- 1Center for Precision Medicine, School of Medicine and School of Biomedical Sciences, Huaqiao University, Xiamen, China
- 2College of Materials Science and Engineering, Huaqiao University, Xiamen, China
RNA methylation is a reversible post-transcriptional modification to RNA and has a significant impact on numerous biological processes. N6-methyladenosine (m6A) is known as one of the most common types of eukaryotic mRNA methylation modifications, and exists in a wide variety of organisms, including viruses, yeast, plants, mice, and humans. Widespread and dynamic m6A methylation is identified in distinct developmental stages in the brain, and controls development of neural stem cells and their differentiation into neurons, glial cells such as oligodendrocytes and astrocytes. Here we summarize recent advances in our understanding of RNA methylation regulation in brain development, neurogenesis, gliogenesis, and its dysregulation in brain tumors. This review will highlight biological roles of RNA methylation in development and function of neurons and glial cells, and provide insights into brain tumor formation, and diagnostic and treatment strategies.
Introduction
N6-methyladenosine (m6A) is the most common and abundant methylation modification in RNA molecules present in eukaryotes (1, 2). More than 150 distinct chemical marks on cellular RNAs have been identified to date, and m6A modifications account for over 80% of all RNA methylations (3). High-throughput m6A sequencing studies have shown that thousands of mRNAs and non-coding RNAs are modified by m6A, which in turn affects gene expression, participates in animal development and pathogenesis of human diseases (4, 5). m6A is the most prevalent internal mRNA modification, with an average of one to three modifications per transcript, and potentially regulates every step in mRNA metabolism to some extent (6).
m6A methylation is catalyzed by an m6A methyltransferase complex (MTC) composed of methyltransferase-like 3 and 14 (METTL3 and METTL14) and their cofactors such as Wilms tumor 1-associated protein (WTAP), termed as “writer” (7–9). Removal of m6A is facilitated by Fat mass and obesity-associated (FTO) and AlkB homolog H5 (ALKBH5), two m6A demethylases that recognize distinct sets of target mRNAs, termed as “eraser” (10, 11). YTHDF1/2/3 and YTHDC1, members of the YT521-B homology (YTH) domain family proteins, are m6A direct “readers,” which affect translation, stability, and splicing of target mRNAs (12) (Figure 1). m6A modification has emerged as a multifaceted controller for gene expression regulation, mediated through its effector proteins—writers, readers, and erasers (6).
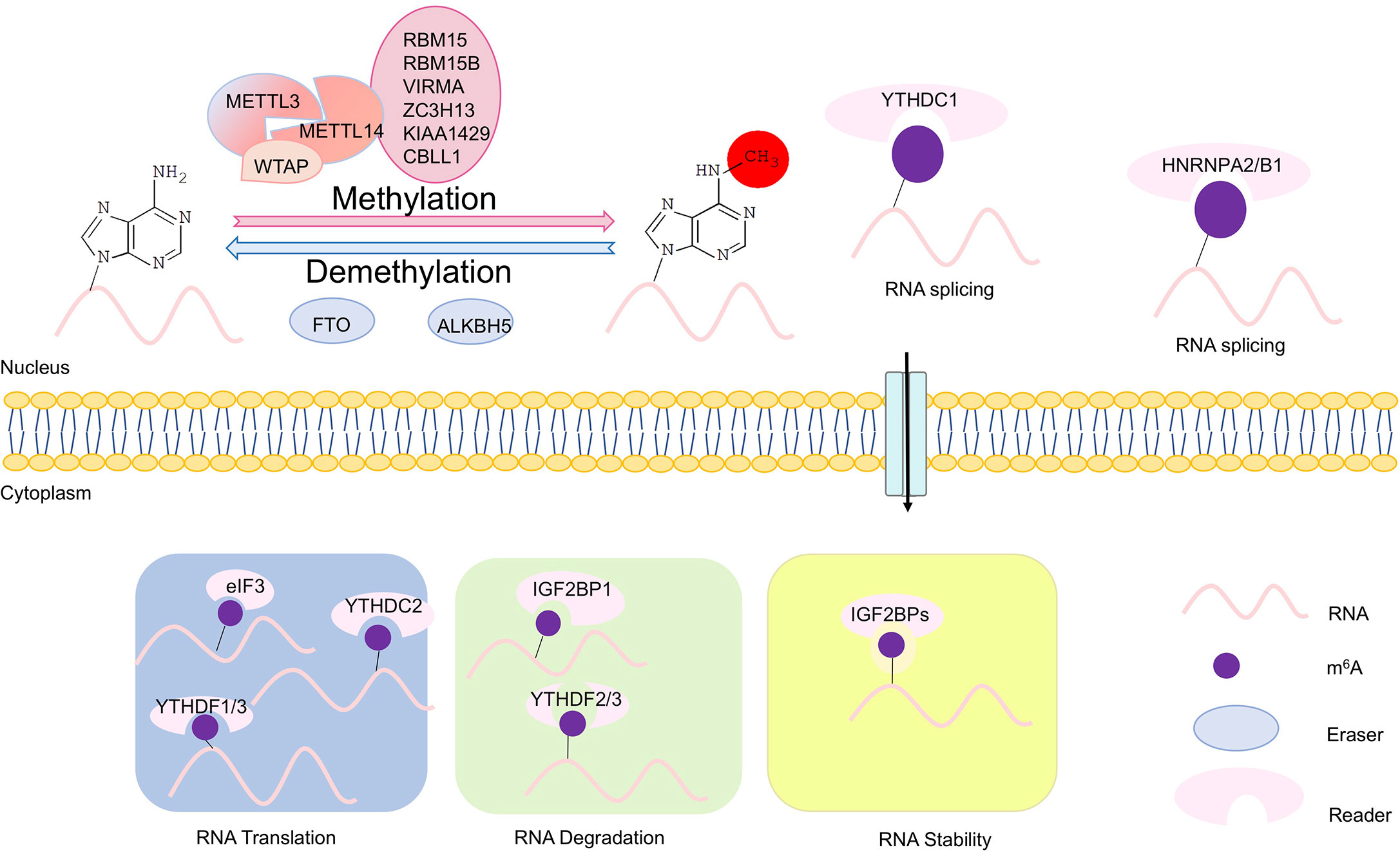
Figure 1 Scheme of m6A modifications. The writers, erasers, and readers of N6-methyladenosine (m6A). The m6A writer complex, which comprises the core methyltransferase-like protein 3 (METTL3) and its adaptors, is located in the nucleus. m6A demethylation is executed by two demethylases FTO and ALKBH5. The m6A erasers also are localized in the nucleus. In the nucleus, m6A can bind specific nuclear reader proteins such as YTHDC1 and HNRNPA2/B1, which may affect RNA splicing and mRNA export. Upon mRNA being exported to the cytoplasm, m6A binds to specific reader proteins, which affects stability, translation and/or localization of mRNAs. In the cytoplasm, translation of m6A modified mRNAs is mediated by the m6A readers YTHDF1, YTHDF3, and YTHDC2, the eukaryotic translation initiation factor eIF3, and METTL3. YTHDF2 and YTHDF3 regulate degradation of m6A modified mRNAs, while the insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) enhances stability m6A modified mRNAs.
