- 1Department of Clinical Oncology, Xijing Hospital, Fourth Military Medical University, Xi’an, China
- 2Department of Internal Medicine, 63650 Military Hospital, Urumqi, China
- 3Department of Respiratory Medicine, Xinjiang Command General Hospital of Chinese People’s Liberation Army, Urumqi, China
- 4Department of Pathology, Tangdu Hospital, Fourth Military Medical University, Xi’an, China
- 5Department of Hematology, Xinjiang Command General Hospital of Chinese People’s Liberation Army, Urumqi, China
- 6Department of Endocrinology, Xijing Hospital, Fourth Military Medical University, Xi’an, China
Altered expression levels of the proteins that regulate N6-methyladenosine (m6A) RNA methylation, including methyltransferase-like 14 (METTL14), are associated with cancer development. Based on our analysis of m6A methylation regulators using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets, we focused on the regulatory role of METTL14 in ovarian cancer. We performed bioinformatics and survival analyses with these datasets and also used METTL14-overexpressing SKOV-3 ovarian cancer cells for in vitro studies. Trophinin associated protein (TROAP) siRNA and treatment with or without actinomycin D was used in the cells for qRT-PCR, western blot, cDNA microarray, cell viability, colony formation, luciferase gene reporter, methylated RNA immunoprecipitation (MeRIP)-qPCR, total RNA methylation, and RNA stability assays. Additionally, ovarian cancer and normal tissue samples were analyzed by immunohistochemistry, qRT-PCR, and western blot assays. The TCGA and GEO data confirmed copy number variations (CNVs) of these m6A RNA methylation regulators in ovarian cancer tissues. Furthermore, reduced METTL14 expression was associated with alterations in CNVs as well as poor patient survival in ovarian cancer. Moreover, the METTL14 and m6A RNA methylation levels were both significantly reduced in ovarian cancer tissues than in normal tissues. Restoration of METTL14 expression suppresses ovarian cancer cell proliferation by inhibition of TROAP expression. Further in vivo and in vitro experiments confirmed that METTL14 is a negative regulator of ovarian cancer cell proliferation via TROAP expression and that m6A RNA methylation regulates TROAP mRNA stability. In conclusion, METTL14 overexpression decreased ovarian cancer proliferation by inhibition of TROAP expression via an m6A RNA methylation-dependent mechanism.
Introduction
N6-methyladenosine (m6A) RNA methylation is the most prevalent RNA modification in eukaryotic cells and regulates many functions, including RNA splicing, translocation, stability, and protein translation (1). This modification is catalyzed by RNA methyltransferases known as writers, such as methyltransferase-like 14 (METTL14), and is removed by demethylases known as erasers, such as fat mass and obesity-associated protein (FTO). Additionally, m6A is recognized by m6A-binding proteins known as readers, such as IGF2BPs, which promote RNA stability and translation (2). Altogether, these proteins determine the dynamic and reversible regulation of m6A RNA modification (3), which can become disrupted if the expression or function of these proteins are altered, resulting in the development of human diseases and cancers (4). Aberrant expression of proteins that regulate m6A RNA methylation, such as METTL14, is associated with the development of various human cancers, including breast cancer (1), hepatocellular carcinoma (5), colorectal cancer (6), pancreatic cancer (7), renal clear cell carcinoma (8), bladder cancer (9), and head and neck squamous cell carcinoma (10). Furthermore, the increased expression of m6A RNA methylation regulators is associated with the tumorigenesis and metastasis of ovarian cancer (11) as well as gastrointestinal tract cancer (12). Therefore, a better understanding of these m6A writers, erasers, and readers will provide the information necessary to clarify the biological functions and potential mechanisms of m6A.
Ovarian cancer has the highest death rate of all gynecological malignancies due to its diagnosis at advanced stages as well as a lack of effective clinical treatments (13). Epigenetic m6A methylation has been reported to occur in ovarian cancer (14). However, ovarian cancer is a heterogeneous disease that is divided into many subtypes with different etiologies, pathogeneses, and prognoses. Therefore, further ovarian cancer studies are warranted to improve our knowledge on ovarian tumorigenesis and to reduce the death rate of this deadly cancer in women. Previous ovarian cancer studies have used different approaches, including analyses of aberrant gene copy number variations (CNVs) or potential molecular mechanisms (15), to better understand the development and progression of ovarian cancer. Therefore, in this study, we assessed 23 regulators of m6A methylation using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets to perform bioinformatics and survival analyses. Based on these analyses, we decided to focus on METTL14 in ovarian cancer tissues and cells. We transfected ovarian cancer SKOV-3 and A2780 cells with a METTL14 expression vector or Trophinin associated protein (TROAP) siRNA. The cells were then treated with or without actinomycin D for various assays to understand the in vivo and in vitro regulatory roles of METTL14 in ovarian cancer as well as the underlying molecular events. The results of our study provide important insights into the m6A RNA modification by METTL14 in the regulation of ovarian cancer.
Materials and Methods
Data Resources and Bioinformatic Analysis
The transcriptomic RNA-seq data of 295 ovarian cancer patients was downloaded from cBioPortal for cancer geonomics (https://www.cbioportal.org/study/summary?id=ov_tcga_pan_can_atlas_2018), combined TCGA pancancer atlas studies and followed with clinical data in website. The data on the CNVs of 23 m6A RNA methylation regulators in ovarian cancer was collected, according to previous studies (16). The datasets on the mRNA expression levels of m6A RNA methylation regulators as well as METTL14 were downloaded from the GEO database (GSE14407 and GSE119168, respectively). Next, the data was analyzed using the GEO database-supported ClueGO+CluePedia tool in Cytoscape for the cluster-envisioning characterization of the biological processes (i.e., dysregulation or downregulation) of these m6A RNA methylation genes in ovarian cancer and the functional categories of the gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using the DAVID bioinformatics resource website (https://david.ncifcrf.gov/). The GO terms of the GEO data were assessed using the PANTHER website (http://www.pantherdb.org/) for functional gene categories and associations between genes and the corresponding GO classifications. The R visualization package GOPlot was then used to obtain a better visualization of the relationships between the genes and the selected functional categories. Next, the correlation between METTL14 and TROAP expression levels was obtained from the GEO dataset (GSE28724), and the datasets and tools provided by KMplot were used to analyze the survival of patients with high and low METTL14 levels (http://kmplot.com/analysis/index). In addition, the Human Protein Atlas database was used to verify the translation of TROAP (http://www.proteinatlas.org/), and the m6A RNA methylation sites were predicted by using the m6AVar prediction website (http://m6avar.renlab.org/index.html).
