- 1Department of Medical Oncology, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, China
- 2The First College for Clinical Medicine, Nanjing University of Chinese Medicine, Nanjing, China
- 3Department of Medical Oncology, Jiangsu Provincial Hospital of Chinese Medicine, Nanjing, China
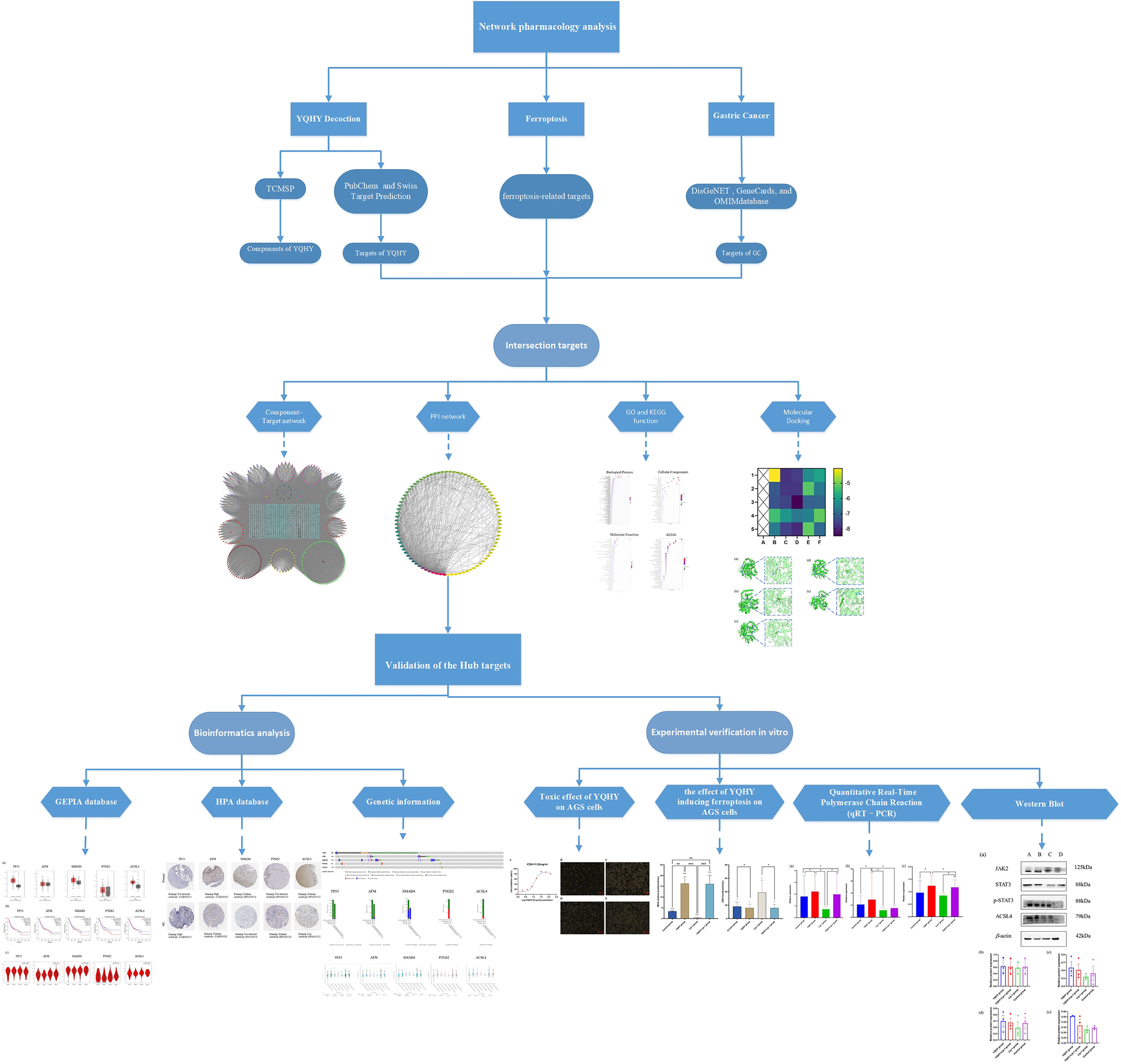
Objective: This study aimed to identify the mechanism of Yiqi Huayu Decoction (YQHY) induced ferroptosis in gastric cancer (GC) by using network pharmacology and experimental validation.
Methods: The targets of YQHY, ferroptosis-related targets, and targets related to GC were derived from databases. Following the protein–protein interaction (PPI) network, the hub targets for YQHY induced ferroptosis in GC were identified. Furthermore, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were used to analyze the hub targets from a macro perspective. We verified the hub targets by molecular docking, GEPIA, HPA, and the cBioPortal database. Finally, we performed cell viability assays, quantitative real-time polymerase chain reaction (qRT-PCR), western blotting, lipid peroxidation, and GSH assays to explore the mechanism of YQHY induced ferroptosis in GC.
Results: We identified the main active compounds and hub targets: Quercetin, DIBP, DBP, Mipax, Phaseol and TP53, ATM, SMAD4, PTGS2, and ACSL4. KEGG enrichment analyses indicated that the JAK2-STAT3 signaling pathway may be a significant pathway. Molecular docking results showed that the main active compounds had a good binding activity with the hub targets. The experimental results proved that YQHY could induce ferroptosis in AGS by increasing the MDA content and reducing the GSH content. qRT–PCR and Western blot results showed that YQHY can induce ferroptosis in GC by affecting the JAK2-STAT3 pathway and the expression of ACSL4.
Conclusions: This study indicated that YQHY can induce ferroptosis in GC by affecting the JAK2–STAT3 pathway and the expression of ACSL4, and induction of ferroptosis may be one of the possible mechanisms of YQHY’s anti-recurrence and metastasis of GC.
1 Introduction
The incidence of GC in China accounts for nearly half of the world (1, 2). At present, surgery is considered the only radical cure method (3). Early GC has a high cure rate, whereas advanced GC is characterized by a high metastasis rate, a high mortality rate, a low curative resection rate, and a poor five-year survival rate (4). The occurrence and development of cancer are closely related to cell death. One of the important characteristics of cancer cells is avoiding death. Thus, it is necessary to explore a new strategy to treat GC by inducing tumor cell death.
Ferroptosis is a new cell death mode discovered in recent years, which is different from autophagy, necrosis, and scorch death. It is the result of ferroptosis-dependent accumulation of lipid peroxides to lethal levels (5). The ferroptosis signature is mainly characterized by the accumulation of iron and lipid reactive oxygen species and inhibition of the cystine/glutamate antiporter (system XC), leading to reduced cystine uptake and reduced glutathione (GSH) synthesis (6). Studies have shown that ferroptosis plays an important role in the development of various cancers (7). There is a close relationship between ferroptosis and GC, as ferroptosis is significantly suppressed in GC, resulting in reduced tumor growth and sensitivity to cisplatin versus paclitaxel (8). The expression of ultralong-chain fatty acid protein 5 and fatty acid desaturase 1 in mesenchymal GC cells is upregulated, leading to ferroptosis sensitization (9). Erastin, a typical ferroptosis inducer, can induce ferroptosis in GC cells. Silencing CDO1 in vitro and in vivo can restore intracellular glutathione levels and prevent ROS production, thus inhibiting ferroptosis in GC cells induced by erastin (10, 11). Therefore, inducing GC cells to undergo ferroptosis could be a potential direction for GC treatment (12).
The pathogenesis of postoperative patients with GC can be summarized as “Qi deficiency and blood stasis and toxin”, with deficiency of vital qi as the foundation and blood stasis and toxin as the standard. YQHY was pioneered by Professor Shen Lin Liu of a nationwide Chinese medicine and is based on Invigorating Qi and invigorating spleen and removing blood stasis as therapeutic points for GC, which is widely used in the clinical treatment of GC. Our previous studies have proven that YQHY can reduce the risk of recurrence and metastasis of patients with GC after stage II and III operations. Compared with chemotherapy alone, the risk of recurrence and metastasis is decreased by 32.8% (P = 0.0042). Especially for patients with stage III disease, the percentage of recurrence and metastasis risk is decreased to 34.7% (P = 0.0072) (13). Traditional Chinese medicine (TCM) has “multicomponent, multitarget, and multipathway” advantages for treating diseases (14). However, the mechanism of YQHY in reducing the risk of recurrence and metastasis of GC is still unclear. Therefore, in this paper, we combined the results of the network pharmacology study and verified them through experiments in vitro, which can provide new ideas for clarifying the pathogenesis of GC and for clinical intervention and treatment. The protocol of our study procedures is shown in Figure 1.
2 Methods
2.1 Network Pharmacology Analysis
2.1.1 Screening the Main Active Compounds of YQHY
The active compounds of YQHY were derived from the TCMSP database (TCMSP, http://lsp.nwu.edu.cn/tcmsp.php). To better screen the active compounds of YQHY, OB ≥30% (15) and DL ≥0.05 (16) were used as screening conditions in this article. After that, a total of 360 active compounds were finally obtained.
2.1.2 Collection of Targets
Through the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), the screened active compounds were transformed into the SMILES structural formula, and the SMILES structural formula was imported into the Swiss Target Prediction website, setting Homo sapiens, to predict all potential targets of the active compounds in YQHY. The relevant targets in GC were obtained by using the DisGeNET, GeneCards, and OMIM databases. We screened targets related to ferroptosis from the FerrDB (http://www.zhounan.org/ferrdb/) database (17). All potential targets and corresponding active compounds of YQHY were input into Cytoscape to construct the “YQHY active compound-target” network.
2.1.3 Screening of Hub Targets for YQHY Induced Ferroptosis in GC
Taking the intersection of the whole decoction targets, GC targets, and ferroptosis targets, we obtained the targets of YQHY induced ferroptosis in GC. The targets were imported into the STRING database to construct a PPI network and visualized using Cytoscape, of which the CytoNCA plug-in was used to calculate the parameters, and the hub targets for YQHY induced ferroptosis in GC were obtained according to BC, CC, and Degree.
2.1.4 GO and KEGG Enrichment Analysis
The targets of YQHY induced ferroptosis in GC were imported into the DAVID database for GO and KEGG enrichment analysis. The final analysis results included
Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). GO enrichment analysis was performed according to P <0.05 and FDR <0.05, and KEGG enrichment analysis was performed according to P <0.01 as the filtering criterion. Top20 were plotted into bubble charts using R language.
