglia. Integrated microglia adopt ramified morphologies, display migration to the site of injury (19, 20, 28) and phagocytose Aβ (92).
Xenotransplantation offers the benefit of transplanting hiPSC-derived microglia into a more mature and structurally developed environment. However, immune-deficiency and expression of human CSF1 or IL34 are crucial for survival of the transplanted human microglia (33, 93), and ablation of host microglia is usually preferable. Excitingly, several recent xenotransplantation studies have detected improved hiPSC-microglial morphologies, with transcriptional profiles showing higher degrees of similarity between xenotransplanted microglia and freshly isolated human microglia than with cultured primary microglia (9, 33, 34). Meanwhile, the pattern of differentially expressed genes revealed a more muted response to LPS in transplanted cells than in in vitro cultured microglia (33). Engrafted microglia associate with Aβ plaques, appear to phagocytose them (19, 33), and upregulate disease-associated microglia markers, including CD9, MERTK, and TREM2 (33). Xenotransplantation is therefore a viable option for studying hiPSC-derived human microglia allowed to mature in an in vivo brain environment, where the issue of altered in vitro gene expression (31, 32) appears to have been resolved, but making this technique amenable to large scale-up will be extremely challenging.
Discussion
Strides towards more physiological hiPSC-microglia culture conditions are being made apace, including the use of defined serum-free media in monoculture, the implementation of co-cultures, whether in 2D format, organoids, or xenotransplantation. One important underdeveloped gap in the repertoire is an in vitro model containing multiple hiPSC types embedded in a defined 3D scaffold, which would both enhance microglial identity and be amenable to advanced approaches, including arrayed drug screening, genome-wide CRISPR screens, high content imaging, cell multiplexing, and single cell transcriptomics. Further work is necessary to identify viscoelastic scaffold materials and ECM components that optimize microglial morphology, adhesion, and function. The process of neuroinflammation is arguably best modeled with all participating cell types present. Loss of function in astrocytes and/or microglia, impacting homeostatic support, or a toxic gain of function in the same cells, leading to the release of neurotoxic factors, would both probably have detrimental effects on neuronal health. Both scenarios are likely occurring in tandem in various neurodegenerative diseases, as well as in normal ageing, where defective phagocytosis as well as pro-inflammatory profiles have recently been reported (94, 95). Many links in the harmful inflammatory cascades that have been demonstrated to originate with microglia, propagate to astrocytes, and ultimately cause neurodegeneration, are not fully elucidated [(77), reviewed by (87)], and there is much more to learn about cross-talk and compensatory mechanisms between these cell types. Ultimately, the trade-off between the physiological relevance of a culture-system and experimental control over conditions must be decided based on the scientific question asked. Various hiPSC-microglia models will continue to be relevant, but the more defined and comparable we can make them, the better we will be able to understand and control the double-edged sword that is microglial function.
Author Contributions
Conceptualization: SS, AH, and SC. Writing—Original Draft: SS and AH. Writing—Review and Editing: SS, AH, WJ, and SC. Funding Acquisition: WJ and SC. Supervision: WJ and SC. All authors contributed to the article and approved the submitted version.
Funding
We thank the following for financial support: The Oxford Martin School for core support to the James Martin Stem Cell Facility (LC0910-004; SC); BBSRC industrial CASE studentship (BB/M011224/1; SS); Eli Lilly and Company (AH, SS). The authors declare that this study received funding from Eli Lilly and Company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The figure was created with BioRender.com.