Tissue Samples and Cell Culture
Normal (n = 20) and cancerous (n = 20) ovarian tissue samples were collected at the Xijing Hospital (Xi’an, China) between May and November 2019. All patients were histologically diagnosed with epithelial ovarian cancer. The tissue specimens were obtained immediately after resection and then divided into three portions for RNA isolation, protein extraction, and storage in liquid nitrogen for further analyses. The experimental protocol was approved by the Ethics Committee of Xijing Hospital, The Fourth Military Medical University. The ovarian cancer SKOV-3 and A2780 cell lines was obtained from the Shanghai Institutes of Biological Sciences (Chinese academy of sciences, Shanghai, China) and maintained in Roswell Park Memorial Institute 1640 Medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (10 U/mL), and streptomycin (50 µg/mL) at 37°C in a 5% CO2 atmosphere.
Plasmid Construction, Lentivirus Production, and Cell Transduction
The pMD-18T plasmid carrying METTL14 cDNA was constructed by PCR amplification with the following primers: 5′-ATGGATAGCCGCTTGCAGGAGATCCGGG-3′ and 5′-TTATCGAGGTGGAAAGCCACCTCTGTG-3′. The resulting product was then subcloned into pLenti-6.3 (Invitrogen, Carlsbad, CA, USA) for lentiviral production. The lentivirus was packaged and used to transduce the SKOV-3 cells, according to the standard protocols recommended by the manufacturer. To generate the ovarian cancer cell line with stable METTL14 expression, SKOV-3 cells were grown and transduced with the lentivirus carrying the METTL14 expression vector, or only the vector as a control, and then treated with puromycin (1.5 ug/mL) for 2 weeks for cell selection. The METTL14 cDNA was cloned into the pC3 plasmid (Invitrogen) for transfections.
Small-Interfering RNA and Cell Transfection
TROAP siRNA (si-TROAP) and Wilms tumor-associated protein (WTAP) siRNA (si-WTAP) was obtained from GenePharma Inc. (Shanghai, China). Briefly, SKOV-3 cells were cultured and transiently transfected with si-TROAP or negative control oligonucleotides at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen) for 24–48 h. The cells were then collected for quantitative reverse transcription (qRT)-PCR and western blot analyses (17). The siRNA sequences were as follows: (TROAP) 5′-GTAGGATTGAGCCTGAGAT-3′, (WTAP) 5′-GGAGGUAGTGGUUACGUAAAU-3′, and (negative control) 5′-UUCUCCGAACGUGUAACGUTT-3′ (18).
Plasmid Construction and Luciferase Reporter Assay
The pMD-18T vectors carrying either the TROAP or METTL14 3′-untranslated region (UTR) were constructed by PCR amplification. The 5′-CCCTGCCCCTGTGGCCCAGCCCTTG-3′ and 5′-CTGAGTGTTTTAACAGTCCAG-3′ primer sequences were used to amplify the 3′-UTR of the 575-bp TROAP fragment containing the m6A methylation site. The product was then subcloned into the pMD-18T vector (Takara bio, Shiga, Japan). The vector was amplified and sequenced before use. Similarly, the METTL14 3′-UTR fragments were also cloned into the pMD-18T vector. These pMD-18T vectors were used to release the inserts, which were then subcloned into pGL3 vectors (Promega, Madison, WI, USA). Additionally, the wild-type METTL14 3′-UTR m6A methylation site (GGA; pGL3-WT-UTR) was mutated to CGT (pGL3-Mut-UTR). After amplification and DNA sequence confirmation, these vectors were used to transfect SKOV-3 cells. Specifically, SKOV-3 cells were seeded into 48-well plates as duplicates, grown for 24 h, and then co-transfected with 1 µg pGL3-WT-UTR (TROAP 3′-UTR containing the m6A methylation site) or pGL3-Mut-UTR as well as 1 µg renilla luciferase vector pRL-TK for 48 h. The total protein lysates were obtained using a kit from Promega and used for the luciferase assay, according to the dual-luciferase reporter assay method (19). Firefly luciferase activity was used as a normalization control for each sample.
qRT-PCR
Total cellular RNA was isolated from cells and tissues using Trizol reagent (Invitrogen), and cDNA was obtained by reverse transcription using SuperScript II reverse transcriptase (Invitrogen), according to the manufacturer’s recommended protocol (20). Next, the relative mRNA expression levels of METTL14, TROAP, and GAPDH (as a normalization control) were assessed using SYBR Premix Ex Taq II (Takara, Dalian, China). The primer sequences were as follows: (METTL14) 5′-GAACACAGAGCTTAAATCCCCA-3′ and 5′-TGTCAGCTAAACCTACATCCCTG-3′, (TROAP) 5′-CCTCCGGGGTGTATCTCCTAC-3′ and 5′-ACGGCGCACGATGTAACAG-3′, (WTAP) 5′-CTTCCCAAGAAGGTTCGATTGA-3′ and 5′-TCAGACTCTCTTAGGCCAGTTAC-3′, (GAPDH) 5′-GGAGCGAGATCCCTCCAAAAT-3′ and 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western Blot Analysis
Western blot analysis was used to determine changes in protein levels, according to a previous study (14). The primary antibodies used were rabbit polyclonal anti-TROAP (Cat. #HPA044102; Sigma-Aldrich, St. Louis, MO, USA), anti-METTL14 (Cat. #ab220031; Abcam, Cambridge, MA, USA), anti-cyclin D1 (Cat. #ab16663; Abcam), anti-survivin (Cat. # ab76424; Abcam), anti-p-AKT (Cat. #ab38449; Abcam), anti-WTAP (Cat. #ab195380; Abcam), and polyclonal rabbit anti-TROAP (Cat. #13634-1-AP; Proteintech, Rosemont, IL 60018, USA). The Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA) was used for protein imaging.
Cell Viability and Colony Formation Assays
Cell viability was measured using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega). Briefly, tumor cells (5 × 103) were seeded into the individual wells of a 96-well plate and cultured for 24 h. The cells were then subjected to the assay, according to the manufacturer’s protocol. Furthermore, the colony formation capacities of SKOV-3 and A2780 cells after transfection were assayed, as previously described (21).