m6A modifications in mRNAs or non-coding RNAs play important roles in virtually all types of bioprocesses including tissue development, self-renewal and differentiation of stem cells, heat shock response, circadian clock control, DNA damage response, and maternal-to-zygotic transition (8, 12). m6A is an important epitranscriptomic mark with high abundance in the central nervous system (CNS), and plays a crucial role in neural development and function (13). Dysregulation of m6A modifications also is associated with tumorigenesis of various cancers, such as gliomas (14).
In this review, we first summarize the recent advance in our understanding of biological functions and underlying molecule mechanisms of m6A regulation in neural development, with an emphasis in neurons and glial cells. We then highlight m6A regulatory roles in formation of brain tumors.
N6-methyladenosine (m6A) Modifications
As the most common and prevalent internal modification in eukaryotic mRNAs, m6A methylation has a significant impact on various physiological events (6, 15).
Components and Functions of m6A Modifications
Modification of m6A on mRNAs is post-transcriptionally installed, erased, and recognized by m6A methyltransferases, demethylases and m6A-specific binding proteins, respectively. Methyltransferases include METTL3/14, WTAP, RBM15/15B, and KIAA1429, also termed as “writers” (1, 7, 9) (Figure 1). METTL3 is the catalytic subunit, and METTL14 is an essential component to facilitate RNA binding (16). m6A methyltransferase is widely conserved among eukaryotic species that range from yeast, plants, and flies to mammals (17, 18). Demethylases consist of FTO and ALKBH5, termed as “erasers” (10, 11, 19). And m6A-specific binding proteins include YTHDF1/2/3 and IGF2BP1, termed as “readers” (20) (Figure 1).
In mammals, m6A is widely distributed in multiple tissues, with a higher expression in the liver, kidney, and brain than in other tissues (21). In the rodent brain, the global level of m6A is developmentally regulated, with expression peaking in the adult brain (22). Studies of m6A modifications have revealed m6A binding sites in over 25% of human transcripts, with enrichment in long exons, near stop codon and 3′ untranslated terminal region (3’-UTR) (2, 21, 22).
m6A modification in eukaryotic mRNAs exhibits substantial contributions to post-transcriptional gene expression regulation, and plays crucial and evolutionarily conserved roles in fundamental cellular processes such as meiosis and cell differentiation in yeast, plants, and mammals (18). m6A methyltransferase is crucial for yeast meiosis, differentiation of mouse embryonic stem cells, and viability of human cells (18, 23, 24). Depletion of the METTL3 homologs in yeast and flies leads to developmental arrest and defects in gametogenesis (18, 25). The m6A demethylase AlkBH5 deficient male mice are characterized by impaired fertility, resulting from apoptosis that affects meiotic metaphase-stage spermatocytes (19). Moreover, m6A modifications improve the stability of mRNAs and can control protein production (15, 26). For example, YTHDF1/2/3 exhibit 5- to 20-fold higher binding affinity for methylated RNAs compared to unmethylated RNAs (12). YTHDF1 and YTHDF3 bind m6A at the 3′ end of transcripts and increase their cap-dependent translation, possibly through a looping interaction with eukaryotic elongation factor 3 (eIF3) (15, 27). And the mRNA-binding protein IGF2BP1 enhances stability and translation of oncogenic mRNAs, including c-Myc, and in turn promotes cell proliferation and tumorigenesis (28).
m6A Modifications in mRNAs
m6A modification appears to directly affect biological activities of RNAs with unclear molecular mechanisms (Figure 1). m6A modification directly recruits m6A-specific proteins of the YTH domain family (29). These proteins contribute methyl-selective RNA binding with an amount of cellular processes, and produce m6A-dependent regulation of pre-mRNA processing, microRNA (miRNA) processing, translation initiation, and mRNA decay (5). Mature mRNAs with m6A methylation are regulated in the cytoplasm by the YTH family proteins. YTHDF1 is associated with initiating ribosomes, and delivers its target mRNAs for enhanced translation efficiency in HeLa cells (15). A second YTH family protein, YTHDF2, directly recruits the CCR4-NOT deadenylase complex and accelerates degradation of methylated transcripts (12, 30).