References
1. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science (80) (2010) 330:841–5. doi: 10.1126/science.1194637
2. Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci (2014) 17:131–43. doi: 10.1038/nn.3599
3. Swinnen N, Smolders S, Avila A, Notelaers K, Paesen R, Ameloot M, et al. Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia (2013) 61:150–63. doi: 10.1002/glia.22421
4. Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science (80) (2011) 333:1456–8. doi: 10.1126/science.1202529
5. Sakai J. Core Concept: How synaptic pruning shapes neural wiring during development and, possibly, in disease. Proc Natl Acad Sci USA (2020) 117:16096–9. doi: 10.1073/pnas.2010281117
6. Matcovitch-Natan O, Winter DR, Giladi A, Aguilar SV, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science (2016) 353(6301):aad8670. doi: 10.1126/science.aad8670
7. Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol (2018) 18:225–42. doi: 10.1038/nri.2017.125
8. Sabogal-Guáqueta AM, Marmolejo-Garza A, de Pádua VP, Eggen B, Boddeke E, Dolga AM. Microglia alterations in neurodegenerative diseases and their modeling with human induced pluripotent stem cell and other platforms. Prog Neurobiol (2020) 190:101805. doi: 10.1016/j.pneurobio.2020.101805
9. Mancuso R, Van Den Daele J, Fattorelli N, Wolfs L, Balusu S, Burton O, et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat Neurosci (2019) 22:2111–6. doi: 10.1038/s41593-019-0525-x
10. Dubbelaar ML, Kracht L, Eggen BJL, Boddeke EWGM. The Kaleidoscope of Microglial Phenotypes. Front Immunol (2018) 9:1753. doi: 10.3389/fimmu.2018.01753
11. Sayed N, Liu C, Wu JC. Translation of Human-Induced Pluripotent Stem Cells from Clinical Trial in a Dish to Precision Medicine. J Am Coll Cardiol (2016) 67:2161–76. doi: 10.1016/j.jacc.2016.01.083
12. Dawson TM, Golde TE, Lagier-Tourenne C. Animal models of neurodegenerative diseases. Nat Neurosci (2018) 21:1370–9. doi: 10.1038/s41593-018-0236-8
13. Rehbach K, Zhang H, Das D, Abdollahi S, Prorok T, Ghosh S, et al. Publicly available hiPSC lines with extreme polygenic risk scores for modeling schizophrenia. bioRxiv (2020). doi: 10.1101/2020.07.04.185348
14. Bennett ML, Bennett FC. The influence of environment and origin on brain resident macrophages and implications for therapy. Nat Neurosci (2020) 23:157–66. doi: 10.1038/s41593-019-0545-6
15. Bian Z, Gong Y, Huang T, Lee CZW, Bian L, Bai Z, et al. Deciphering human macrophage development at single-cell resolution. Nature (2020) 582:571–6. doi: 10.1038/s41586-020-2316-7
16. Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, et al. Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Rep (2017) 18:391–405. doi: 10.1016/j.celrep.2016.12.041
17. Buchrieser J, James W, Moore MD. Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages. Stem Cell Rep (2017) 8:334–45. doi: 10.1016/j.stemcr.2016.12.020
18. Hasselmann J, Blurton-Jones M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia (2020) 68:721–39. doi: 10.1002/glia.23781
19. Abud EM, Ramirez RN, Martinez ES, Healy LM, Cecilia HH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron (2017) 94:278–93. doi: 10.1016/j.neuron.2017.03.042.iPSC-derived
20. Brownjohn PW, Smith J, Solanki R, Lohmann E, Houlden H, Hardy J, et al. Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem Cell Rep (2018) 10:1294–307. doi: 10.1016/j.stemcr.2018.03.003
21. Claes C, Van Den Daele J, Boon R, Schouteden S, Colombo A, Monasor LS, et al. Human stem cell–derived monocytes and microglia-like cells reveal impaired amyloid plaque clearance upon heterozygous or homozygous loss of TREM2. Alzheimer’s Dement (2019) 15:453–64. doi: 10.1016/j.jalz.2018.09.006
22. Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep (2017) 8:1516–24. doi: 10.1016/j.stemcr.2017.04.023
23. Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci (2017) 20:753–9. doi: 10.1038/nn.4534
24. Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, et al. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity (2017) 47:183–98.e6. doi: 10.1016/j.immuni.2017.06.017
25. Garcia-reitboeck P, Phillips A, Piers TM, Houlden H, Hardy J, Pocock JM, et al. Human Induced Pluripotent Stem Cell-Derived Mutations Show Specific Deficits in Phagocytosis Article Human Induced Pluripotent Stem Cell-Derived Microglia-Like Cells Harboring TREM2 Missense Mutations Show Specific Deficits in Phagocytosis. CellReports (2018) 24:2300–11. doi: 10.1016/j.celrep.2018.07.094
26. McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, Blurton-Jones M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener (2018) 13:1–13. doi: 10.1186/s13024-018-0297-x
27. Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Rep (2017) 8:1727–42. doi: 10.1016/j.stemcr.2017.05.017
28. Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med (2016) 22:1358–67. doi: 10.1038/nm.4189
29. Grubman A, Vandekolk TH, Schröder J, Sun G, Hatwell-Humble J, Chan J, et al. A CX3CR1 Reporter hESC Line Facilitates Integrative Analysis of In-Vitro-Derived Microglia and Improved Microglia Identity upon Neuron-Glia Co-culture. Stem Cell Rep (2020) 14:1018–32. doi: 10.1016/j.stemcr.2020.04.007
30. Gutbier S, Wanke F, Dahm N, Rümmelin A, Zimmermann S, Christensen K, et al. Large-scale production of human IPSC-derived macrophages for drug screening. Int J Mol Sci (2020) 21:1–23. doi: 10.3390/ijms21134808
31. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science (80) (2017) 356:1248–59. doi: 10.1126/science.aal3222
32. Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron (2017) 94:759–73. doi: 10.1016/j.neuron.2017.04.043
33. Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, et al. Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron (2019) 103:1016–33.e10. doi: 10.1016/j.neuron.2019.07.002
34. Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, et al. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc Natl Acad Sci USA (2019) 116:25293–303. doi: 10.1073/pnas.1913541116
35. Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, et al. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat Commun (2020) 11:1–16. doi: 10.1038/s41467-020-15411-9
36. Haenseler W, Rajendran L. Concise Review: Modeling Neurodegenerative Diseases with Human Pluripotent Stem Cell-Derived Microglia. Stem Cells (2019) 37:724–30. doi: 10.1002/stem.2995
37. Miron VE, Priller J. Investigating Microglia in Health and Disease : Challenges and Opportunities. Trends Immunol (2020) 41(9):785–93. doi: 10.1016/j.it.2020.07.002
38. Speicher AM, Wiendl H, Meuth SG, Pawlowski M. Generating microglia from human pluripotent stem cells: Novel in vitro models for the study of neurodegeneration. Mol Neurodegener (2019) 14:1–16. doi: 10.1186/s13024-019-0347-z
39. Hanger B, Couch A, Rajendran L, Srivastava DP, Vernon AC. Emerging Developments in Human Induced Pluripotent Stem Cell-Derived Microglia: Implications for Modelling Psychiatric Disorders With a Neurodevelopmental Origin. Front Psychiatry (2020) 11:789. doi: 10.3389/fpsyt.2020.00789
40. Kleman AM, Yuan JY, Aja S, Ronnett GV, Landree LE. Physiological glucose is critical for optimized neuronal viability and AMPK responsiveness in vitro. J Neurosci Methods (2008) 167:292–301. doi: 10.1016/j.jneumeth.2007.08.028
41. Rocktäschel P, Sen A, Cader MZ. High glucose concentrations mask cellular phenotypes in a stem cell model of tuberous sclerosis complex. Epilepsy Behav (2019) 101:106581. doi: 10.1016/j.yebeh.2019.106581
42. Andreone BJ, Przybyla L, Llapashtica C, Rana A, Davis SS, van Lengerich B, et al. Alzheimer’s-associated PLCγ2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nat Neurosci (2020) 23:927–38. doi: 10.1038/s41593-020-0650-6
43. Giulian D, Baker TJ. Characterization of ameboid microglia isolated from developing mammalian brain. J Neurosci (1986) 6:2163–78. doi: 10.1523/jneurosci.06-08-02163.1986