Methylated RNA Immunoprecipitation-qPCR
MeRIP-qPCR was performed to quantify the m6A-modified levels of TROAP. Specifically, total cellular RNA was isolated from SKOV-3 cells using Trizol regent. The RNA samples (100 μg for each reaction) were incubated with 3 μg anti-m6A (Cat. #208577; Abcam, Cambridge, MA, USA) or anti-IgG (Cat. #172730; Abcam) that were conjugated with A/G magnetic beads in an immunoprecipitation buffer containing RNase inhibitor and protease inhibitors. Next, RNA from the mixtures was collected through centrifugation, cleaned using a phenol-chloroform solution, and eluted with elution buffer. The RNA samples (10 ng for each reaction) were then subjected to reverse transcription using Superscript III random hexamers (Invitrogen, Shanghai, China), and the cDNA was further analyzed by qPCR. The TROAP primer sequences that were used for MeRIP-qPCR were 5′-TTGCGGCGTCTCACCGTTCAACCT-3′ and 5′-GCCTCCATTAAGAGGGACACACTGG-3′. TROAP mRNA levels were quantified using the 2-ΔΔCt associated sample addition method for fold enrichment.
RNA Stability Assay
SKOV-3 cells with or without METTL14 overexpression were treated with actinomycin D at a final concentration of 5 ug/mL for 0.5, 1, 2, or 4 h. The cells were then collected for total RNA isolation for qRT-PCR analysis. The turnover rate and half-life of mRNAs were then determined according to a previous study (22).
Tissue Microarray and Immunohistochemistry Assays
A TMA containing 12 healthy endometrial tissues and 58 ovarian cancer tissues was purchased from Servicebio (Wuhan, China; Cat. #OC-1601). Immunohistochemistry was used to assess the expression of METTL14. Briefly, the TMA sections were deparaffinized in xylene, rehydrated in a series of ethanol solutions, and then subjected to antigen retrieval in 10 mM sodium citrate/0.05% Tween 20 (pH 6). The TMA sections were then incubated with anti-METTL14 antibody (Cat. #ab220030; Abcam), and the vectastain elite ABC HRP and DAB substrate kits were used for immunohistochemical detection (Vector laboratories, CA, US), according to the manufacturer’s recommended protocols. Hematoxylin was used to stain the cell nuclei. Next, the immunostained TMA sections were reviewed, photographed, and quantified using the ImageJ plugin IHC profiler to determine the grade of epithelial component staining (0, no staining; 1, low positive staining; 2, positive staining; 3, high positive staining).
Immunohistochemistry and Fluorescence In Situ Hybridization
IHC performed with ovarian cancer tissue microarray and animal tissue specimens. For IHC, the samples were incubated with primary antibody, followed by incubation with HRP-conjugated secondary antibody. METTL14 FISH was performed to analyze the copy numbers of the METTL14 gene in ovarian cancer and normal tissue samples. Briefly, tissue sections were deparaffinized in xylene, rehydrated in a series of ethanol solutions, and then subjected to FISH with the METTL14 FISH probe (1–2.5 nM) in a buffer containing 2× saline-sodium citrate, 25% formamide, 10% dextran sulfate, and 1 mg/mL yeast in a humidified chamber for 3–4 h at 42°C, according to the manufacturer’s recommended protocol. The specific FISH signal was detected as brown punctate dots. Images were captured using a Nikon 90i microscope-equipped orca ER camera (Hamamatsu Photonics, Hamamatsu, Japan) with a 60×, 1.4 N.A. VC objective lens and were quantified using the Volocity visualization software (Perkin elmer, Waltham, MA, USA).
Animal Experiments
Six-week-old BALB/c female nude mice were housed in a pathogen-free animal facility under normal conditions. All animal experimental procedures were approved by the Animal Care Committee of the Fourth Military Medical University. SKOV-3 cells (1 ×106) stable transfected with METTL14 or control lentivirus, were injected subcutaneously into the right flank of BALB/c nude mice. Tumor growth was monitored every 5 days for a total period of 25-40 days.
Statistical Analysis
Continuous variable data were summarized as the mean ± standard error of the mean from at least three independent experiments and analyzed using Student’s t-tests. Associations of the changes in the expression levels of m6A RNA methylation regulators with the clinicopathological or distinguishing molecular features of cancer patients were assessed using the χ2 or Fisher’s exact tests. One-way analysis of variance (ANOVA) or Student’s t-tests were performed to analyze individuals who were clustered by clinical or molecular-pathological criteria in relation to the ovarian cancer signature incidence rates for specific clinical conditions. Kaplan-Meier curves and the log-rank test were used to analyze the significance of m6A RNA methylation, CNVs, and mutations in association with overall patient survival. Survival information was acquired by patient follow-up, and the information from the last follow-up was used for these analyses. All statistical analyses were performed using Prism version 7.0 or R version 3.4.1 (https://www.r-project.org/) of GraphPad (San Diego, CA, USA). A P-value ≤ 0.05 was considered to be statistically significant.
Results
METTL14 Was Downregulated in Ovarian Cancer and Predicted Poor Patient Survival
In this study, we assessed the TCGA datasets for the mutations of each individual m6A RNA methylation regulator in ovarian cancer, including AlkB homolog 5 (ALKBH5), E74 like ETS transcription factor 3 (ELF3), FMRP translational regulator 1 (FMR1), FTO, Heterogeneous nuclear ribonucleoprotein (HNRNP)A2/B1, HNRNPC, Insulin like growth factor 2 mRNA binding protein (IGF2BP) 1, IGF2BP2, IGF2BP3, Vir like m6A methyltransferase associated (Also known as KIAA1429), Leucine rich pentatricopeptide repeat containing (LRPPRC), METTL3, METTL14, RNA binding motif protein (RBM) 15, RBM15B, RBMX, WTAP, YTH N6-methyladenosine RNA-binding protein (YTHDF)1, YTHDF2, YTHDF3, YTH domain-containing (YTHDC)1, YTHDC2 and Zinc finger CCCH-type containing 13 (ZC3H13). We analyzed the CNVs of these m6A RNA methylation regulators in 295 ovarian cancer patients who completed the recommended diagnostic next-generation sequencing-based gene panel test.