Moreover, some RNA transcripts exhibit increased half-lives upon m6A methylation. The well-established RNA stabilizer protein (HuR)/microRNA pathway mediates m6A-upregulated RNA stability (8). m6A modifications can assist protein binding either by destabilizing the helix around it, in turn allowing protein access, or by causing a conformation change to place m6A in a single-stranded context (31).
m6A Modifications in Translational Regulations
Interestingly, the methyltransferase complex may also function as a protein scaffold in RNA-processing and metabolism (19, 32). Translation regulation by m6A occurs during initiation and elongation. Sequences in the 5’-UTR of mRNAs are important for ribosome recruitment and translation initiation (33). m6A residues within 5’-UTR can act as an m6A-induced ribosome engagement site (MIRES), which promotes cap-independent translation of mRNAs (34). Moreover, eukaryotic elongation factor 3 family (eIF3a/b/h) can function as m6A readers, and physically interact with METTL3 to enhance translation by forming densely packed polyribosomes through recognizing m6A modifications at the 5’-UTR of mRNAs (35, 36). METTL3, independent of METTL14, is associated with chromatin and localized to the transcriptional start sites of active genes that have the CAATT-box binding protein CEBPZ present. Promoter-bound METTL3 can induce m6A modifications within the coding region of the associated mRNA transcript, and enhance its translation by relieving ribosome stalling (34, 37). In addition, METTL13-mediated methylation of eukaryotic elongation factor 1A (eEF1A) increases translation elongation and enhances protein synthesis to promote tumorigenesis (38).
Based on above biochemical and genetic evidence, m6A methylation plays a broad role in many aspects of bioprocesses by direct modifications on mRNAs, and through regulating RNA transcription and translation.
m6A Modifications in nervous system development
Proper development of the brain is critical for its function. Deficits in neural development have been implicated in many brain disorders. In the adult mouse brain, almost half of stably expressed RNAs are methylated, indicating important roles of m6A in brain development and function.
m6A Modifications in Development of the Cerebral Cortex
The cerebral cortex controls social interactions, decision-making, behavioral output, and other complex cognitive behaviors (39). In the developing cortex, m6A modifications are enriched in transcripts involved in neurogenesis and neuronal differentiation (40, 41). Studies have shown that Mettl14 deletion leads to a significant reduction of m6A levels in cortical mRNAs in vivo and in cultured cortical neural progenitors (40). Mettl14 deletion in the embryonic mouse brain causes prolonged cell cycle in cortical radial glia cells (RGCs), results in delayed neurogenesis and gliogenesis (40) (Figure 2).
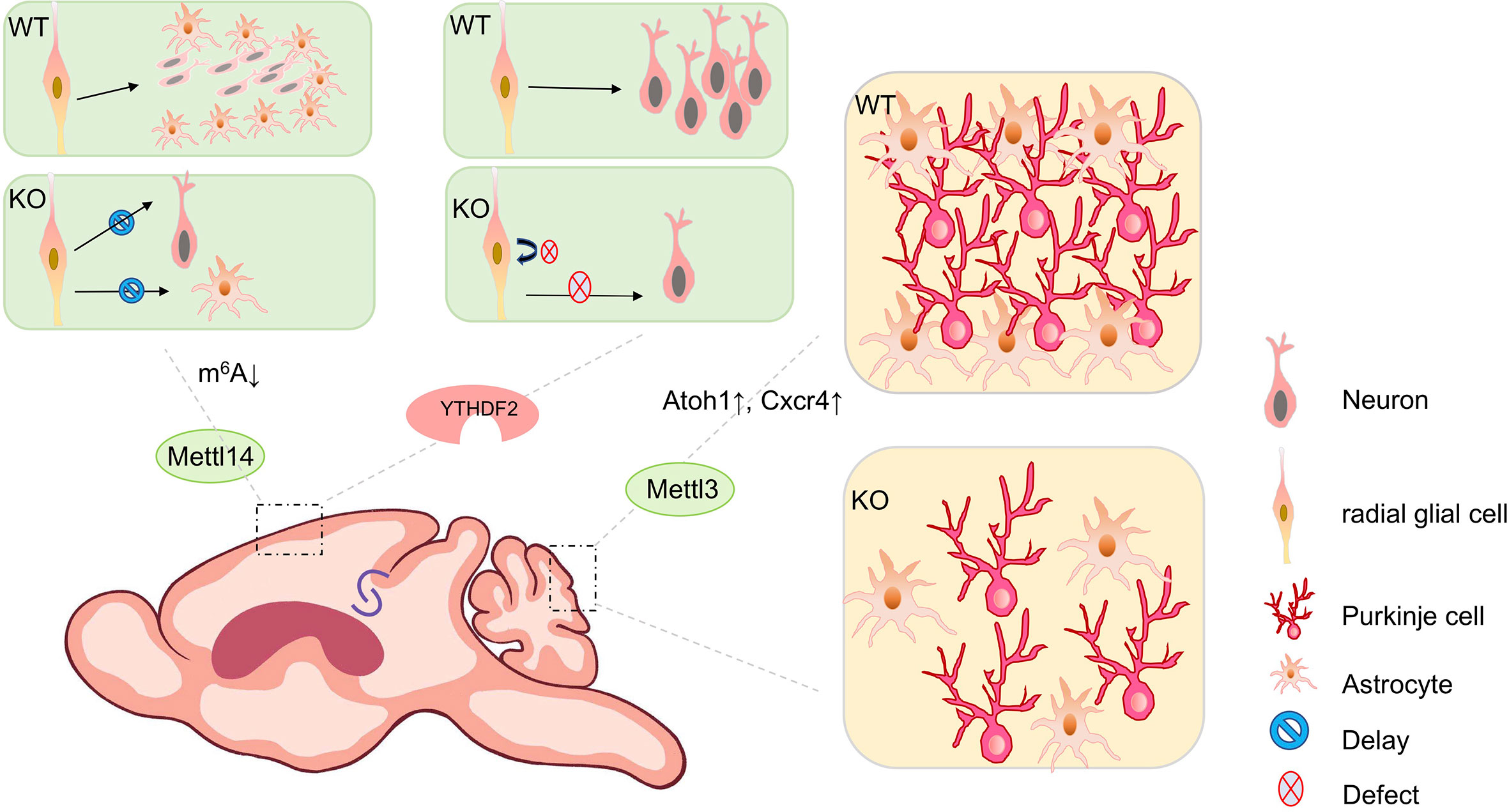
Figure 2 The biological impact of m6A modifications in mouse brain development. m6A modifications play a key role throughout brain development during mouse embryonic and postnatal stages. Mettl14 conditional knockout mouse (cKO) in the mouse embryonic brain causes prolonged cell cycle in cortical radial glial cells, results in delayed neurogenesis and gliogenesis, compared to wild type (WT) mice. Conditional depletion of Ythdf2 in mice causes decreased self-renewal of neural stem/progenitor cells (NSPCs) and defects in spatiotemporal generation of neurons in the embryonic cortex. Knocking out Mettl3 in the mouse embryonic brain causes cerebellar hypoplasia. Ectopic expression of Mettl3 leads to a disorganized laminal structure of both Purkinje cells and glial cells. Key developmental genes such as Atoh1 and Cxcr4 are abnormally upregulated due to the extended mRNA half-lives induced by m6A depletion.