44. Witting A, Möller T. Microglia Cell Culture: A Primer for the Novice. In: In Vitro Neurotoxicology. Totowa, NJ: Springer Protocols by Humana Press (2011). p. 49–66. doi: 10.1007/978-1-61779-170-3
45. Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, et al. Structure and Function of Lipopolysaccharide Binding Protein. Science (80) (1990) 249:1429–32. doi: 10.1126/science.2402637
46. Liu JJ, Mustafa S, Barratt DT, Hutchinson MR. Corticosterone preexposure increases NF-κB translocation and sensitizes IL-1β responses in BV2 microglia-like cells. Front Immunol (2018) 9:3. doi: 10.3389/fimmu.2018.00003
47. Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci USA (2015) 112(25):2725–34. doi: 10.1073/pnas.1504393112
48. Abutbul S, Shapiro J, Szaingurten-Solodkin I, Levy N, Carmy Y, Baron R, et al. TGF-β signaling through SMAD2/3 induces the quiescent microglial phenotype within the CNS environment. Glia (2012) 60:1160–71. doi: 10.1002/glia.22343
49. Elmore MRP, Najafi AR, Koike MA, Nazih N, Spangenberg EE, Rice RA, et al. CSF1 receptor signaling is necessary for microglia viability, which unmasks a cell that rapidly repopulates the microglia- depleted adult brain. Neuron (2014) 82:380–97. doi: 10.1016/j.neuron.2014.02.040.CSF1
50. Nieweg K, Schaller H, Pfrieger FW. Marked differences in cholesterol synthesis between neurons and glial cells from postnatal rats. J Neurochem (2009) 109:125–34. doi: 10.1111/j.1471-4159.2009.05917.x
51. Loving BA, Bruce KD. Lipid and Lipoprotein Metabolism in Microglia. Front Physiol (2020) 11:393. doi: 10.3389/fphys.2020.00393
52. McIlvain G, Schwarb H, Cohen NJ, Telzer EH, Johnson CL. Mechanical properties of the in vivo adolescent human brain. Dev Cognit Neurosci (2018) 34:27–33. doi: 10.1016/j.dcn.2018.06.001
53. Lam D, Enright HA, Cadena J, Peters SKG, Sales AP, Osburn JJ, et al. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci Rep (2019) 9:1–15. doi: 10.1038/s41598-019-40128-1
54. Sadtler K, Wolf MT, Ganguly S, Moad CA, Chung L, Majumdar S, et al. Divergent immune responses to synthetic and biological scaffolds. Biomaterials (2019) 192:405–15. doi: 10.1016/j.biomaterials.2018.11.002
55. Chaudhuri O, Cooper-white J, Janmey PA, Mooney DJ, Shenoy VB. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature (2020) 584:535–46. doi: 10.1038/s41586-020-2612-2
56. Jensen C, Teng Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front Mol Biosci (2020) 7:33. doi: 10.3389/fmolb.2020.00033
57. Frampton JP, Hynd MR, Shuler ML, Shain W. Fabrication and optimization of alginate hydrogel constructs for use in 3D neural cell culture. Biomed Mater (2011) 6(1):015002. doi: 10.1088/1748-6041/6/1/015002
58. Jeffery AF, Churchward MA, Mushahwar VK, Todd KG, Elias AL. Hyaluronic acid-based 3D culture model for in vitro testing of electrode biocompatibility. Biomacromolecules (2014) 15(6):2157–65. doi: 10.1021/bm500318d
59. Koss KM, Churchward MA, Jeffery AF, Mushahwar VK, Elias AL, Todd KG. Improved 3D hydrogel cultures of primary glial cells for in vitro modelling of neuroinflammation. J Vis Exp (2017) 2017:e56615. doi: 10.3791/56615
60. Leite DM, Baskovic BZ, Civita P, Neto C, Gumbleton M, Pilkington GJ. A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. FASEB J (2020) 34(1):1710–27. doi: 10.1096/fj.201901858RR
61. Haw RTY, Tong CK, Yew A, Lee HC, Phillips JB, Vidyadaran S. A three-dimensional collagen construct to model lipopolysaccharide-induced activation of BV2 microglia. J Neuroinflammation (2014) 11:134. doi: 10.1186/1742-2094-11-134
62. Cho HJ, Verbridge SS, Davalos RV, Lee YW. Development of an in vitro 3D brain tissue model mimicking in vivo-like pro-inflammatory and pro-oxidative responses. Ann Biomed Eng (2017) 46:877–87. doi: 10.1007/s10439-018-2004-z
63. Park J, Wetzel I, Marriott I, Dréau D, D’Avanzo C, Kim DY, et al. Cho H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci (2018) 21:941–51. doi: 10.1038/s41593-018-0175-4