We found a significant association between the CNVs of the 23 m6A RNA methylation regulators, including METTL14, and their mRNA expression levels (Figure 1A and Supplemental Figure S1). Our data demonstrated that increased copy numbers coincided with elevated mRNA expression levels, whereas decreased copy numbers coincided with reduced mRNA expression levels. To verify this relationship between CNVs and mRNA levels, we evaluated the expression of 10 key m6A RNA methylation regulators in normal and cancerous ovarian tissues. Because a lot of shallow deletion was shown in METTL14, we hypothesized that the reduction in its copy number would result in decreased METTL14 mRNA expression levels. Among the 10 key m6A RNA methylation regulators, the GEO data only showed a reduction in the METTL14 mRNA levels in the ovarian cancer tissues (Figure 1B), which was associated with a lower METTL14 copy number. Through further analysis of the comparative genomic hybridization data using high-resolution microarrays, we found lower METTL14 copy numbers in ovarian cancer tissues than in normal ovarian tissues (Figure 1C). Immunohistochemical analysis of the METTL14 expression pattern using the ovarian cancer tissue array was consistent with the online database data, showing reduced METTL14 expression in ovarian cancer tissues (Figures 1D, E). Moreover, Kaplan-Meier analysis showed that patients with reduced METTL14 expression had a lower overall survival (Figure 1F).
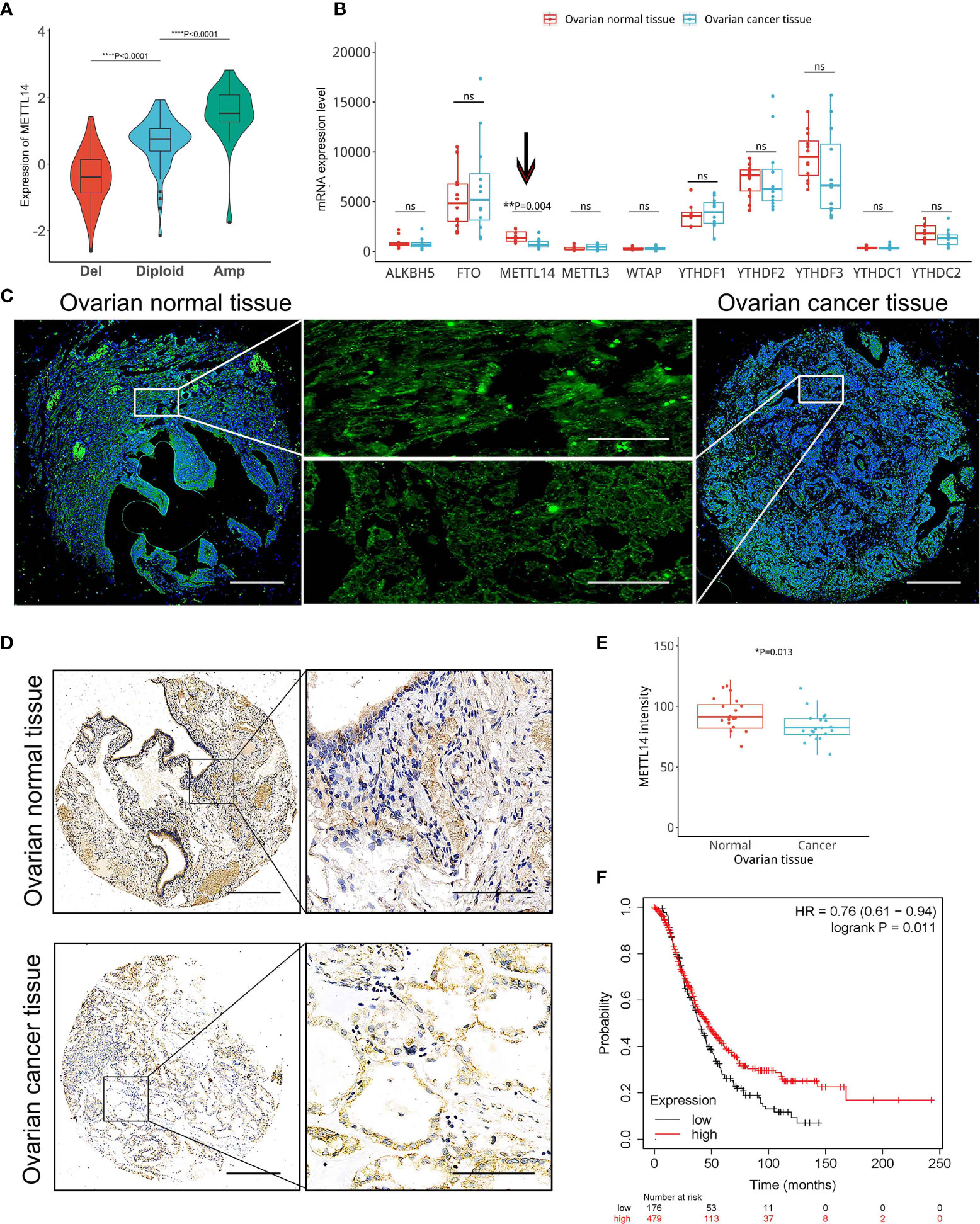
Figure 1 Correlation among reduced METTL14 copy number variations (CNVs) and decreased METTL14 mRNA and protein expression levels in ovarian cancer tissues. (A) The TCGA dataset and available tools were used for the correlation analysis of the METTL14 copy number (deletion, diploid and amplification) and mRNA expression levels (****P < 0.0001 by ANOVA). (B) Expression of METTL14 in ovarian cancer tissues vs. normal tissues in the GEO database (GSE14407 with METTL14, probe ID 235552_at, **P = 0.004). ns, not significant. (C) FISH images of METTL14 copy numbers in normal and ovarian cancer tissues. Magnification: 50 × (left and right) and 400 × (middle). (D) Immunohistochemistry of METTL14 expression in the tissue microarray of normal and ovarian cancer tissues. Magnification: 50 × (left) and 400 × (right). (E) Summarized data of (D) (*P=0.013). Graphical data represented as the mean ± SD. (F) Kaplan-Meier overall survival curves stratified by METTL14 expression levels in 655 ovarian cancer patients. The mean METTL14 mRNA value was used to assign patients to two subgroups (ID 236060_at). The log-rank test was used to calculate the differences between the two groups (*P = 0.011).
Downregulated METTL14 Induced Aberrant m6A Modification and May Be Correlated With Tumor Cell Proliferation
The m6A modification is mainly catalyzed by m6A methyltransferases and demethylases (23). Therefore, we speculated that abnormal m6A modification could be induced by an imbalance between key m6A methyltransferases or demethylases. Thus, we assessed the global m6A levels and found that the m6A levels were significantly lower in ovarian cancer tissues than in normal ovarian tissues (Figure 2A). Because METTL14 is an m6A RNA methylation writer, its reduced expression led to a decrease in its writer methyltransferase function. Therefore, we first assessed the METTL14 m6A methyltransferase levels using the GEO database (GSE119168) and found its levels to be significantly decreased in ovarian cancer tissues (Figure 2B). Our linear data analysis also showed a positive correlation between METTL14 mRNA levels and the global m6A levels in ovarian tissues (Figure 2C). These results indicated that downregulated METTL14 levels resulted in the low m6A RNA methylation levels in the ovarian cancer tissues. Further GEO database (GSE119168) analysis showed that most genes with low levels of m6A RNA methylation in ovarian cancer tissues were enriched in cell cycle, cell proliferation progression, mRNA stability, mRNA process and other pathways (Figures 2D–F). These results suggest that METTL14 may function as a negative regulator of tumor cell proliferation via its m6A methyltransferase function.