2.1.5 Molecular Docking
Molecular docking is mainly used for structural docking of small molecules with target proteins and evaluating their binding affinities with defined binding sites (18). A negative docking binding energy suggests efficient autonomous binding of the small molecule to the target protein. It is generally believed that the lower the energy the conformation in which a ligand binds to a receptor is stabilized, the more likely the effect will be. In this study, the main active compounds and hub targets of YQHY induced ferroptosis in GC were molecularly docked. The TCMSP database was used to download the structure diagram of the main active compounds, which were saved in SDF format. Open Babel GUL software was used to convert the SDF format into the PDB format. We used the PDB database to download the 3D structure diagram of hub targets, and saved it in PDB format, then imported them into AutoDock software for molecular docking. Docking results were visualized using PyMol software.
2.2 Bioinformatics Analysis
The hub targets were input into the online tool GEPIA (http://gepia.cancer-pku.cn/index.html) (19) to verify their mRNA expression level, pathological stages, and overall survival (OS) in TCGA-STAD. The protein expression of hub targets was investigated in the HPA database (https://www.proteinatlas.org/) (20). The cBioPortal tool (http://www.cbioportal.org/) (21) was used to discover the genetic information and the correlation between mRNA expression of hub targets.
2.3 Experimental Validation In Vitro
2.3.1 Preparation of YQHY Freeze-Dried Powder
The YQHY decoction consists of Huangqi, Dangshen, Chenpi, Banxia, Baizhu, Baishao, Danggui, Sanleng, Ezhu, Sheshecao, Shijianchuan, Fulin, Muxiang, Sharen, and Gancao. YQHY medicine was dried for 24 h, pulverized, and sieved with an 80 mesh sieve. The decoction was mixed, filtered, and concentrated to 250 ml. The concentrated solution was added to a special freeze-drying bottle, quickly frozen with liquid nitrogen, and dried at −50°C in FreezeDryers. The lyophilized powder was removed at 48 h and stored under cryogenic drying after crushing.
2.3.2 Cell Viability Assay
Different concentrations of YQHY were added to the experimental group, and F12K culture medium was added to the zeroing group and control group. The reaction was carried out at 37°C and 5% carbon dioxide for 24 h. After CCK8 solution was added to each well to react for 1 h, the OD of each sample was measured by setting up the microplate reader at 490 nm. IC50 was calculated using GraphPad Prism software.
2.3.3 Lipid Peroxidation and GSH Assays
The MDA and GSH contents in cell lysates were assessed using lipid peroxidation and GSH assay kits, respectively, according to the manufacturer’s (Abcam) instructions.
2.3.4 Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
According to the results of network pharmacology, we selected ACSL4, JAK2, and STAT3 to explore the mechanism by which YQHY induces ferroptosis in GC. Total RNA was extracted from cultured cells using TRIzol and phenol–chloroform phase separation according to the manufacturer’s instructions and then reverse transcribed using the RevertAid RT Reverse Transcription Kit (Invitrogen, K1691). The primer sequences used for qRT-PCR are listed in Table 1. qRT–PCR was performed using a StepOne Real-Time PCR system (Applied Biosystems).
2.3.5 Western Blot
The protein expression of JAK2, STAT3, p-STAT3, and ACSL4 was detected by Western blot. Total proteins were obtained using RIPA buffer, and protein concentrations were measured using a BCA protein assay kit. The protein samples were blotted onto PVDF membranes. Next, the membranes were incubated at 4°C overnight with primary antibodies against JAK2, STAT3, p-STAT3, ACSL4, and β-actin, followed by HRP-conjugated secondary antibodies. The grayscale values were quantified by Image Lab software.
2.3.6 Statistical Analysis
The experimental results were statistically analyzed by GraphPad Prism software. Single-factor ANOVA was used to compare the differences in experimental data among groups. A P-value less than 0.05 was considered significant.
3 Results
3.1 Network Pharmacology Analysis
3.1.1 Screening the Main Active Compounds of YQHY
Through the screening of the TCMSP database, a total of 360 active compounds were obtained, namely, 23 Huangqi, 27 Dangshen, 10 Chenpi, 20 Banxia,16 Baizhu, 17 Baishao, 22 Danggui, 10 Sanleng, 21 Ezhu, 7 Sheshecao, 4 Shijianchuan, 16 Fulin, 30 Muxiang, 54 Sharen, and 83 Gancao.
3.1.2 Collection of Targets and Construction of the “YQHY compound-target” Network
After the prediction of the database and deletion of duplicate values, a total of 1,373 potential targets of active compounds, 15,842 targets of GC, and 205 targets of ferroptosis were obtained. A total of 87 common targets were obtained by combining the three target sets. At the same time, a “YQHY compound-target” network was constructed (Figure 2), in which quercetin, DIBP, DBP, Mipax, and Phaseol were considered the main active components for YQHY induced ferroptosis in GC. Then the PPI network was constructed according to the degree value (Figure 3).
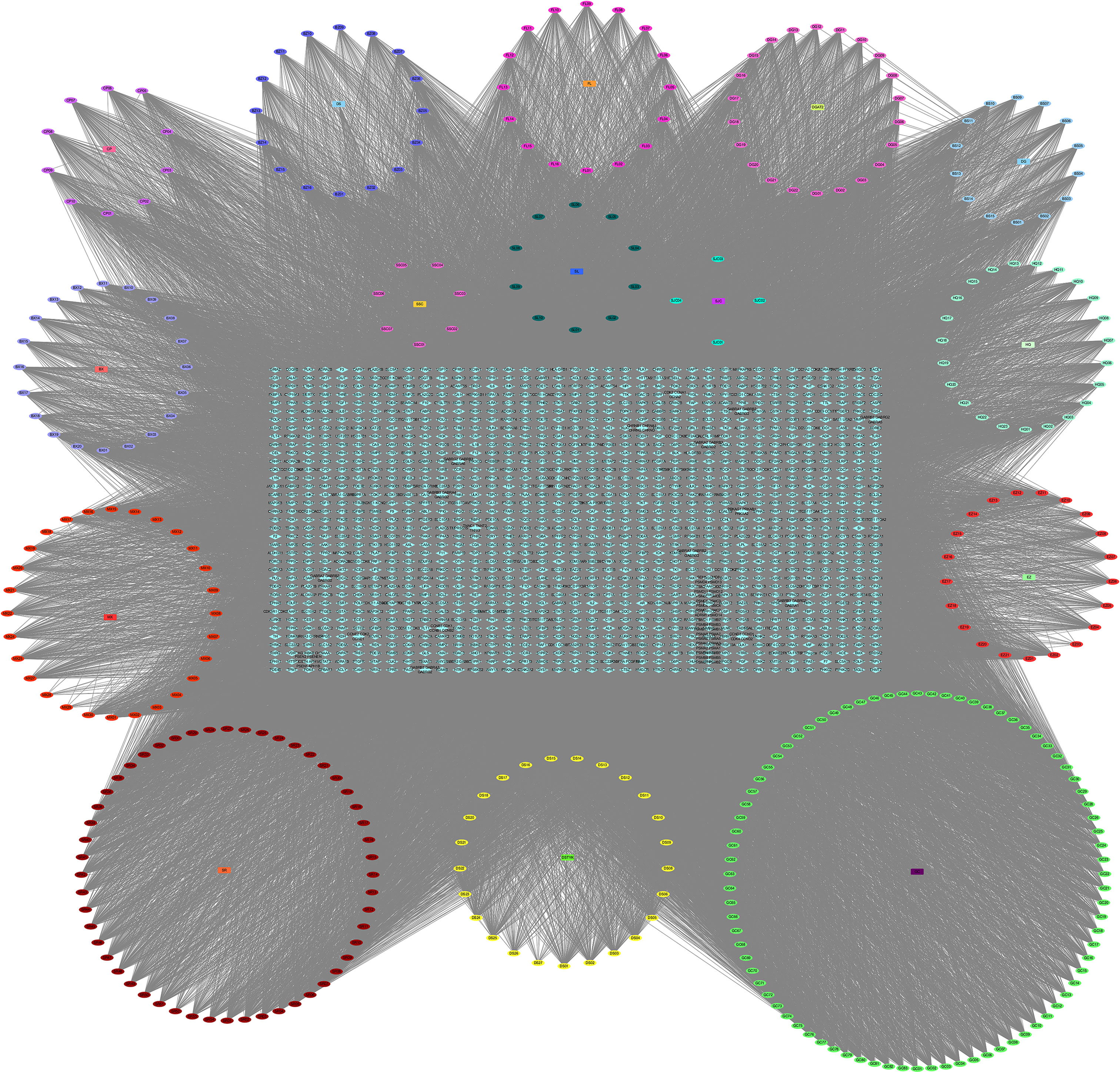
Figure 2 YQHY Compound-Target Network. The circle represents the YQHY active compound, the rectangle represents the herb, and the blue diamond represents the target.

Figure 3 Protein–protein interaction Network. The darker the color, the greater the Degree value. The lighter the yellow color, the smaller the degree value.
3.1.3 Screening of Hub Targets for YQHY Induced Ferroptosis in GC
According to the ranking of BC, CC, and degree calculated by the CytoNCA plug-in, TP53, ATM, SMAD4, PTGS2, and ACSL4 were identified as the hub targets of YQHY induced ferroptosis in GC. Meanwhile, a PPI network diagram of hub targets was constructed.
3.1.4 GO and KEGG Enrichment Analysis
GO enrichment analysis showed that the biological functions mainly involved positive regulation of transcription from the RNA polymerase II promoter, negative regulation of apoptotic processes, and oxidation–reduction processes. Cell components mainly included the cytoplasm, cytosol, and nucleus. Molecular functions mainly included protein binding, identical protein binding, and DNA binding. KEGG functional enrichment analysis showed that the hub targets were mainly enriched in microRNAs in cancer and the JAK2-STAT3 signaling pathway (Figure 4), suggesting that the JAK2-STAT3 pathway is one of the main potential signaling pathways of YQHY induced ferroptosis in GC.