64. Pöttler M, Zierler S, Kerschbaum HH. An artificial three-dimensional matrix promotes ramification in the microglial cell-line, BV-2. Neurosci Lett (2006) 410:137–40. doi: 10.1016/j.neulet.2006.09.082
65. Song Q, Jiang Z, Li N, Liu P, Liu L, Tang M, et al. Anti-inflammatory effects of three-dimensional graphene foams cultured with microglial cells. Biomaterials (2014) 35:6930–40. doi: 10.1016/j.biomaterials.2014.05.002
66. Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Costa VS, et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci USA (2015) 112:12516–21. doi: 10.1073/pnas.1516645112
67. Jiang Z, Song Q, Tang M, Yang L, Cheng Y, Zhang M, et al. Enhanced migration of neural stem cells by microglia grown on a three- dimensional graphene scaffold ACS. Appl. Mater. Interfaces (2016) 8(38):25069–77. doi: 10.1021/acsami.6b06780
68. Chronopoulou L, Togna AR, Guarguaglini G, Masci G, Giammaruco F, Togna GI, et al. Self-assembling peptide hydrogels promote microglial cells proliferation and NGF production. Soft Matter (2012) 8:5784–90. doi: 10.1039/c2sm25528f
69. Pires LR, Rocha DN, Ambrosio L, Pêgo AP. The role of the surface on microglia function: Implications for central nervous system tissue engineering. J R Soc Interface (2015) 12:1–10. doi: 10.1098/rsif.2014.1224
70. Liu L, Koo Y, Akwitti C, Russell T, Gay E, Laskowitz DT, et al. Three-dimensional (3D) brain microphysiological system for organophosphates and neurochemical agent toxicity screening. PLoS One (2019) 14:e0224657. doi: 10.1371/journal.pone.0224657
71. Blazquez R, Pukrop T. 3D coculture model of the brain parenchyma–metastasis interface of brain metastasis. In: Methods in Molecular Biology. New York, NY: Springer Protocols by Humana Press Inc. (2017). p. 213–22. doi: 10.1007/978-1-4939-7021-6_16
72. Hermida MA, Kumar JD, Schwarz D, Laverty KG, Di Bartolo A, Ardron M, et al. Three dimensional in vitro models of cancer: Bioprinting multilineage glioblastoma models. Adv Biol Regul (2020) 75:100658. doi: 10.1016/j.jbior.2019.100658
73. Sridar S, Churchward MA, Mushahwar VK, Todd KG, Elias AL. Peptide modification of polyimide-insulated microwires: Towards improved biocompatibility through reduced glial scarring. Acta Biomater (2017) 60:154–66. doi: 10.1016/j.actbio.2017.07.026
74. Qin H, Wilson CA, Sun JL, Zhao X, Benveniste EN. LPS induces CD40 gene expression through the activation of NF-κB and STAT-1α in macrophages and microglia. Blood (2005) 106:3114–22. doi: 10.1182/blood-2005-02-0759
75. Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol (1990) 27:229–37. doi: 10.1016/0165-5728(90)90073-V
76. Amadio S, De Ninno A, Montilli C, Businaro L, Gerardino A, Volonté C. Plasticity of primary microglia on micropatterned geometries and spontaneous long-distance migration in microfluidic channels. BMC Neurosci (2013) 14:1–12. doi: 10.1186/1471-2202-14-121
77. Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature (2017) 541:481–7. doi: 10.1038/nature21029
78. Guttenplan KA, Liddelow SA. Astrocytes and microglia: Models and tools. J Exp Med (2019) 216:71–83. doi: 10.1084/jem.20180200
79. Wright GJ, Puklavec MJ, Willis AC, Hoek RM, Sedgwick JD, Brown MH, et al. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity (2000) 13:233–42. doi: 10.1016/S1074-7613(00)00023-6
80. Biber K, Neumann H, Inoue K, Boddeke HWGM. Neuronal “On” and “Off” signals control microglia. Trends Neurosci (2007) 30:596–602. doi: 10.1016/j.tins.2007.08.007
81. Mott RT, Ait-Ghezala G, Town T, Mori T, Vendrame M, Zeng J, et al. Neuronal expression of CD22: Novel mechanism for inhibiting microglial proinflammatory cytokine production. Glia (2004) 46:369–79. doi: 10.1002/glia.20009
82. Maciejewski-Lenoir D, Chen S, Feng L, Maki R, Bacon KB. Characterization of fractalkine in rat brain cells: migratory and activation signals for CX3CR-1-expressing microglia. J Immunol (1999) 163:1628–35.