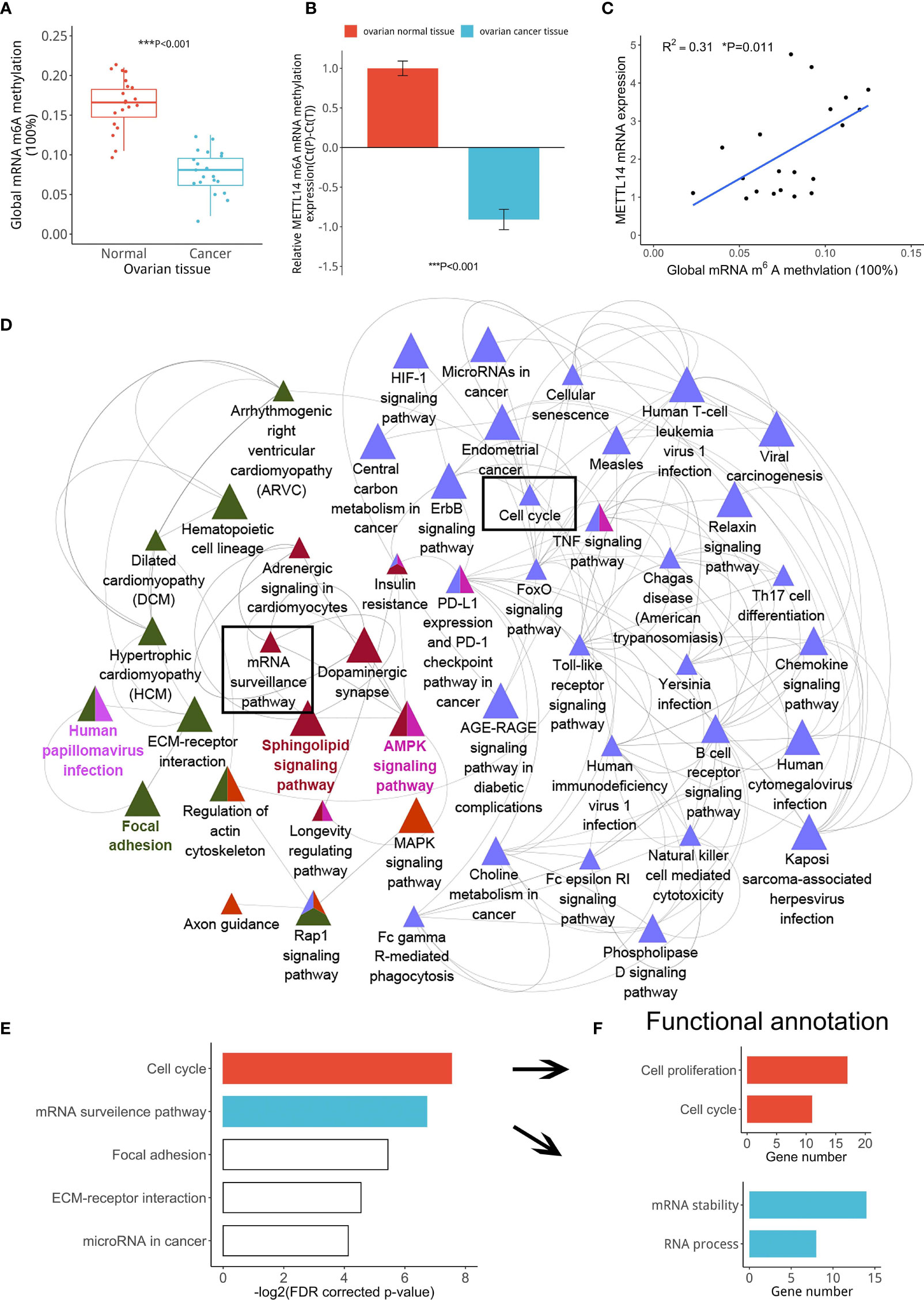
Figure 2 Correlation among reduced METTL14 expression and ovarian cancer cell proliferation. (A) Global mRNA m6A methylation levels from ovarian cancer or normal tissues (***P < 0.001). (B) MeRIP-seq data showing the METTL14 mRNA levels in the GEO database (GSE119168) with the RNA probe (ID: 118685469) (***P < 0.001). (C) Global mRNA m6A methylation levels of ovarian cancer tissues were found to be associated with METTL14 mRNA levels (*P = 0.011). Graphical data represented as the mean ± SD. (D) Pathway enrichment analysis using the ClueGO and CluePedia plugins found in the Cytoscape software. Cluster analysis showing the most enriched pathways according to the m6A RNA methylation expression levels using the GSE119168 data. (E) Classification of the gene categories from cluster analysis mainly included the cell cycle, mRNA surveillance pathway, and related signaling pathways. (F) KEGG gene functional annotation analysis using DAVID classified the gene categories and signaling pathways from two biological processes.
METTL14 Overexpression Inhibited the Proliferation of Ovarian Cancer Cells In Vitro
We transduced SKOV-3 cells with the lentivirus containing the METTL14 expression vector to evaluate the effect of METTL14 overexpression on the regulation of ovarian cancer cell proliferation in vitro. We confirmed that the METTL14 expression levels were increased after lentiviral transduction by qPCR and western blot (Figures 3A, B). We further demonstrated that the cell viability (Figure 3C) and colony formation capacity of the tumor cells were reduced with METTL14 overexpression (Figures 3D, E). Further distribution analysis by flow cytometry showed that SKOV-3 cells were arrested at the G1 phase of the cell cycle in response to METTL14 overexpression. In addition, the percentages of cells at the S and G2/M phases were decreased accordingly (Figure 3F). The similar results of colony formation assay and flow cytometry in A2780 cells (Figures 3G–I) further suggest that METTL14 may function as a negative regulator of ovarian cancer cell proliferation.