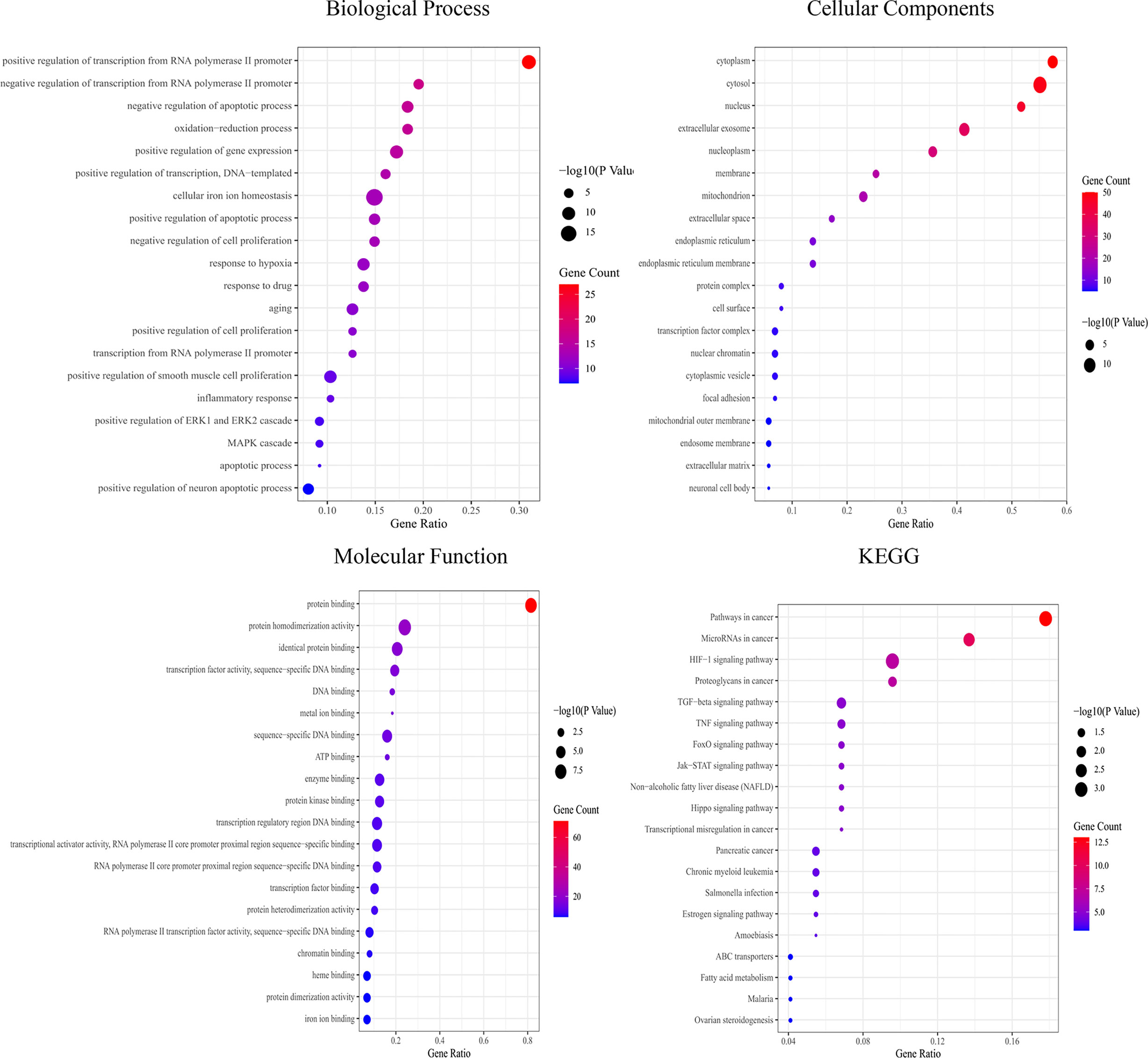
Figure 4 Bubble diagram for GO and KEGG enrichment analysis. The bubble size represents the number of enriched genes, and the bubble color difference represents the significant magnitude of target gene enrichment.
3.1.5 Molecular Docking
A binding energy <−5 kcal mol−1 indicates good binding activity. The results of molecular docking were plotted as a heatmap (Figure 5). It can be seen from the figure that DIBP, DBP, Phaseol with TP53, SMAD4, and ACSL4 all had good binding activities. We selected the molecular docking results of ACSL4 with the main active compounds, and visual analysis was carried out by PyMol software (Figure 6).
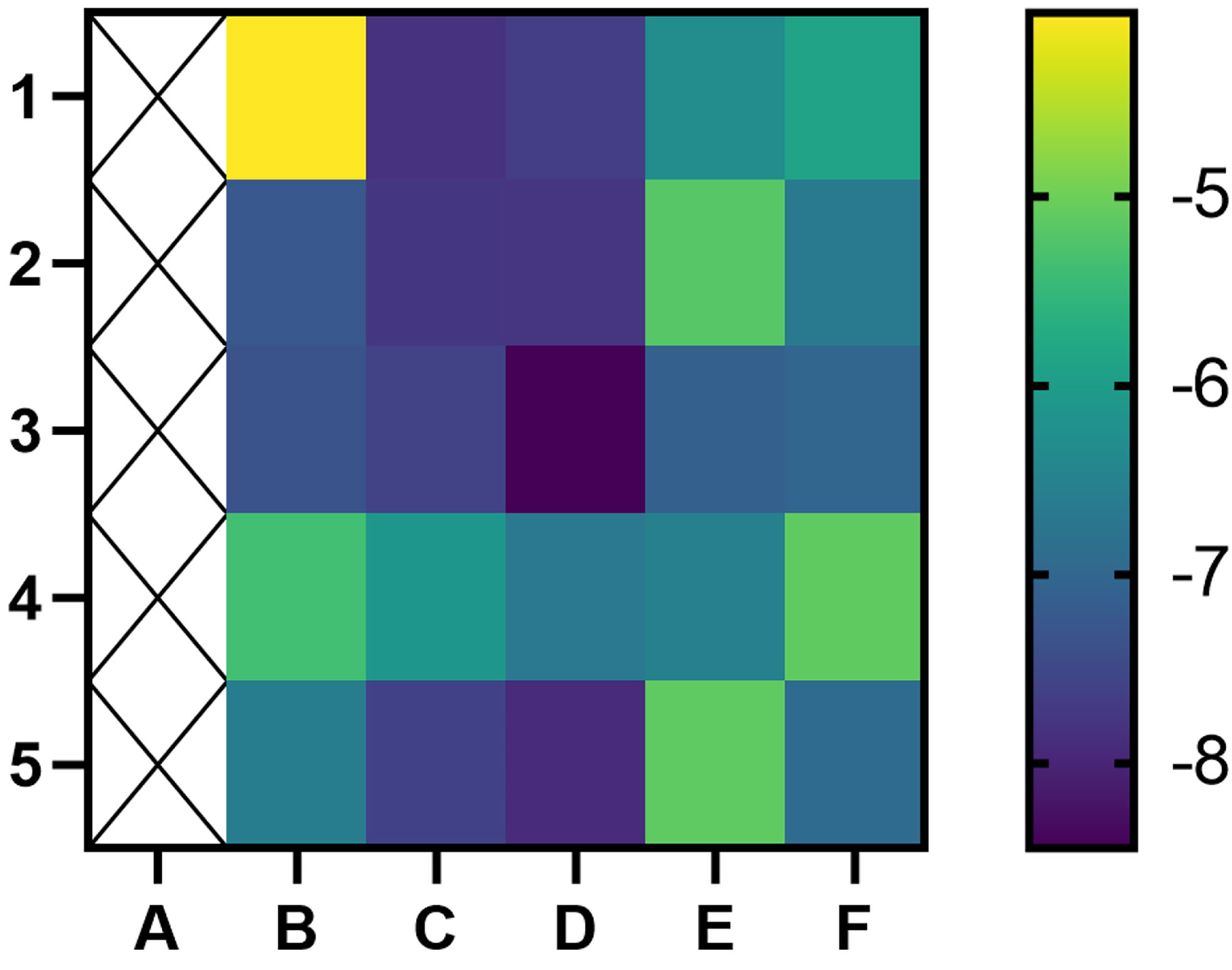
Figure 5 Heatmap of binding between main active components and hub targets. The darker the color, the better the binding activity. (A) represent blank, (B–F) represent TP53, ATM, SMAD4, PTGS2, and ACSL4, respectively, while 1, 2, 3, 4, and 5 represent Quercetin, DIBP, DBP, Mipax, and Phaseol, respectively.
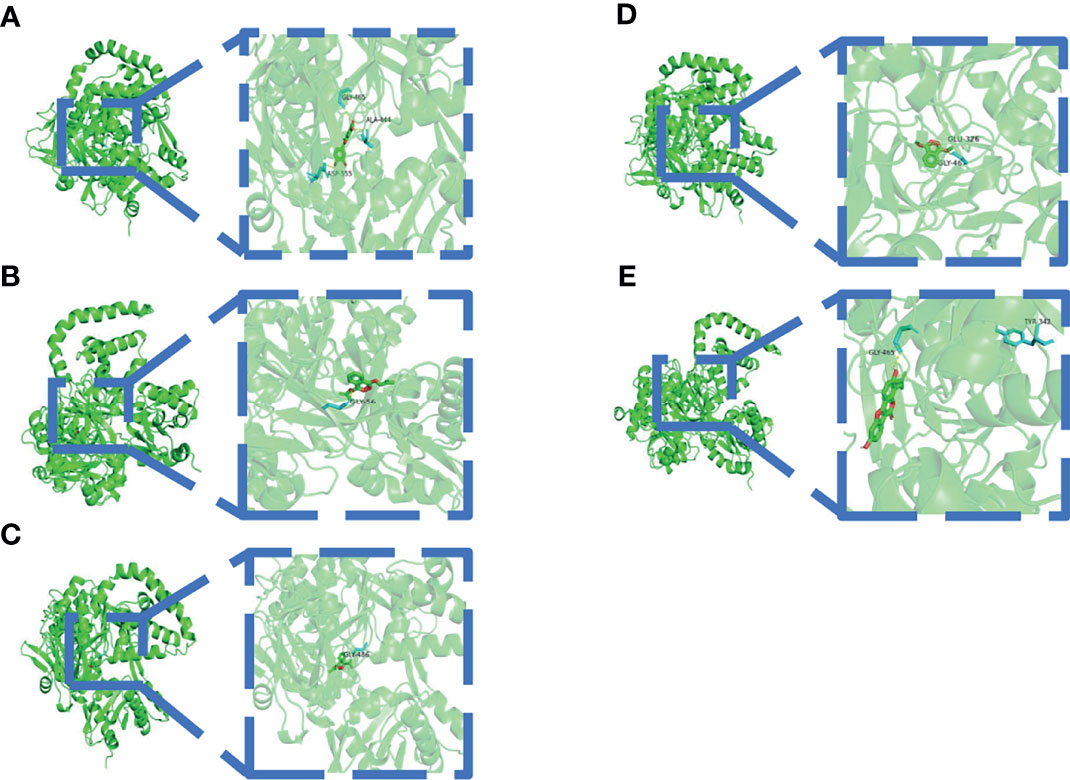
Figure 6 Schematic diagram of docking between active compounds and ACSL4. (A–E) represent the molecular binding of ACSL4 with Quercetin, DIBP, DBP, Mipax, and Phaseol, respectively.
3.2 Bioinformatics Analysis
The results of the GEPIA database showed that the mRNA levels of TP53 and ACSL4 were significantly highly expressed in GC tissues (Figure 7A). Survival analysis of the hub targets (Figure 7B) showed that the prognostic value of PTGS2 was significantly different (P <0.05). We analyzed the relationship between hub target mRNA levels and the pathological stage of GC. The results showed that the levels of ATM changed significantly with pathological stage and increased significantly in stage IV (Figure 7C).