Moreover, Fto knockout mice show a significant increase of m6A levels in transcripts of hippocampus (42). Altered expression of genes with m6A modifications contributes to impaired adult neurogenesis (42, 43). In addition, conditional depletion of Ythdf2 in mice causes decreased self-renewal of neural stem/progenitor cells (NSPCs) and defects in spatiotemporal generation of neurons in the embryonic cortex (44). Ythdf1 knockout mice exhibit impaired hippocampal synaptic transmission and long-term potentiation (13). Ythdf1 re-expression in hippocampus in adult Ythdf1 knockout mice rescues behavioral and synaptic defects, while hippocampus-specific acute knockdown of Ythdf1 or Mettl3 recapitulates the hippocampal deficiency (13) (Figure 2).
m6A Modifications in Cerebellar Development
Studies have shown that m6A levels are higher in the cerebellum than in the cerebral cortex, and a substantial number of cerebellar RNAs exhibits developmentally regulated methylation (45). m6A writers (METTL3, METTL14, and WTAP) and erasers (ALKBH5 and FTO) are highly expressed at the early stage of cerebellar development by postnatal day 7 (P7), and show a gradual reduction towards the maturation of cerebellar neurons by P60 (45). From P7 to P60, numbers of temporal-specific m6A peaks in start codon regions of RNA transcripts are greatly increased, while they are decreased in the coding sequence (CDS) and stop codon regions, which suggests that m6A modification status might be associated with cerebellar development (45).
Knocking out Mettl3 in the mouse embryonic brain causes cerebellar hypoplasia, due to drastically enhanced apoptosis of newborn cerebellar granule cells (CGCs) in the external granular layer (EGL) (46) (Figure 2). Key developmental genes such as Atoh1 and Cxcr4 are abnormally upregulated due to the extended mRNA half-lives induced by m6A depletion (46). Ectopic expression of Mettl3 leads to a disorganized laminal structure of both Purkinje cells and glial cells (45). Moreover, deletion of the eraser gene Alkbh5 causes increased nuclear export of hypermethylated RNAs, and abnormal proliferation and differentiation in the cerebellum (45). In addition, the cerebellum of Fto-deficient mouse is smaller than that of wild-type mouse (42).
m6A Modifications in Synaptogenesis and Axon Guidance
m6A modifications also contribute to neuronal growth and regeneration as well as to the local regulation of synaptic functions (22, 47). Synaptic m6A epitranscriptome (SME), which is functionally enriched in synthesis and modulation of tripartite synapses, has been identified in mouse adult forebrains using low-input m6A-sequencing of synaptosomal RNAs (48). The synaptic m6A peak distribution along mRNAs shows characteristic accumulation at the stop codon (22, 40).
Increased adenosine methylation in a subset of mRNAs important for neuronal signaling, including many in the dopaminergic (DA) signaling pathway has been found in the midbrain and striatum of Fto-knockout mice (43). Inhibition of FTO leads to increased m6A modifications and decreased local translation of axonal GAP-43 mRNA, which eventually represses axon elongation (49). Moreover, knockdown of Ythdf1 in hippocampal neurons reduces the cell surface expression of AMPA receptor subunit GluA1 and causes altered spine morphology and reduced excitatory synaptic transmission (48). Mutation of m6A sites in Robo3.1 mRNA or YTHDF1 knockdown or knockout leads to reduction of Robo3.1 protein, but not Robo3.1 mRNA, indicating that YTHDF1-mediated translation of m6A-modified Robo3.1 mRNA controls pre-crossing of axon guidance in the spinal cord (50). In addition, YTHDF3-knockdown neurons display a decreased percentage of spines containing a postsynaptic density (PSD) and surface GluA1 expression, indicating synaptic deficits in both structure and transmission (48).
In summary, these studies demonstrate important functions of m6A modifications in the nervous system. Mechanistic roles of m6A in regulating proliferation and differentiation of neural progenitors remain unclear. Whether such a mechanism is widespread within the brain will be an important area of future research.
m6A Modifications in Glial Cell Development
Glial cells, including oligodendrocytes and astrocytes, which are derived from the neuroepithelium in the CNS, and microglia, which are derived from mesodermal hematopoietic cells, make up 10–20% of the cells in the Drosophila nervous system and at least 50% of the cells in the human brain (51).
m6A Regulations in Gliogenesis
Embryonic neurogenesis and gliogenesis involve NSC proliferation, differentiation of NSCs into various neural and glial cell types, and their migration to their final destinations in the nervous system.