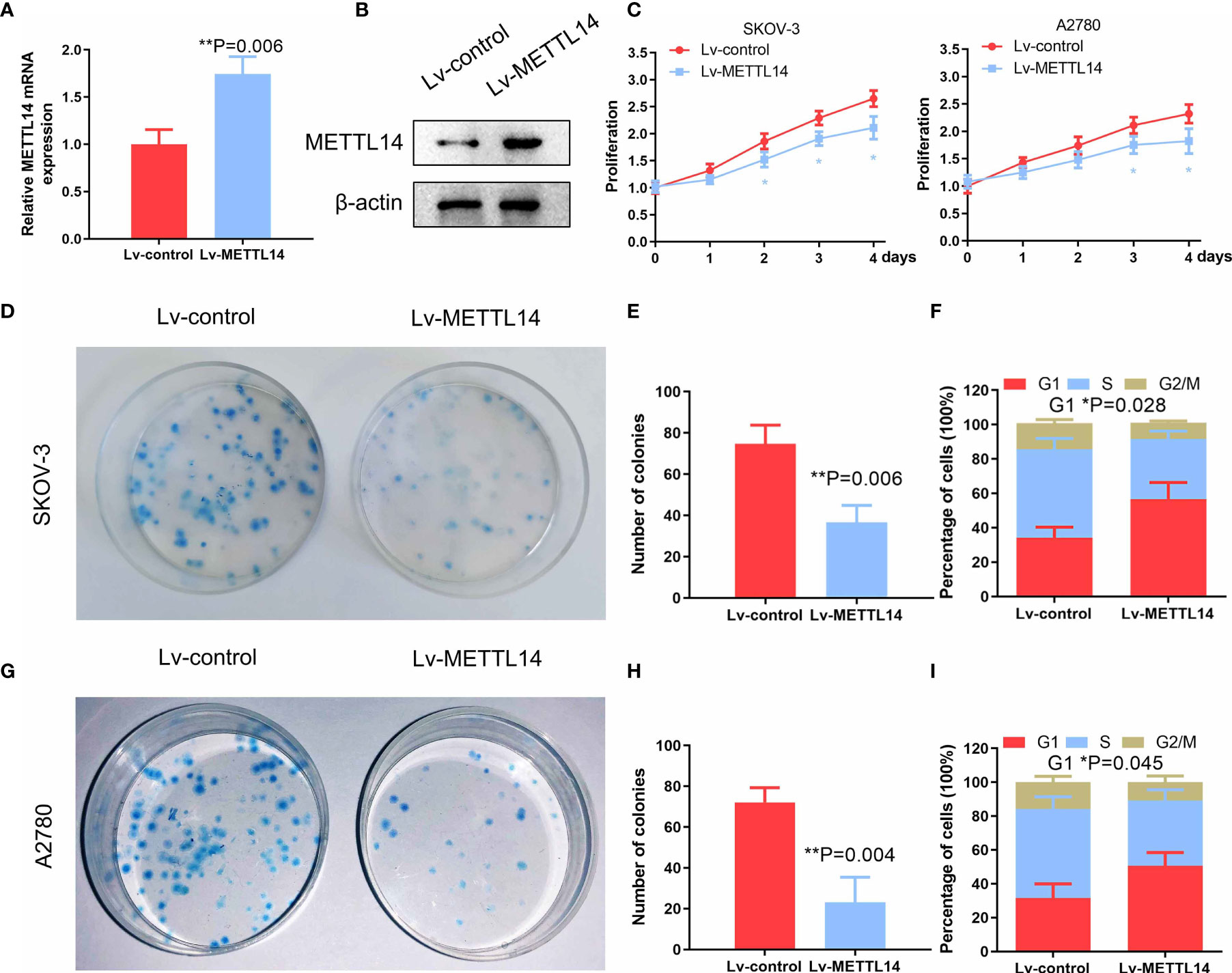
Figure 3 Inhibitory effects of METTL14 overexpression by lentiviral transduction on ovarian cancer cell proliferation and colony formation. (A) METTL14 mRNA levels of ovarian cancer cells after transduction with Lv-control and Lv-METTL14 (**P = 0.006). (B) Western blot of METTL14 protein levels of ovarian cancer cells after transduction with Lv-control and Lv-METTL14. (C) Lv-control and Lv-METTL14 transduced SKOV-3 and A2780 cells were seeded into 96-well plates, cultured for 4 days, and then subjected to an MTS cell proliferation assay (*P < 0.05). (D, E) Lv-control and Lv-METTL14 transduced SKOV-3 cells were seeded into 3.5-cm plates, cultured for 14 days, and then subjected to a colony formation assay (**P = 0.006). (F) Lv-control and Lv-METTL14 transduced SKOV-3 cells were cultured in serum-free medium for 24 h, cultured in complete growth medium for an additional 16 h, and then collected for flow cytometry to determine cell cycle distribution (*P = 0.028). (G–I) Lv-control and Lv-METTL14 transduced A2780 cells, subjected to a colony formation assay (**P = 0.004) and cell cycle distribution. Bar graph displaying the percentages of cells within the G1, S, and G2/M phases (*P = 0.045). Graphical data represented as the mean ± SD.
METTL14 Downregulated TROAP Expression in Ovarian Cancer Cells and Tissues
Since METTL14 had a profound impact on ovarian cancer cell proliferation, we reasoned that the specific genes regulated by METTL14 should predominantly function in this process. We firstly analyzed the inversely correlated genes with METTL14 from the GEO database (GSE14407) (Supplementary data1). According to cytoscape gene ontology analysis, these genes were involved in several biological processes, including cell proliferation (Supplementary Figure S2). From these data, we selected candidate genes involved in the regulation of cell proliferation for further analysis by qRT-PCR and then constructed a gene-gene correlation matrix using the expression patterns of these genes. Interestingly, we identified a negative correlation between the expression of METTL14 and the cell proliferative gene TROAP (Figure 4A). We postulated that the METTL14-TROAP axis may be responsible for the regulation of ovarian cancer cell proliferation. mRNA expression profiles in GEO database (GSE28724) indicated that suppressed METTL14 expression inversely correlated with TROAP (Figures 4B, C). Thus, we performed qRT-PCR and wester blotting and observed a dramatic increased TROAP expression in ovarian cancer tissues (Figures 4D, E). Furthermore, ectopic expression of METTL14 significantly reduced TROAP expression in SKOV-3 cells compared with the control cells (Figures 4F, G). To investigate whether these changes in protein expression levels occur in tumor tissues, TROAP immunohistochemical staining of normal and ovarian cancer tissues were validated within Human Protein Atlas project (Figure 4H). Altogether, these data suggest that METTL14 functions as a negative regulator of TROAP in ovarian cancer cells and tissues.