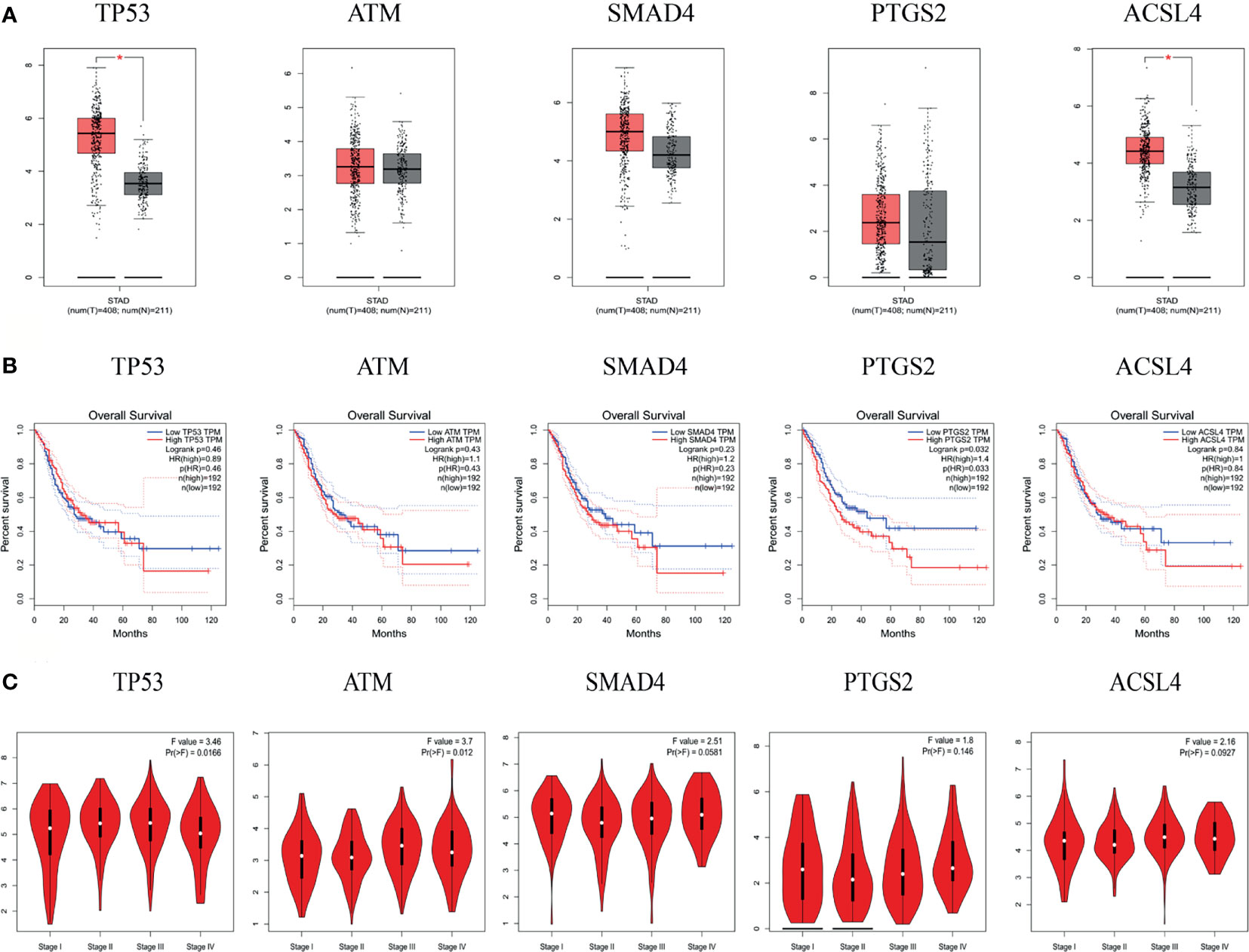
Figure 7 mRNA expression level, pathological stage and OS in the GEPIA database. (A) Box plots showing the mRNA expression levels of TP53, ATM, SMAD4, PTGS2, and ACSL4. Red represents Tumor, Gray represents normal. (B) The line charts show the OS of hub genes in GEPIA. The survival curve comparing the patients with high (red) and low (blue) expression in GC. (C) The violin diagram shows the stage plot of mRNA expression level and pathological stage in the GEPIA database.
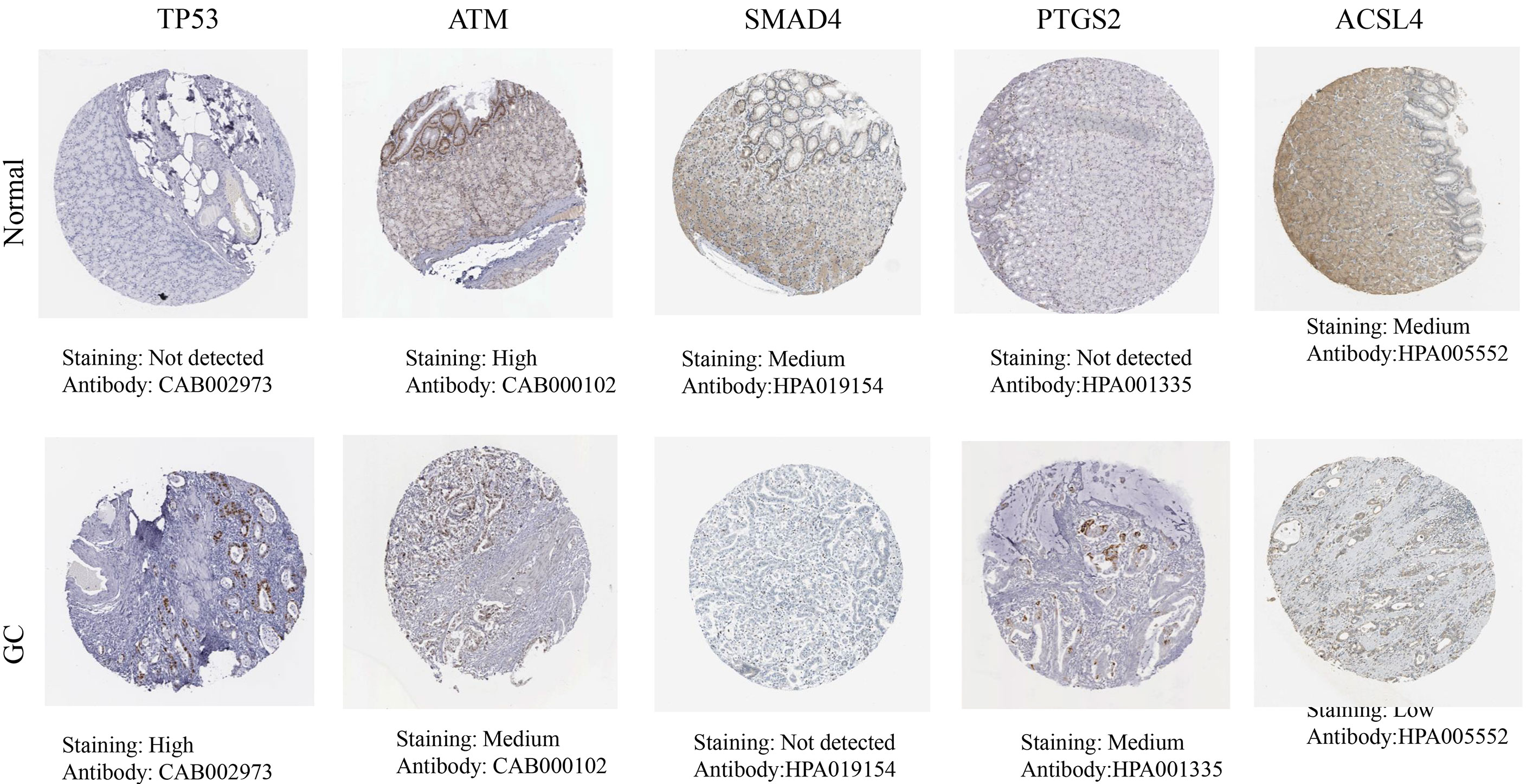
The HPA database results showed that the hub targets were expressed to different degrees in normal gastric tissues. Compared with normal gastric tissues, the expression levels of TP53 and PTGS2 were increased in GC tissues, while the expression of ATM, SMAD4, and ACSL4 was decreased in GC tissues (Figure 8).
The cBioPortal tool showed that 278 of 434 patients (64%) had genetic mutations in these five targets (Figure 9A). An overview of the genetic variation of five targets was also analyzed (Figure 9B). Figure 9C shows the number of gene mutations in different types of GC.