83. Szepesi Z, Manouchehrian O, Bachiller S, Deierborg T. Bidirectional Microglia–Neuron Communication in Health and Disease. Front Cell Neurosci (2018) 12:323. doi: 10.3389/fncel.2018.00323
84. Neumann H, Wekerle H. Neuronal Control of the Immune Response in the Central Nervous System: Linking Brain Immunity to Neurodegeneration. J Neuropathol Exp Neurol (1998) 57:1–9. doi: 10.1097/00005072-199801000-00001
85. Nimmerjahn A, Kirchhoff F, Helmchen F. Neuroscience: Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science (80) (2005) 308:1314–8. doi: 10.1126/science.1110647
86. Liddelow SA, Marsh SE, Stevens B. Microglia and astrocyte interactions in health and disease: Dynamic Duo or Partners in Crime? Trends Immunol (2020) 41:1–16. doi: 10.1016/j.it.2020.07.006
87. Liddelow SA, Barres BA. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity (2017) 46:957–67. doi: 10.1016/J.IMMUNI.2017.06.006
88. Goshi N, Morgan RK, Lein PJ, Seker E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J Neuroinflammation (2020) 17:155. doi: 10.1186/s12974-020-01819-z
89. Nimmervoll B, White R, Yang JW, An S, Henn C, Sun JJ, et al. LPS-induced microglial secretion of TNFα increases activity-dependent neuronal apoptosis in the neonatal cerebral cortex. Cereb Cortex (2013) 23:1742–55. doi: 10.1093/cercor/bhs156
90. Wang X, Chen S, Ma G, Ye M, Lu G. Involvement of proinflammatory factors, apoptosis, caspase-3 activation and Ca2+ disturbance in microglia activation-mediated dopaminergic cell degeneration. Mech Ageing Dev (2005) 126:1241–54. doi: 10.1016/j.mad.2005.06.012
91. Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun (2018) 9:1–14. doi: 10.1038/s41467-018-06684-2
92. Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron (2018) 98:1141–54.e7. doi: 10.1016/j.neuron.2018.05.008
93. Mathews S, Branch Woods A, Katano I, Makarov E, Thomas MB, Gendelman HE, et al. Human Interleukin-34 facilitates microglia-like cell differentiation and persistent HIV-1 infection in humanized mice. Mol Neurodegener (2019) 14:12. doi: 10.1186/s13024-019-0311-y
94. Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci (2020) 23:194–208. doi: 10.1038/s41593-019-0566-1
Keywords: microglia, human, induced pluripotent stem cells, in vitro models, media composition, 3D scaffolds, co-culture, physiological relevance
Citation: Hedegaard A, Stodolak S, James WS and Cowley SA (2020) Honing the Double-Edged Sword: Improving Human iPSC-Microglia Models. Front. Immunol. 11:614972. doi: 10.3389/fimmu.2020.614972
Received: 07 October 2020; Accepted: 04 November 2020;
Published: 08 December 2020.
Edited by:
Amanda Sierra, Achucarro Basque Center for Neuroscience, SpainReviewed by:
Mathew Blurton-Jones, University of California, Irvine, United StatesSimona Lange, Roche, Switzerland
Copyright © 2020 Hedegaard, Stodolak, James and Cowley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sally A. Cowley, sally.cowley@path.ox.ac.uk
†These authors have contributed equally to this work
 Anne Hedegaard
Anne Hedegaard Szymon Stodolak
Szymon Stodolak William S. James
William S. James Sally A. Cowley
Sally A. Cowley