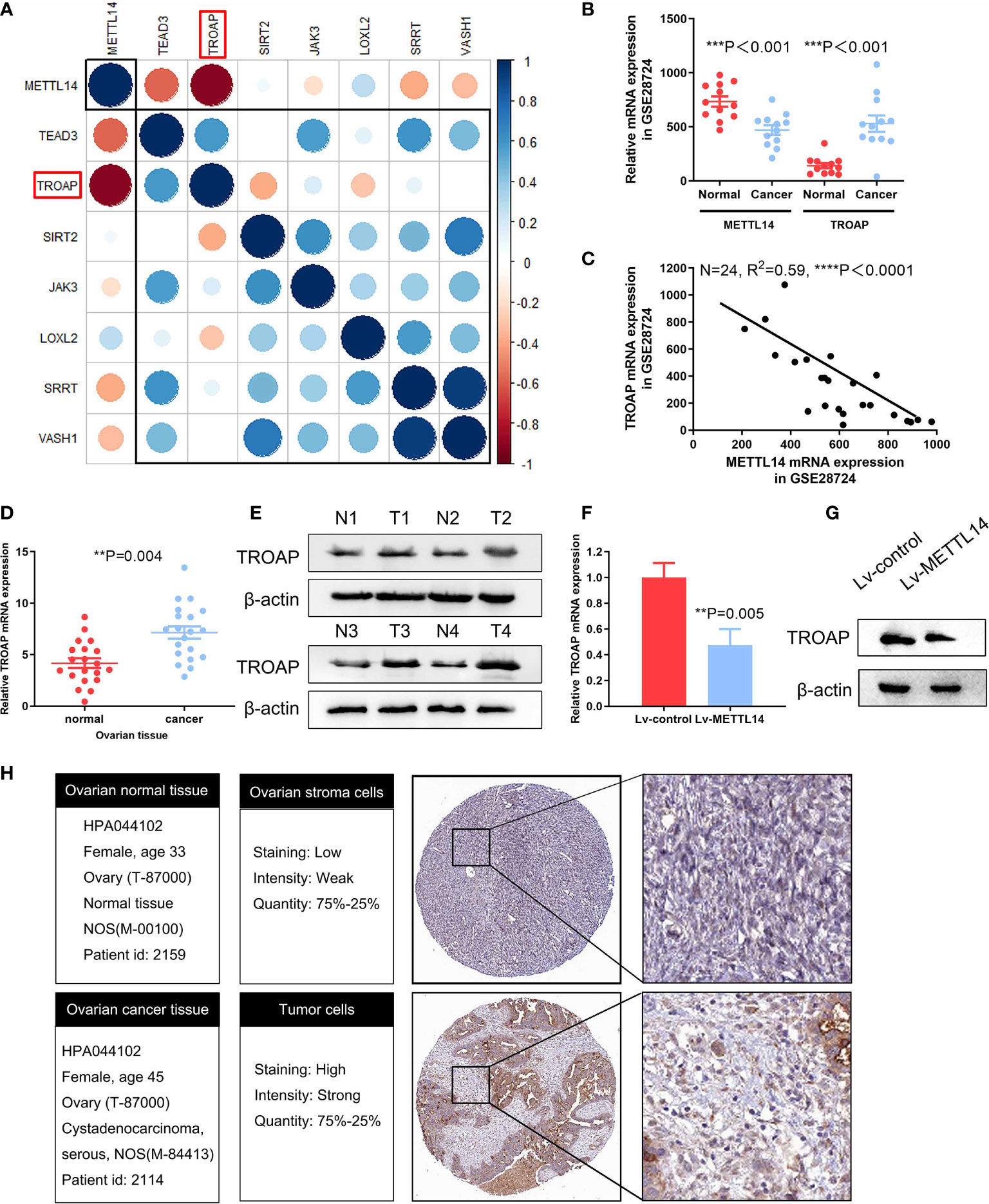
Figure 4 Confirmation of TROAP as a downstream target of METTL14. (A) Correlation matrix of candidate genes for each biological process phenotype stratified by expression levels of cell proliferation-related genes, which were measured by qRT-PCR. Blue indicates a positive correlation, and red indicates a negative correlation. In addition, brightness is proportional to correlation strength. TROAP is indicated in the red box. (B) Graph displaying the negative correlation between TROAP (probe ID: 202265_at) and METTL14 (probe ID: 241689_at) expression in 34 types of ovarian cancer cell lines from the GEO dataset (GSE28724) (***P < 0.001). (C) Expression levels of TROAP are negatively correlated with METTL14 in human ovarian cancer lines GEO dataset (GSE28724) (****P < 0.0001). (D, E) Analysis of TROAP at the mRNA level (D) and protein level (E) in normal and cancer ovarian tissue. (F) TROAP mRNA expression levels in METTL14-overexpressing SKOV-3 cells (**P = 0.005). Graphical data represented as the mean ± SD. (G) Western blot of TROAP protein expression levels. (H) Comparison of relative TROAP expression between ovarian cancer and normal tissues in The Human Protein Atlas database.
METTL14 Inhibited Ovarian Cancer Cell Proliferation by Targeting TROAP
Because we found TROAP to be a functional downstream target of METTL14 in SKOV-3 cells, we next evaluated if the reintroduction of TROAP into the METTL14-overexpressing cells could reverse the inhibitory effects of METTL14 on cell proliferation. To test this hypothesis, firstly, we stably transfected SKOV-3 cells with TROAP plasmid, increasing the mRNA and protein expression of TROAP compared with control cells (Figures 5A, B). We then transfected exogenous TROAP into the METTL14-overexpressing ovarian cancer cells. As shown in Figures 5C, D, the expression of TROAP in the METTL14-overexpressing cells significantly increased after TROAP transfection. Although METTL14 overexpression decreased the proliferation of SKOV-3 cells (Figure 5E), the overexpression of TROAP reversed this inhibitory effect on cell proliferation in the SKOV-3 and A2780 cells (Figure 5E and Supplementary Figure S3).