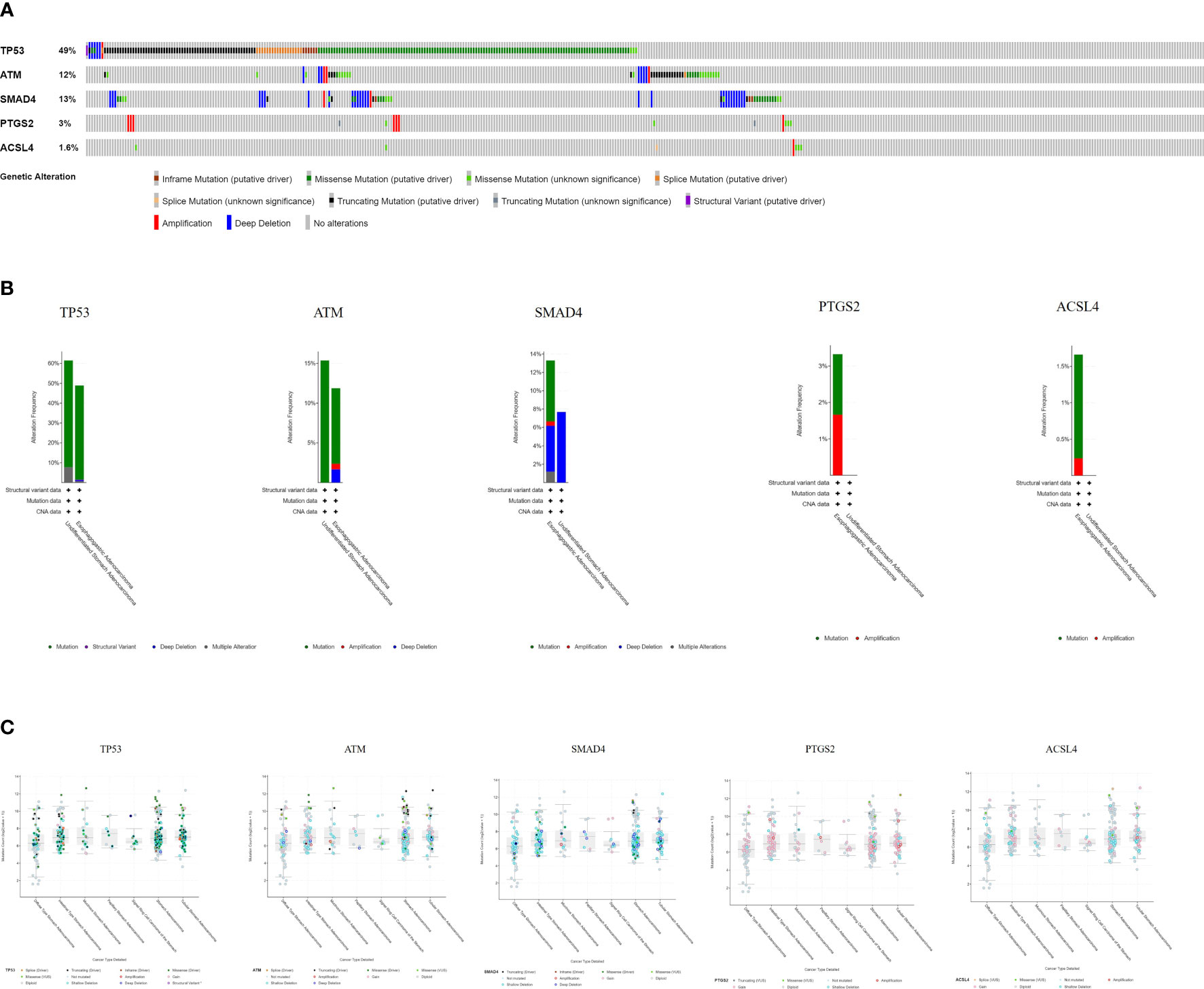
Figure 9 Genetic information of hub targets. (A) Data from TCGA of gastric adenocarcinoma showed that 278 of 434 patients (64%) had genetic mutations in these five targets. (B) The diagram shows the genetic variation of five targets. (C) The diagram shows the number of gene mutations in different types of GC.
3.3 Experimental Validation In Vitro
3.3.1 Cell Viability Assay
It can be concluded that with increasing concentrations of YQHY, the toxic effect on AGS gradually intensified, and the inhibition rate of AGS showed a gradually increasing trend (Figure 10). According to the curve in the figure, we found that the IC50 of YQHY on AGS was 11.20 mg/ml, which means that approximately 50% of AGS was inhibited after exposure to 11.20 mg/ml YQHY for 24 h. This result suggested that YQHY had a toxic effect on AGS, and we chose this concentration at half inhibition rate for subsequent experiments.
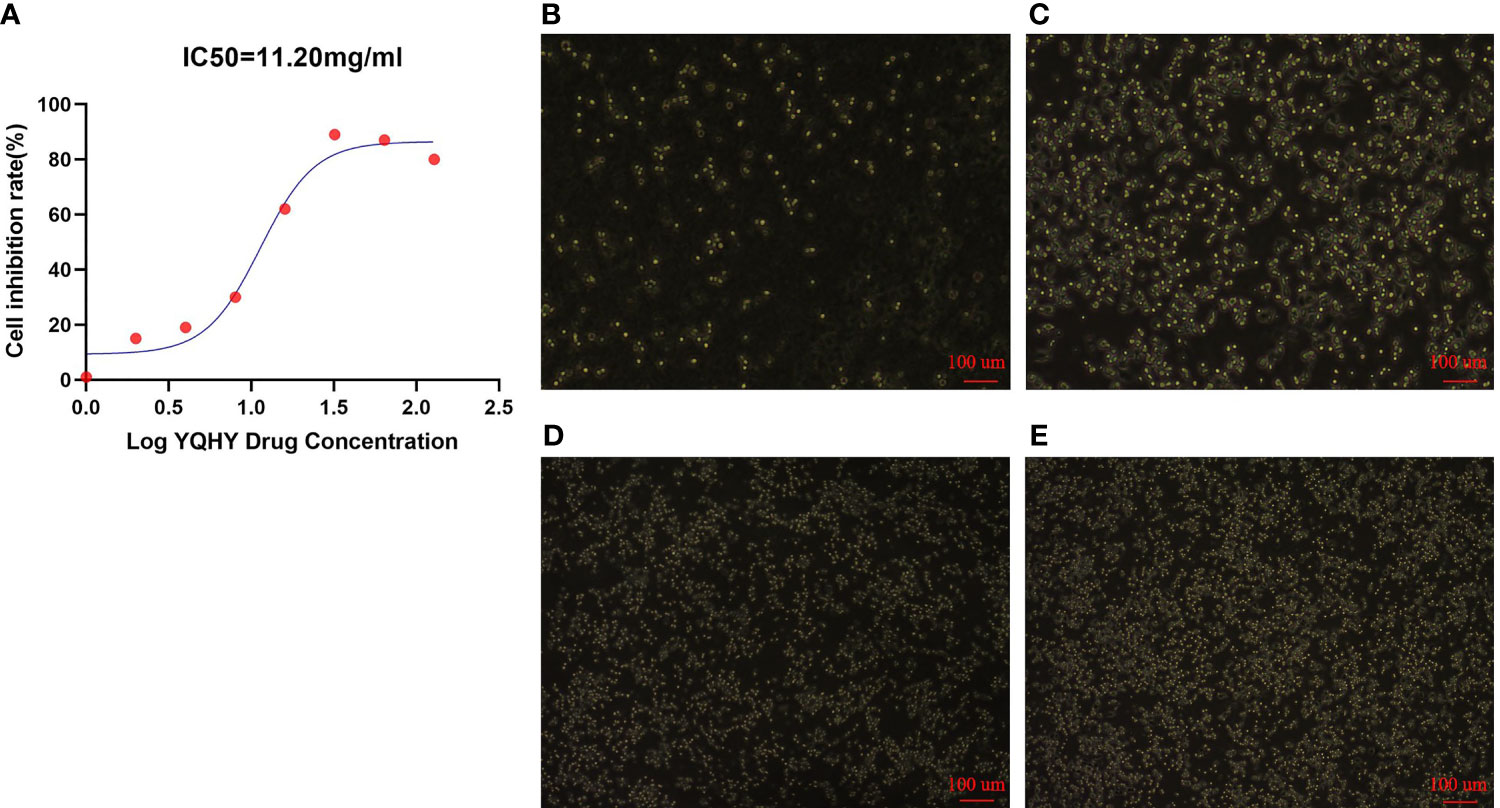
Figure 10 The graph of YQHY inhibiting AGS growth. (A) The IC50 of YQHY on AGS cells was 11.20 mg/ml, which means that approximately 50% of AGS cells were inhibited after exposure to 11.20 mg/ml YQHY for 24 h. (B–E) show the growth state of AGS cells in different groups after 24 h of dose. Among them, (B) represents the YQHY group, (C) represents the YQHY + liproxstatin-1 group, (D) represents the control group, and (E) represents the liproxstatin-1 group.
3.3.2 Lipid Peroxidation and GSH Assays
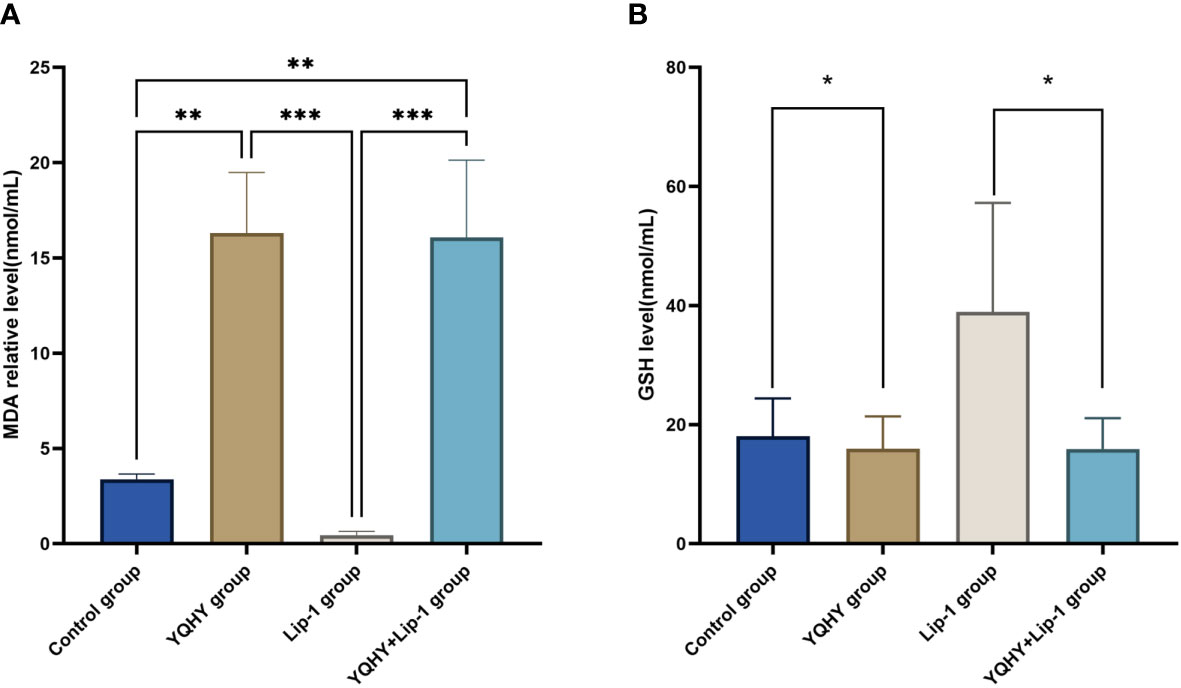
The MDA content in the YQHY group was the highest compared with the other three groups (P <0.05) (Figure 11A), and YQHY reversed the ferroptosis inhibition induced by the ferroptosis inhibitor liproxstatin-1. The GSH content in the YQHY group was the lowest compared with the other three groups (P <0.05) (Figure 11B), and the GSH content in the YQHY + liproxstatin-1 group was lower than that in the control group, indicating that YQHY can reduce the intracellular GSH content and induce ferroptosis in AGS and that YQHY can reverse ferroptosis due to liproxstatin-1 inhibition.
3.3.3 Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
We used qRT-PCR to measure the effect of YQHY, liproxstatin-1, and YQHY + liproxstatin-1 on the mRNA expression of JAK2, STAT3, and ACSL4. The results showed that the mRNA expression levels of JAK2, STAT3, and ACSL4 were higher in the YQHY group than in the control group (P <0.05). After the addition of liproxstatin-1, the mRNA expression of JAK2, STAT3, and ACSL4 was decreased compared with that in the YQHY group (P <0.05). The mRNA expression of JAK2, STAT3 and ACSL4 in the liproxstatin-1 group was significantly decreased relative to the control group (P <0.05) (Figure 12).