In the developing mouse cortex, NSCs or RGCs initially give rise to neurons in embryonic stages, and later switch to produce glial cells in early postnatal stages (52). Recent studies have shown that epigenetic mechanisms are involved in the precise spatiotemporal gene expression program, which controls transition in the developmental competence of progenitor cells in the sequential generation of neural and glial progeny and the maintenance of their differentiated identities (53). Several studies have investigated the mechanisms by which m6A regulates RGC differentiation. Reduction of m6A level decreases RGC proliferation, resulting in delayed neurogenesis and gliogenesis (40). Mettl3 depletion not only inhibits neuronal proliferation and differentiation, but also interferes differentiation of NSCs towards the glial lineage (54).
m6A Regulation of Oligodendrocytes
Transcripts that encode a number of histone modifiers are dynamically marked by m6A in oligodendrocytes precursor cells (OPCs) and oligodendrocytes, suggesting that m6A RNA modifications may play a role in regulating the expression of epigenetic modifiers in distinct oligodendrocyte lineages (55). Inactivating an m6A writer component METTL14 results in unchanged numbers of OPCs, decreased numbers of oligodendrocytes and hypomyelination in the CNS (56). A number of RNA transcripts that encode transcription factors implicated in oligodendrocytes lineage progression is dynamically marked by m6A at different stages of the oligodendrocyte lineage. Mettl14 ablation disrupts postmitotic oligodendrocyte maturation and has distinct effects on transcriptomes of OPCs and oligodendrocytes (56). Moreover, loss of Mettl14 in oligodendrocyte lineage cells causes aberrant splicing of myriad RNA transcripts, including those that encode the essential paranodal component neurofascin 155 (NF155) (56). These results indicate a time-specific post-transcriptional regulatory role of m6A in OPCs and oligodendrocytes.
Moreover, studies have shown that m6A reader PRRC2A controls OPC generation, proliferation, and fate determination. Deletion of Prrc2a in mouse OPCs leads to hypomyelination and consequent locomotive and cognitive defects, without affecting neurogenesis (57). PRRC2A binds and stabilizes the methylated transcript of oligodendrocyte transcription factor 2 (Olig2), a key oligodendroglial lineage determination transcription factor, in an m6A-dependent manner (57).
m6A Regulation of Astrocytes
Studies have shown that astrocytes and neurons are derived from a common neuroepithelial precursor (58). Mettl3 regulates lineage commitment during NSC differentiation, with a preference towards a neuronal fate. Mettl3-mediated m6A modification reduces the percentage of new born astrocytes (54). Knockout of Mettl14 in the mouse developing nervous system results in a significant decrease in the number of S100b+ astrocytes (40). Knocking down Mettl3 causes reduced astrocyte numbers in the developing cerebellum (45). Alkbh5 deficiency leads to reduced dendritic arborization of Purkinje cells, concomitant with an increase in disorganization of the radial fibers in astrocytes (45).
In summary, these results indicate that m6A modifications are critical for proper temporal progression of gliogenesis including oligodendrocytes and astrocytes.
m6A Modifications in Primary Brain tumors
Brain tumors are categorized into various types based on their nature, origin, rate of growth, and progression stage (59). Primary brain tumors can be broadly classified as malignant or non-malignant (benign) tumors, and graded from I to IV using a classification scheme specified by the World Health Organization (WHO) (60). Glioblastoma (GBM), a grade IV glioma, is the most prevalent (80% of all brain tumors) malignant and lethal intrinsic tumor in the CNS (61, 62).
RNA modifications, especially m6A modifications, have been shown to be essential for tumor development (63, 64). In particular, m6A modifications seem to play pivotal roles since both m6A writers and erasers contribute to the tumorigenesis of glioblastoma, especially glioma stem cells (GSCs) (62). Studies have shown that as the WHO grade is increased, the expression of WTAP, RBM15, YTHDF, and ALBKH5 is increased, while the expression of FTO is decreased in glioma (65).
m6A Writers Play an Oncogenic Role in Glioblastoma
Studies have shown that high expression of METTL3 is associated with clinical aggressiveness of malignant gliomas. METTL3 plays an oncogenic role by modulating nonsense-mediated mRNA decay (NMD) of splicing factors and alternative splicing of BCLX and NCOR2 isoform switches in glioblastoma (66). Silencing METTL3 or overexpressing dominant-negative mutant form of METTL3 suppresses growth and self-renewal of GSCs. METTL3 maintains the stability of a specific set of transcripts, such as apoptosis pathways and glial differentiation genes including SRSF1/2/3/6/11, CASP3/7, CASPB, DFFB, BMP2, LIF, IL1B, and HES1 in glioblastoma (66) (Figure 3). It appears that the oncogenic ability of METTL3 is dependent upon its methyltransferase catalytic domain. Knockdown of METTL14 expression reduces m6A levels in transcripts in GSCs, however, knockout of METTL14 has no effect on tumorigenesis of glioblastoma, suggesting that catalytic activity in METTL3 might be crucial in tumorigenesis (21, 67).