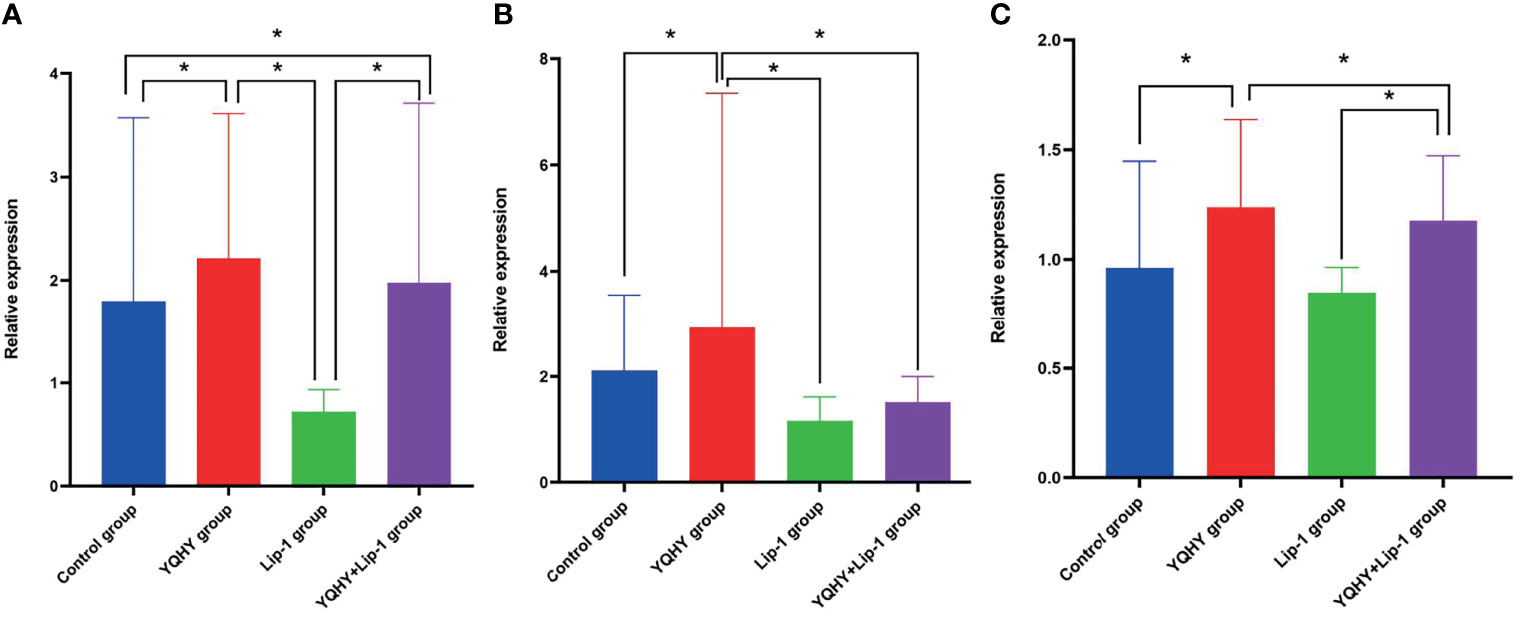
Figure 12 Effect of YQHY on JAK2, STAT3, and ACSL4 mRNA. (A) represents the relative mRNA expression of JAK2, (B) represents the relative mRNA expression of STAT3, and (C) represents the relative mRNA expression of ACSL4. (*P < 0.05).
3.3.4 Western Blot
Compared with the control group, the protein expression of JAK2, STAT3, p-STAT3, and ACSL4 in the YQHY group was higher than those in the other three groups, while the expression level of each protein in the liproxstatin-1 group was lower. It could be concluded that YQHY can induce ferroptosis in AGS by regulating the JAK2-STAT3 pathway and the expression of ACSL4 (Figure 13).
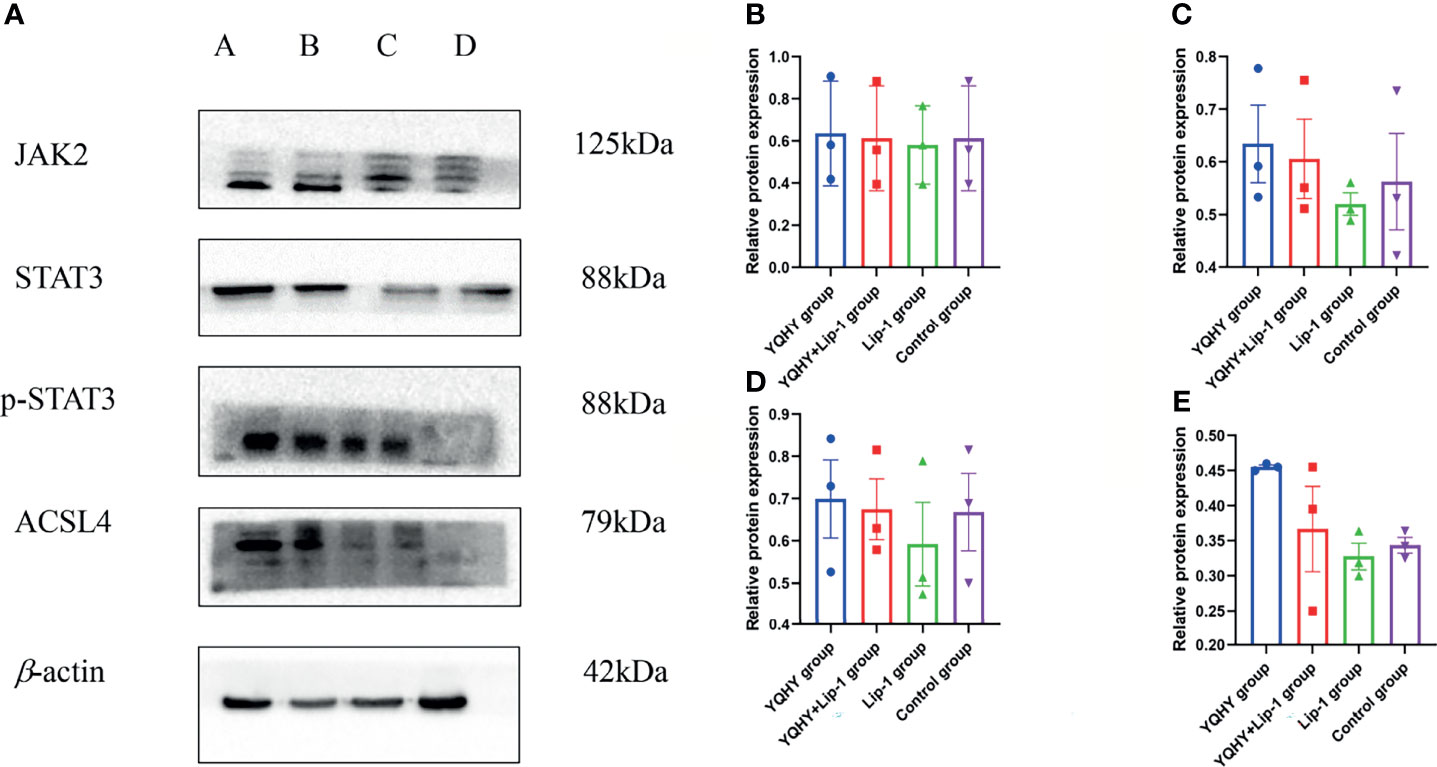
Figure 13 Effect of YQHY on JAK2, STAT3, p-STAT3, and ACSL4 protein. (A) A represents the YQHY group, B represents the YQHY + liproxstatin-1 group, C represents the liproxstatin-1 group, and D represents the control group. (B) represents the relative protein expression of JAK2, (C) represents the relative protein expression of STAT3, (D) represents the relative protein expression of p-STAT3, and (E) represents the relative protein expression of ACSL4.
4 Discussion
Recently, the incidence and mortality of GC have gradually increased, which is related to diet, lifestyle, genetics, and other factors. Due to distant metastasis and recurrence, most patients have a poor prognosis (22). According to clinical studies, YQHY has a clear antirecurrence and metastasis effect in GC (23). Our team wanted to further investigate whether this is achieved by inducing ferroptosis in GC and whether the influence of the JAK2–STAT3 signaling pathway and ACSL4 expression is one of the mechanisms of YQHY-induced ferroptosis in GC.
Through the network pharmacology study, Quercetin, DIBP, DBP, Mipax, and Phaseol were identified as the main active compounds of YQHY-induced ferroptosis in GC. Quercetin is a flavonoid, which exists in many edible and medicinal plants (24). It has various pharmacological activities, such as antioxidant, anti-inflammatory, and antitumor activities. Quercetin can be used as an antioxidant activity ROS scavenger and metal chelating agent to protect gastric epithelial cells from oxidative damage (25, 26). Many studies have shown that quercetin has antiproliferative and antiangiogenic effects in various cancers (27), such as lung cancer (28), breast cancer (29), colon cancer (30), and GC (31). Quercetin was found to arrest cell division by inducing cell cycle arrest at G0/G1 or G2/M phase in BGC-823 gastric cancer cells through protein analysis of Bax, Bcl-2, and caspases (32). Phasol is rich in flavonol glycosides and coumarin. Coumarin can induce apoptosis of Jurkat cells by inducing mitochondrial depolarization, which may contribute to cell death by inducing cell cycle arrest at the G1 phase, reducing BCL2 levels, and increasing PARP-1 cleavage (33), but there have been no reports on its association with GC. DIBP, DBP, and Mipax were the main compounds we identified for the treatment of GC, but there is no related report.
We analyzed the biological functions and signaling pathways of YQHY and found that TP53, ATM, SMAD4, PTGS2, and ACSL4 were the hub targets of YQHY induced ferroptosis in GC. TP53 is a tumor suppressor gene that has attracted wide attention at present. Mutant TP53 has an influence on the proliferation, migration, survival, and invasion of tumors and the drug resistance of chemotherapy drugs (34). When TP53 is mutated, the original tumor suppressor function is affected, which further increases the apoptosis process of gastric mucosa cells and impairs the damage repair function of the cells, thus inducing the transition of normal cells to cancer cells and increasing the incidence of GC (35). ATM is a cell housekeeping gene encoding ATM protein kinase, which plays a key role in the early signal transduction of cell cycle checkpoints (36, 37). ATM is a potential marker for GC (38), and previous studies (39) have confirmed that HP infection can cause upregulated expression of ATM. SMAD4 is a tumor suppressor gene discovered in recent years. Patients with GC often have mutations or loss of expression of SMAD4 (40). Reduced expression of SMAD4 in GC is one of the main reasons for its abnormal proliferation and migration (41). PTGS2 is closely related to apoptosis and promotes tumor progression (42). Lin (43) demonstrated that PTGS2 mediated cisplatin-induced BCL2 expression and subsequent apoptosis resistance through a PGE2/EP4/MAPK (ERK1/2, P38)-dependent mechanism. The ACSL family mainly catalyzes fatty acids with 12–20 carbon chains (44). Evidence has shown that ACSLs are enzymes crucial for the body’s response to fatty acid metabolism (45). Mammalian ACSLs have five different isozymes, namely, ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6, which are different in intracellular functions in their tissue expression specificity (46). Studies have shown that ACSL4 is involved in various biological processes, such as proliferation, apoptosis, migration, and invasion of tumor cells. It is also a biomarker and contributor of ferroptosis (47). Ferroptosis contributes to the antitumor function of several tumor suppressors, such as p53, BAP1, and fumarase (48). ACSL4 was differentially expressed in tumor tissues compared with adjacent carcinoma normal tissues. For example, an increase in ACSL4 expression is related to the differentiation of colon adenocarcinoma (49). One of these reports indicated that ACSL4 is part of the mechanism responsible for the promotion of breast cancer cell proliferation, invasion, and migration (50). However, ACSL4 is underexpressed in GC tissue and represents an undifferentiated or poorly differentiated malignant phenotype (51). Therefore, the expression of ACSL4 can inhibit the migration and invasion of GC, which illustrates that ACSL4 can be a potential therapeutic target as a tumor suppressor in GC.
The enrichment analysis of the KEGG suggested that the JAK2–STAT3 pathway was one of the main potential signaling pathways of YQHY induced ferroptosis in GC. The JAK2–STAT3 signaling pathway is a typical oncogenic signaling pathway (52). JAK2 serves as a signaling hub that integrates extracellular signals from interleukin receptors and oncogenic receptor tyrosine kinases into STAT3, which phosphorylates STAT3 at Y705 and homodimerizes with p-STAT3 to induce its nuclear translocation and transcriptional activity through interaction with its phosphorylated Y705 site and SH2 domain (53, 54). STAT3 binds to the promoters of its target genes to induce tumor cell migration, growth, and differentiation and plays an important role in the development of a variety of tumors (55).
Molecular docking results showed that the main active compounds of YQHY had good binding activity with the hub targets, and then we identified the hub targets in different databases. We found that the mRNA levels of TP53 and ACSL4 were significantly expressed in GC tissues, and the prognostic value of PTGS2 was significantly different (P b<0.05). The levels of ATM changed significantly with pathological stage and increased significantly in stage IV. The cBioPortal tool showed that 278 of 434 patients (64%) had genetic mutations in these five targets. The results of the above analysis were mostly consistent with previous literature reports.
To further clarify the relationship between YQHY and ferroptosis, we selected ACSL4, which is closely related to ferroptosis, as the target of this topic, and the JAK2–STAT3 pathway as the pathway of this topic to explore whether YQHY could induce ferroptosis in GC by influencing the expression of ACSL4, JAK2, and STAT3. The results showed that YQHY had a toxic effect on AGS, and with increasing concentration, the inhibition rate of AGS showed a gradually increasing trend. The increase in MDA and the decrease in GSH synthesis are the main characteristics of ferroptosis. We found that the MDA content in the YQHY group was the highest compared with the other three groups (P <0.05), and YQHY reversed the ferroptosis inhibition induced by the ferroptosis inhibitor liproxstatin-1. The GSH content in the YQHY group was the lowest compared with the other three groups (P <0.05), and the GSH content in the YQHY + liproxstatin-1 group was lower than that in the control group, indicating that YQHY can reduce the intracellular GSH content and induce ferroptosis in AGS and that YQHY can reverse ferroptosis due to liproxstatin-1 inhibition.
The qRT-PCR results showed that the mRNA expression levels of JAK2, STAT3, and ACSL4 were higher in the YQHY group than in the control group. After the addition of liproxstatin-1, the mRNA expression of JAK2, STAT3, and ACSL4 was decreased compared with that in the YQHY group. The mRNA expression of JAK2, STAT3 and ACSL4 in the liproxstatin-1 group was significantly decreased relative to the control group.Western blot analysis confirmed that compared with the control group, the expression of JAK2, STAT3, p-STAT3, and ACSL4 protein in the YQHY group was higher than those in the other three groups, while the expression level of each protein in the liproxstatin-1 group was lower. It could be concluded that YQHY can induce ferroptosis in AGS by regulating the JAK2-STAT3 pathway and the expression of ACSL4. This illustrates that influencing the JAK2-STAT3 signaling pathway and expression of ACSL4 is one of the mechanisms of inducing ferroptosis in GC, and induction of ferroptosis may be one of the possible mechanisms of YQHY’s anti-recurrence and metastasis of GC.
5 Conclusion
The material basis of YQHY induced ferroptosis in GC is based on active compounds such as Quercetin, DIBP, DBP, Mipax, and Phaseol. The related mechanism was characterized by multiple targets and pathways. YQHY can induce ferroptosis in GC by affecting the JAK2–STAT3 pathway and the expression of ACSL4, and induction of ferroptosis may be one of the possible mechanisms of YQHY’s anti-recurrence and metastasis of GC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
PS designed the research. SS analyzed the data and wrote the paper. FW, SG, WH, JL, JZ, PG, SR, XC, and YL selected the materials. All authors read and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (nos. 81673918). Pilot GC project of clinical collaboration of traditional Chinese medicine and Western medicine on major difficult diseases in the state administration of traditional Chinese medicine; the 2019” Construction Project of Evidence-based Capacity for Traditional Chinese Medicine” (2019XZZX-ZL003) in state administration of traditional Chinese medicine; the Open Program of the Third Phase of the Program of Traditional Chinese Medicine (TCM) Advantageous Subjects (ZYX03KF020); and the Science and Technology Project of Jiangsu Provincial Administration of Traditional Chinese Medicine (ZD201803).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.820059/full#supplementary-material
Abbreviations
GC, gastric cancer; YQHY, Yiqi Huayu Decoction; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; OB, Oral bioavailability; DL, Drug-likeness; GO, Gene Ontology; BP, Biological process; CC, Cellular component; MF, Molecular function; FDR, Error Detection Rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, Protein–protein interaction; OS, Overall Survival; qRT-PCR, Quantitative Real-Time Polymerase Chain Reaction.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Zhang T, Chen H, Yin X, He Q, Man J, Yang X, et al. Changing Trends of Disease Burden of Gastric Cancer in China From 1990 to 2019 and Its Predictions: Findings From Global Burden of Disease Study. Chin J Cancer Res (2021) 33(1):11–26. doi: 10.21147/j.issn.1000-9604.2021.01.02
3. Yusefi AR, Bagheri Lankarani K, Bastani P, Radinmanesh M, Kavosi Z. Risk Factors for Gastric Cancer: A Systematic Review. Asian Pac J Cancer Prev (2018) 19(3):591–603. doi: 10.22034/APJCP.2018.19.3.591
4. Nie S, Yang G, Lu H. Current Molecular Targeted Agents for Advanced Gastric Cancer. Onco Targets Ther (2020) 13:4075–88. doi: 10.2147/OTT.S246412
5. Liang C, Zhang X, Yang M, Dong X. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater (2019) 31(51):e1904197. doi: 10.1002/adma.201904197
6. Liu Z, Zhao Q, Zuo ZX, Yuan SQ, Yu K, Zhang Q, et al. Systematic Analysis of the Aberrances and Functional Implications of Ferroptosis in Cancer. iScience (2020) 23(7):101302. doi: 10.1016/j.isci.2020.101302
7. Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a New Form of Cell Death: Opportunities and Challenges in Cancer. J Hematol Oncol (2019) 12(1):34. doi: 10.1186/s13045-019-0720-y
8. Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, et al. CAF Secreted miR-522 Suppresses Ferroptosis and Promotes Acquired Chemo-Resistance in Gastric Cancer. Mol Cancer (2020) 19(1):43. doi: 10.1186/s12943-020-01168-8
9. Lee JY, Nam M, Son HY, Hyun K, Jang SY, Kim JW, et al. Polyunsaturated Fatty Acid Biosynthesis Pathway Determines Ferroptosis Sensitivity in Gastric Cancer. Proc Natl Acad Sci U.S.A. (2020) 117(51):32433–42. doi: 10.1073/pnas.2006828117