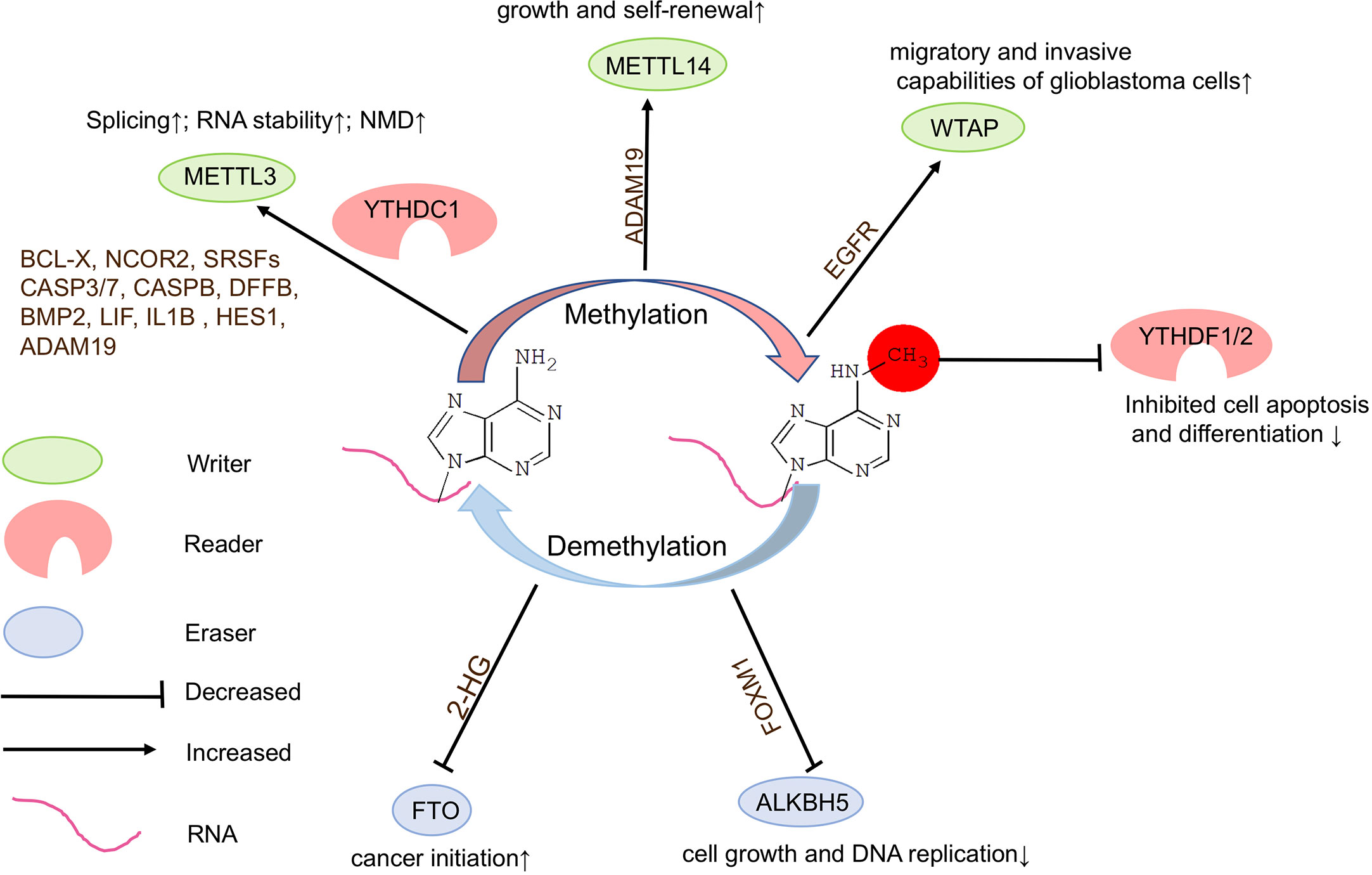
Figure 3 Functions of m6A modifications in brain tumors. N6-methyladenosine (m6A) modifications play a crucial role in brain tumorigenesis. Inadequate or dysregulated expression of writers, erasers, and readers of m6A is associated with brain tumor formation.
In addition, low levels of METTL3 or METTL14 lead to decreased m6A modifications on ADAM19 and increased level of ADAM19 in GSCs, ultimately causing glioma (67). The elevated sphere-formation rate induced by knockdown of Mettl3 or Mettl14 in GSCs can be reversed by knockdown of ADAM19, suggesting that ADAM19 acts as a target of m6A RNA methylation to regulate GSC self-renewal. It appears that knockdown of Mettl3 or Mettl14 dramatically promotes human GSC growth, self-renewal, and tumorigenesis (67). These controversial discoveries suggest that the role of METTL3 in glioblastoma requires further studies based on large amount of tumor samples and well-designed experimental systems.
Moreover, WTAP, an important component of the m6A methyltransferase complex, can regulate migratory and invasive capabilities of glioblastoma cells by increasing expression of epidermal growth factor receptor (EGFR) (68) (Figure 3).
Suppressing m6A Erasers May Inhibit Tumorigenesis
In glioma, mutations occur only in 0.1% of cases for m6A ereasers ALKBH5 and no mutations have been reported in FTO (69). Knockdown of Alkbh5 inhibits cell growth and decreased DNA replication in GSCs, and causes decreased Foxm1 transcription, and extended survival with a lower rate of tumor formation in mice (70) (Figure 3). These results demonstrate that the demethylation activity of ALKBH5 is critical to represses GSC-induced tumorigenesis.
Moreover, mutation of isocitrate dehydrogenase 1 (IDH1) occurs frequently, which results in accumulation of the metabolic byproduct 2-hydroxy-glutarate (2-HG) in glioma. 2-HG can inhibit FTO activity, and in turn increase global m6A modifications and contribute to cancer initiation (66) (Figure 3). In addition, treatment of GSCs with an FTO inhibitor MA2 suppresses GSC-initiated tumorigenesis and prolongs the lifespan of GSC-engrafted mice (67). Studies also have shown that FTO may play an oncogenic role via maintaining the stability of transcripts of avian myelocytomatosis viral oncogene homolog (c-Myc) and CCAAT enhancer binding protein alpha (CEBPA) in glioma, especially IDH1/2 mutant glioma (71).
m6A Readers Promote Progression of Glioblastoma
Studies have shown that YTHDF1 and YTHDF2 mRNA expression levels are positively correlated with malignancy of gliomas, with significant increases in higher grade gliomas, suggesting a role for these m6A readers in glioma progression (65, 69) (Figure 3). YTHDF2 may recognize specific methylated mRNAs, lead to their decay and subsequently to decreased cell apoptosis and differentiation, and in turn promote glioblastoma growth and de-differentiation, and also stabilize MYC and VEGFA transcripts in GSCs in an m6A-dependent manner (12, 72).
Moreover, a major splicing factor serine and arginine rich splicing factor 3 (SRSF3) is frequently upregulated in clinical glioma specimens (73). Knockdown of YTHDC1 leads to accumulation of NMD of SRSF3 mRNAs in glioblastoma cells, which can accelerate the proliferation of tumor cells (66).
In summary, altered m6A modifications are associated with the occurrence and development of glioblastoma, likely through regulating self-renewal of glioma stem cells. It appears that both m6A writers and erasers play an oncogenic role, and m6A readers function in progression in development of glioblastoma. However, inconsistent results indicate complicity of m6A modifications in brain tumor formation, likely through regulating distinct downstream genes.
Conclusions and Perspectives
Brain development is based on coordinated spatiotemporal cell fate decisions, and tightly regulated gene expression. Accumulating studies have shown that m6A methylation plays an important role in brain development and even in brain tumorigenesis. A major challenge is to identify specific target RNAs for m6A modifications in specific cell types and at different developmental stages. Recent improvements to m6A mapping methods will undoubtedly facilitate studies of activity-dependent changes to the epitranscriptome within distinct RNA populations in the brain. Moreover, an interesting research will be to determine whether changes to the mRNA modification landscape are causing factors or a consequence of activity-dependent regulation of gene expression.
How m6A methylation regulates brain tumor formation remains obscure. Taking the advantage of technical development of m6A methylation analysis at the single-cell level, mechanistic understanding of RNA methylation in different cell types will be revealed. Interestingly, in the late stage of glioma, high m6A modification levels may increase epigenetic reprogramming of non-GSCs into GSCs, whereas knockdown of METTL3 may reduce the ratio of GSCs in glioblastoma (66). Thus, a more profound breakthrough in the role of m6A methylation in brain tumor diagnostics and treatment strategy also should be developed.
Author Contributions
JW and TS wrote the manuscript and produced figures. YQS modified the manuscript. TS edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Scientific Research Funds of Huaqiao University (Z16Y0017, TS), and the National Natural Science Foundation of China (31771141 and 81701132).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep (2014) 8(1):284–96. doi: 10.1016/j.celrep.2014.05.048
2. Li J, Yang X, Qi Z, Sang Y, Liu Y, Xu B, et al. The role of mRNA m(6)A methylation in the nervous system. Cell Biosci (2019) 9(1). doi: 10.1186/s13578-019-0330-y
3. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res (2018) 46(D1):D303–7. doi: 10.1093/nar/gkx1030
4. Hsu PJ, Shi H, He C. Epitranscriptomic influences on development and disease. Genome Biol (2017) 18(1):197. doi: 10.1186/s13059-017-1336-6
5. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
6. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell (2019) 74(4):640–50. doi: 10.1016/j.molcel.2019.04.025
7. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
8. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol (2014) 16(2):191–8. doi: 10.1038/ncb2902
9. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
10. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol (2017) 18(1):31–42. doi: 10.1038/nrm.2016.132
11. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
12. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
13. Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature (2018) 563(7730):249–53. doi: 10.1038/s41586-018-0666-1
14. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res (2018) 28(5):507–17. doi: 10.1038/s41422-018-0034-6
15. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
16. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Corrigendum: Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature (2017) 542(7640):260. doi: 10.1038/nature21073
17. Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A (1984) 81(18):5667–71. doi: 10.1073/pnas.81.18.5667
18. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev (2015) 29(13):1343–55. doi: 10.1101/gad.262766.115
19. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
20. Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res (2019) 47(1):375–90. doi: 10.1093/nar/gky1012
21. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
22. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
23. Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell (2013) 155(6):1409–21. doi: 10.1016/j.cell.2013.10.047
24. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science (New York NY) (2015) 347(6225):1002–6. doi: 10.1126/science.1261417
25. Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A (2011) 108(36):14855–60. doi: 10.1073/pnas.1111577108
26. Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell (2014) 56(1):5–12. doi: 10.1016/j.molcel.2014.09.001
27. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
28. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
29. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
30. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
31. Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc (2015) 137(5):2107–15. doi: 10.1021/ja513080v
32. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
33. Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature (2015) 517(7532):33–8. doi: 10.1038/nature14010
34. Zhang C, Fu J, Zhou Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front Immunol (2019) 10:922. doi: 10.3389/fimmu.2019.00922
35. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature (2018) 561(7724):556–60. doi: 10.1038/s41586-018-0538-8
36. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
37. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature (2017) 552(7683):126–31. doi: 10.1038/nature24678
38. Liu S, Hausmann S, Carlson SM, Fuentes ME, Francis JW, Pillai R, et al. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell (2019) 176(3):491–504.e421. doi: 10.1016/j.cell.2018.11.038
39. Boles NC, Temple S. Epimetronomics: m6A Marks the Tempo of Corticogenesis. Neuron (2017) 96(4):718–20. doi: 10.1016/j.neuron.2017.11.002
40. Yoon K-J, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell (2017) 171(4):877–89.e817. doi: 10.1016/j.cell.2017.09.003
41. Du K, Zhang L, Lee T, Sun T. m(6)A RNA Methylation Controls Neural Development and Is Involved in Human Diseases. Mol Neurobiol (2019) 56(3):1596–606. doi: 10.1007/s12035-018-1138-1
42. Li L, Zang L, Zhang F, Chen J, Shen H, Shu L, et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet (2017) 26(13):2398–411. doi: 10.1093/hmg/ddx128
43. Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Bronneke HS, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci (2013) 16(8):1042–8. doi: 10.1038/nn.3449
44. Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol (2018) 19(1):69. doi: 10.1186/s13059-018-1436-y
45. Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol (2018) 19(1):68. doi: 10.1186/s13059-018-1435-z
46. Wang CX, Cui GS, Liu X, Xu K, Wang M, Zhang XX, et al. METTL3-mediated m6A modification is required for cerebellar development. PloS Biol (2018) 16(6):e2004880. doi: 10.1371/journal.pbio.2004880
47. Flamand MN, Meyer KD. The epitranscriptome and synaptic plasticity. Curr Opin Neurobiol (2019) 59:41–8. doi: 10.1016/j.conb.2019.04.007
48. Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci (2018) 21(7):1004–14. doi: 10.1038/s41593-018-0173-6
49. Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res (2018) 46(3):1412–23. doi: 10.1093/nar/gkx1182
50. Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res (2019) 47(9):4765–77. doi: 10.1093/nar/gkz157
51. Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature (2010) 468(7321):214–22. doi: 10.1038/nature09611
52. Yao B, Christian KM, He C, Jin P, Ming GL, Song H, et al. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci (2016) 17(9):537–49. doi: 10.1038/nrn.2016.70
53. Chang M, Lv H, Zhang W, Ma C, He X, Zhao S, et al. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol (2017) 7(9):170166. doi: 10.1098/rsob.170166
54. Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X, et al. m(6)A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genomics Proteomics Bioinformatics (2019) 17(2):154–68. doi: 10.1016/j.gpb.2018.12.007
55. Zhou H, Wang B, Sun H, Xu X, Wang Y. Epigenetic Regulations in Neural Stem Cells and Neurological Diseases. Stem Cells Int (2018) 2018:6087143. doi: 10.1155/2018/6087143
56. Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng YL, Elbaz B, et al. m(6)A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron (2020) 105(2):293–309 e295. doi: 10.1016/j.neuron.2019.12.013
57. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res (2019) 29(1):23–41. doi: 10.1038/s41422-018-0113-8
58. Rasband MN. Glial Contributions to Neural Function and Disease. Mol Cell Proteomics (2016) 15(2):355–61. doi: 10.1074/mcp.R115.053744
59. Johnson DR, Guerin JB, Giannini C, Morris JM, Eckel LJ, Kaufmann TJ. 2016 Updates to the WHO Brain Tumor Classification System: What the Radiologist Needs to Know. Radiographics (2017) 37(7):2164–80. doi: 10.1148/rg.2017170037
60. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro-oncology (2018) 20(suppl_4):iv1–iv86. doi: 10.1093/neuonc/noy131
61. Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro-oncology (2019) 21(Suppl 5):v1–v100. doi: 10.1093/neuonc/noz150
62. Dong Z, Cui H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers (Basel) (2020) 12(3):736. doi: 10.3390/cancers12030736
63. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
64. Thapar R, Bacolla A, Oyeniran C, Brickner JR, Chinnam NB, Mosammaparast N, et al. RNA Modifications: Reversal Mechanisms and Cancer. Biochemistry (2019) 58(5):312–29. doi: 10.1021/acs.biochem.8b00949
65. Chai RC, Wu F, Wang QX, Zhang S, Zhang KN, Liu YQ, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (2019) 11(4):1204–25. doi: 10.18632/aging.101829
66. Li F, Yi Y, Miao Y, Long W, Long T, Chen S, et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res (2019) 79(22):5785–98. doi: 10.1158/0008-5472.can-18-2868
67. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep (2017) 18(11):2622–34. doi: 10.1016/j.celrep.2017.02.059
68. Jin DI, Lee SW, Han ME, Kim HJ, Seo SA, Hur GY, et al. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci (2012) 103(12):2102–9. doi: 10.1111/cas.12022
69. Ceccarelli M, Barthel Floris P, Malta Tathiane M, Sabedot Thais S, Salama Sofie R, Murray Bradley A, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell (2016) 164(3):550–63. doi: 10.1016/j.cell.2015.12.028
70. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31(4):591–606.e596. doi: 10.1016/j.ccell.2017.02.013
71. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell (2018) 172(1-2):90–105.e123. doi: 10.1016/j.cell.2017.11.031
72. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov (2020). doi: 10.1158/2159-8290.cd-20-0331
Keywords: N6-methyladenosine (m6A), brain development, neural stem cell, glial cell, brain tumor, glioma
Citation: Wang J, Sha Y and Sun T (2021) m6A Modifications Play Crucial Roles in Glial Cell Development and Brain Tumorigenesis. Front. Oncol. 11:611660. doi: 10.3389/fonc.2021.611660
Received: 29 September 2020; Accepted: 11 January 2021;
Published: 24 February 2021.
Edited by:
Shicheng Guo, University of Wisconsin-Madison, United StatesReviewed by:
Huabing Li, Shanghai Jiao Tong University School of Medicine, ChinaKai Li, Peking-Tsinghua Center for Life Sciences, China
Briana Prager, Cleveland Clinic, United States
Copyright © 2021 Wang, Sha and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Sun, taosun@hqu.edu.cn
 Jing Wang1,2
Jing Wang1,2 Tao Sun
Tao Sun