10. Hao S, Yu J, He W, Huang Q, Zhao Y, Liang B, et al. Cysteine Dioxygenase 1 Mediates Erastin-Induced Ferroptosis in Human Gastric Cancer Cells. Neoplasia (2017) 19(12):1022–32. doi: 10.1016/j.neo.2017.10.005
11. Sha W, Hu F, Xi Y, Chu Y, Bu S. Mechanism of Ferroptosis and Its Role in Type 2 Diabetes Mellitus. J Diabetes Res (2021) 2021:9999612. doi: 10.1155/2021/9999612
12. Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, et al. NFS1 Undergoes Positive Selection in Lung Tumours and Protects Cells From Ferroptosis. Nature (2017) 551(7682):639–43. doi: 10.1038/nature24637
13. Shu P, Tang H, Zhou B, Wang R, Xu Y, Shao J, et al. Effect of Yiqi Huayu Jiedu Decoction on Stages II and III Gastric Cancer: A Multicenter, Prospective, Cohort Study. Med (Baltimore) (2019) 98(47):e17875. doi: 10.1097/MD.0000000000017875
14. Zhang R, Zhu X, Bai H, Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol (2019) 10:123. doi: 10.3389/fphar.2019.00123
15. Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, et al. A Novel Chemometric Method for the Prediction of Human Oral Bioavailability. Int J Mol Sci (2012) 13(6):6964–82. doi: 10.3390/ijms13066964
16. Li J, Zhao P, Li Y, Tian Y, Wang Y. Systems Pharmacology-Based Dissection of Mechanisms of Chinese Medicinal Formula Bufei Yishen as an Effective Treatment for Chronic Obstructive Pulmonary Disease. Sci Rep (2015) 5:15290. doi: 10.1038/srep15290
17. Zhou N, Bao J. FerrDb: A Manually Curated Resource for Regulators and Markers of Ferroptosis and Ferroptosis-Disease Associations. Database (Oxford) (2020) 2020:80. doi: 10.1093/database/baaa021
18. Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci (2019) 20(18):11–3. doi: 10.3390/ijms20184331
19. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
20. Digre A, Lindskog C. The Human Protein Atlas-Spatial Localization of the Human Proteome in Health and Disease. Protein Sci (2021) 30(1):218–33. doi: 10.1002/pro.3987
21. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6(269):pl1. doi: 10.1126/scisignal.2004088
22. Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci (2020) 21(11):57. doi: 10.3390/ijms21114012
23. Huang W, Tang H, Wen F, Lu X, Li Q, Shu P. Jianpi-Yangwei Decoction Inhibits DNA Damage Repair in the Drug Resistance of Gastric Cancer by Reducing FEN1 Expression. BMC Complement Med Ther (2020) 20(1):196. doi: 10.1186/s12906-020-02983-8
24. Reyes-Farias M, Carrasco-Pozo C. The Anti-Cancer Effect of Quercetin: Molecular Implications in Cancer Metabolism. Int J Mol Sci (2019) 20(13):34. doi: 10.3390/ijms20133177
25. Massi A, Bortolini O, Ragno D, Bernardi T, Sacchetti G, Tacchini M, et al. Research Progress in the Modification of Quercetin Leading to Anticancer Agents. Molecules (2017) 22(8):121. doi: 10.3390/molecules22081270
26. Tang SM, Deng XT, Zhou J, Li QP, Ge XX, Miao L. Pharmacological Basis and New Insights of Quercetin Action in Respect to its Anti-Cancer Effects. BioMed Pharmacother (2020) 121:109604. doi: 10.1016/j.biopha.2019.109604
27. Almatroodi SA, Alsahli MA, Almatroudi A, Verma AK, Aloliqi A, Allemailem KS, et al. Potential Therapeutic Targets of Quercetin, a Plant Flavonol, and Its Role in the Therapy of Various Types of Cancer Through the Modulation of Various Cell Signaling Pathways. Molecules (2021) 26(5):69. doi: 10.3390/molecules26051315
28. Wang Q, Chen Y, Lu H, Wang H, Feng H, Xu J, et al. Quercetin Radiosensitizes non-Small Cell Lung Cancer Cells Through the Regulation of miR-16-5p/WEE1 Axis. IUBMB Life (2020) 72(5):1012–22. doi: 10.1002/iub.2242
29. Ezzati M, Yousefi B, Velaei K, Safa A. A Review on Anti-Cancer Properties of Quercetin in Breast Cancer. Life Sci (2020) 248:117463. doi: 10.1016/j.lfs.2020.117463
30. Li Y, Wang Z, Jin J, Zhu SX, He GQ, Li SH, et al. Quercetin Pretreatment Enhances the Radiosensitivity of Colon Cancer Cells by Targeting Notch-1 Pathway. Biochem Biophys Res Commun (2020) 523(4):947–53. doi: 10.1016/j.bbrc.2020.01.048
31. Lei CS, Hou YC, Pai MH, Lin MT, Yeh SL. Effects of Quercetin Combined With Anticancer Drugs on Metastasis-Associated Factors of Gastric Cancer Cells: In Vitro and In Vivo Studies. J Nutr Biochem (2018) 51:105–13. doi: 10.1016/j.jnutbio.2017.09.011
32. Yang X, Zhang W, Zhao Z, Li N, Mou Z, Sun D, et al. Quercetin Loading CdSe/ZnS Nanoparticles as Efficient Antibacterial and Anticancer Materials. J Inorg Biochem (2017) 167:36–48. doi: 10.1016/j.jinorgbio.2016.11.023
33. Thuy NTT, Lee JE, Yoo HM, Cho N. Antiproliferative Pterocarpans and Coumestans From Lespedeza Bicolor. J Nat Prod (2019) 82(11):3025–32. doi: 10.1021/acs.jnatprod.9b00567
34. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep (2019) 28(5):1370–1384 e1375. doi: 10.1016/j.celrep.2019.08.061
35. Stoehr R, Weisser R, Wendler O, Giedl J, Daifalla K, Gaisa NT, et al. P53 Codon 72 Polymorphism and Risk for Squamous Cell Carcinoma of the Penis: A Caucasian Case-Control Study. J Cancer (2018) 9(22):4234–41. doi: 10.7150/jca.26050
36. Rimkus SA, Wassarman DA. A Pharmacological Screen for Compounds That Rescue the Developmental Lethality of a Drosophila ATM Mutant. PloS One (2018) 13(1):e0190821. doi: 10.1371/journal.pone.0190821
37. Zheng C, Zhou Y, Huang Y, Chen B, Wu M, Xie Y, et al. Effect of ATM on Inflammatory Response and Autophagy in Renal Tubular Epithelial Cells in LPS-Induced Septic AKI. Exp Ther Med (2019) 18(6):4707–17. doi: 10.3892/etm.2019.8115
38. Nie K, Zheng Z, Wen Y, Shi L, Xu S, Wang X, et al. Construction and Validation of a TP53-Associated Immune Prognostic Model for Gastric Cancer. Genomics (2020) 112(6):4788–95. doi: 10.1016/j.ygeno.2020.08.026
39. Santos JC, Gambeloni RZ, Roque AT, Oeck S, Ribeiro ML. Epigenetic Mechanisms of ATM Activation After Helicobacter Pylori Infection. Am J Pathol (2018) 188(2):329–35. doi: 10.1016/j.ajpath.2017.10.005
40. Ma C, Wang X, Yang F, Zang Y, Liu J, Wang X, et al. Circular RNA Hsa_Circ_0004872 Inhibits Gastric Cancer Progression via the miR-224/Smad4/ADAR1 Successive Regulatory Circuit. Mol Cancer (2020) 19(1):157. doi: 10.1186/s12943-020-01268-5
41. Ruoming W, Zhen Y, Tengteng Z, Jisheng H. Tumor Suppressor microRNA-31 Inhibits Gastric Carcinogenesis by Targeting Smad4 and SGPP2. Cancer Gene Ther (2015) 22(12):564–72. doi: 10.1038/cgt.2015.41
42. Desai SJ, Prickril B, Rasooly A. Mechanisms of Phytonutrient Modulation of Cyclooxygenase-2 (COX-2) and Inflammation Related to Cancer. Nutr Cancer (2018) 70(3):350–75. doi: 10.1080/01635581.2018.1446091
43. Lin XM, Li S, Zhou C, Li RZ, Wang H, Luo W, et al. Cisplatin Induces Chemoresistance Through the PTGS2-Mediated Anti-Apoptosis in Gastric Cancer. Int J Biochem Cell Biol (2019) 116:105610. doi: 10.1016/j.biocel.2019.105610
44. Quan J, Bode AM, Luo X. ACSL Family: The Regulatory Mechanisms and Therapeutic Implications in Cancer. Eur J Pharmacol (2021) 909:174397. doi: 10.1016/j.ejphar.2021.174397
45. Cruz-Gil S, Sanchez-Martinez R, Gomez de Cedron M, Martin-Hernandez R, Vargas T, Molina S, et al. Targeting the Lipid Metabolic Axis ACSL/SCD in Colorectal Cancer Progression by Therapeutic miRNAs: miR-19b-1 Role. J Lipid Res (2018) 59(1):14–24. doi: 10.1194/jlr.M076752
46. Tang Y, Zhou J, Hooi SC, Jiang YM, Lu GD. Fatty Acid Activation in Carcinogenesis and Cancer Development: Essential Roles of Long-Chain Acyl-CoA Synthetases. Oncol Lett (2018) 16(2):1390–6. doi: 10.3892/ol.2018.8843
47. Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a Biomarker and Contributor of Ferroptosis. Biochem Biophys Res Commun (2016) 478(3):1338–43. doi: 10.1016/j.bbrc.2016.08.124
48. Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, et al. Intercellular Interaction Dictates Cancer Cell Ferroptosis via NF2-YAP Signalling. Nature (2019) 572(7769):402–6. doi: 10.1038/s41586-019-1426-6
49. Sanchez-Martinez R, Cruz-Gil S, Garcia-Alvarez MS, Reglero G, Ramirez de Molina A. Complementary ACSL Isoforms Contribute to a non-Warburg Advantageous Energetic Status Characterizing Invasive Colon Cancer Cells. Sci Rep (2017) 7(1):11143. doi: 10.1038/s41598-017-11612-3
50. Sha R, Xu Y, Yuan C, Sheng X, Wu Z, Peng J, et al. Predictive and Prognostic Impact of Ferroptosis-Related Genes ACSL4 and GPX4 on Breast Cancer Treated With Neoadjuvant Chemotherapy. EBioMedicine (2021) 71:103560. doi: 10.1016/j.ebiom.2021.103560
51. Ye X, Zhang Y, Wang X, Li Y, Gao Y. Tumor-Suppressive Functions of Long-Chain Acyl-CoA Synthetase 4 in Gastric Cancer. IUBMB Life (2016) 68(4):320–7. doi: 10.1002/iub.1486
52. Yuan K, Ye J, Liu Z, Ren Y, He W, Xu J, et al. Complement C3 Overexpression Activates JAK2/STAT3 Pathway and Correlates With Gastric Cancer Progression. J Exp Clin Cancer Res (2020) 39(1):9. doi: 10.1186/s13046-019-1514-3
53. Liang L, Hui K, Hu C, Wen Y, Yang S, Zhu P, et al. Autophagy Inhibition Potentiates the Anti-Angiogenic Property of Multikinase Inhibitor Anlotinib Through JAK2/STAT3/VEGFA Signaling in non-Small Cell Lung Cancer Cells. J Exp Clin Cancer Res (2019) 38(1):71. doi: 10.1186/s13046-019-1093-3
54. Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, et al. The JAK2/STAT3/CCND2 Axis Promotes Colorectal Cancer Stem Cell Persistence and Radioresistance. J Exp Clin Cancer Res (2019) 38(1):399. doi: 10.1186/s13046-019-1405-7
Keywords: ferroptosis, GC, network pharmacology, molecular docking, bioinformatics, Yiqi Huayu decoction
Citation: Song S, Wen F, Gu S, Gu P, Huang W, Ruan S, Chen X, Zhou J, Li Y, Liu J and Shu P (2022) Network Pharmacology Study and Experimental Validation of Yiqi Huayu Decoction Inducing Ferroptosis in Gastric Cancer. Front. Oncol. 12:820059. doi: 10.3389/fonc.2022.820059
Received: 06 December 2021; Accepted: 17 January 2022;
Published: 14 February 2022.
Edited by:
Jian Chen, Guilin Medical University, ChinaReviewed by:
Hongguang Ding, Guangdong Provincial People’s Hospital, ChinaAli Hafez El-Far, Damanhour University, Egypt
Bichun Li, Yangzhou University, China
Copyright © 2022 Song, Wen, Gu, Gu, Huang, Ruan, Chen, Zhou, Li, Liu and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Shu, shupengsp@njucm.edu.cn
 Siyuan Song
Siyuan Song Fang Wen2
Fang Wen2 Wenjie Huang
Wenjie Huang