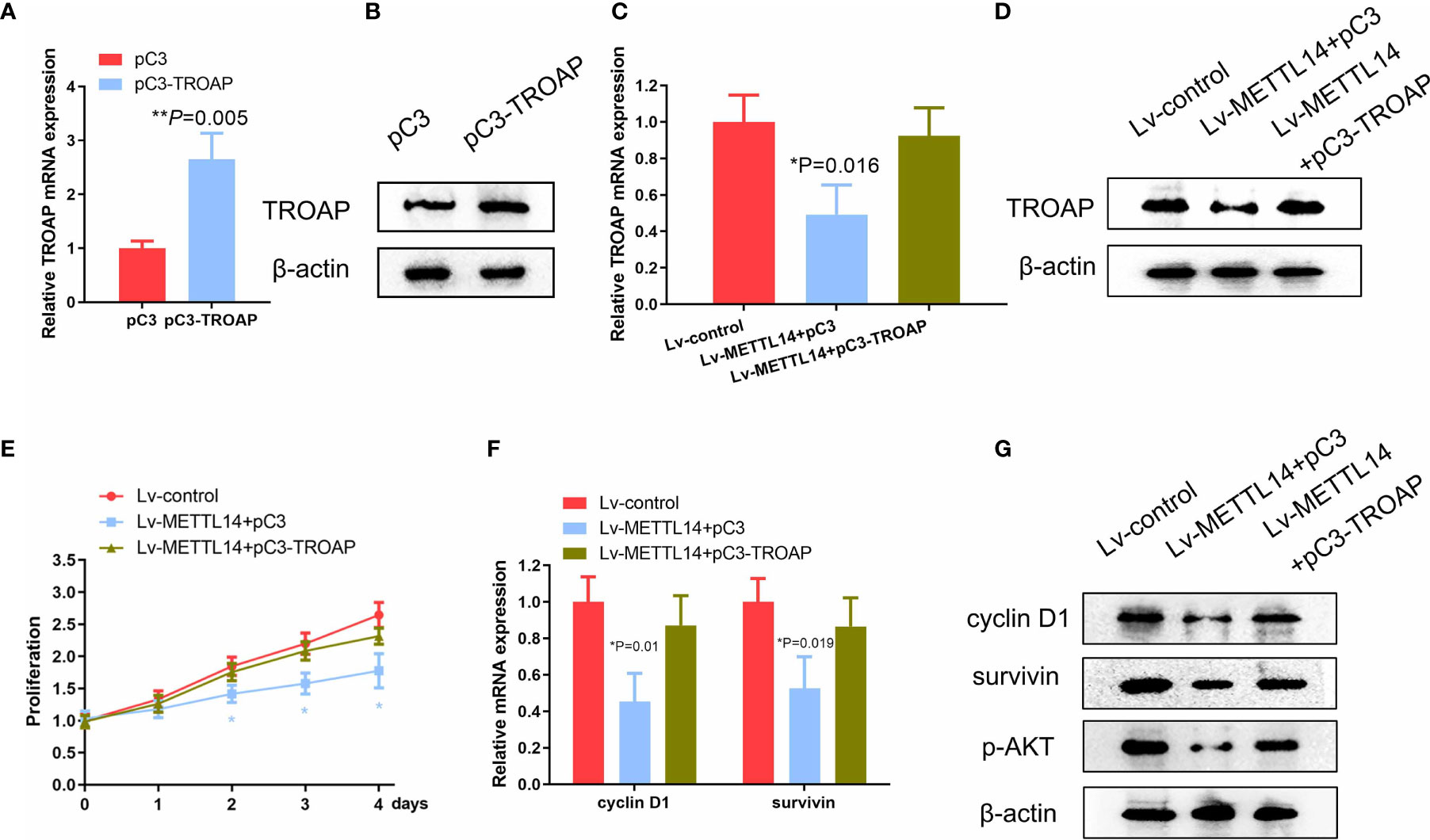
Figure 5 METTL14 suppresses the proliferation of ovarian cancer cell via TROAP. (A, B) TROAP mRNA and protein expression transduced with pC3 or pC3-TROAP (**P=0.005). (C, D) Analyses of TROAP mRNA and protein expression levels in METTL14-overexpressing SKOV-3 cells transduced with pC3 or pC3-TROAP (*P = 0.016). (E) MTS assay of METTL14-overexpressing SKOV-3 cells transduced with pC3 or pC3-TROAP. Values at the indicated timepoints are provided as the mean absorbance with a standard deviation of 6 wells (*P < 0.05). (F) Cyclin D1 and survivin mRNA levels in METTL14-overexpressing SKOV-3 cells with or without TROAP ectopic expression (*P = 0.01, *P = 0.019). Graphical data represented as the mean ± SD. (G) Analysis of expression levels of cyclin D1, survivin, and p-AKT protein by using western blot.
We next evaluated the expression of TROAP target genes, such as cyclin D1, survivin, and p-AKT, because their downregulation may also be involved in the inhibition of ovarian cancer cell proliferation due to METTL14 (24–26). As expected, the exogenous expression of METTL14 downregulated the expression levels of cyclin D1, survivin, and p-AKT. However, transfection with TROAP increased both the mRNA and protein levels of these genes compared with control cells (Figures 5F, G). Taken together, these data suggest that METTL14 may function as a negative regulator of ovarian cancer cell proliferation via inhibition of TROAP and its downstream targets.
METTL14 Decreased TROAP Stability Through m6A Methylation
To further verify whether METTL14-induced downregulation of TROAP expression is dependent on m6A mRNA methylation in ovarian cancer tissues and cells, we assessed the m6A methylation rate in ovarian cancer cells after transfection with the METTL14 expression vector. Our results showed that METTL14 overexpression significantly elevated the rate of m6A RNA methylation in ovarian cancer SKOV-3 cells (Figure 6A). To determine the possible effects of the elevated m6A methylation on TROAP expression, we determined the m6A methylation site of TROAP mRNA using the m6AVar online tool, which predicts the sequence-based m6A alteration sites. We found an GGA m6A sequence motif in the TROAP 3′-UTR proximal to the stop codon (Figure 6B). In addition, METTL14 overexpression induced the accumulation of m6A methylation in this region of the TROAP mRNA (Figure 6C). Next, the half-life of TROAP mRNA was measured in SKOV-3 cells after transfection with the METTL14 expression vector and treatment with actinomycin D. We found that METTL14 had a strong decay effect on the TROAP mRNA (Figure 6D). To further verify that METTL14 functions at the m6A methylation site of the TROAP 3′-UTR to reduce the expression of its mRNA, we constructed luciferase reporter vectors carrying either the wild-type or mutated TROAP 3′-UTR (Figure 6E) and found that METTL14 overexpression resulted in the decreased luciferase activity of the wild-type TROAP 3′-UTR, whereas mutation of this m6A methylation site from A to T almost completely blocked the inhibitory effect of METTL14 overexpression (Figure 6F).
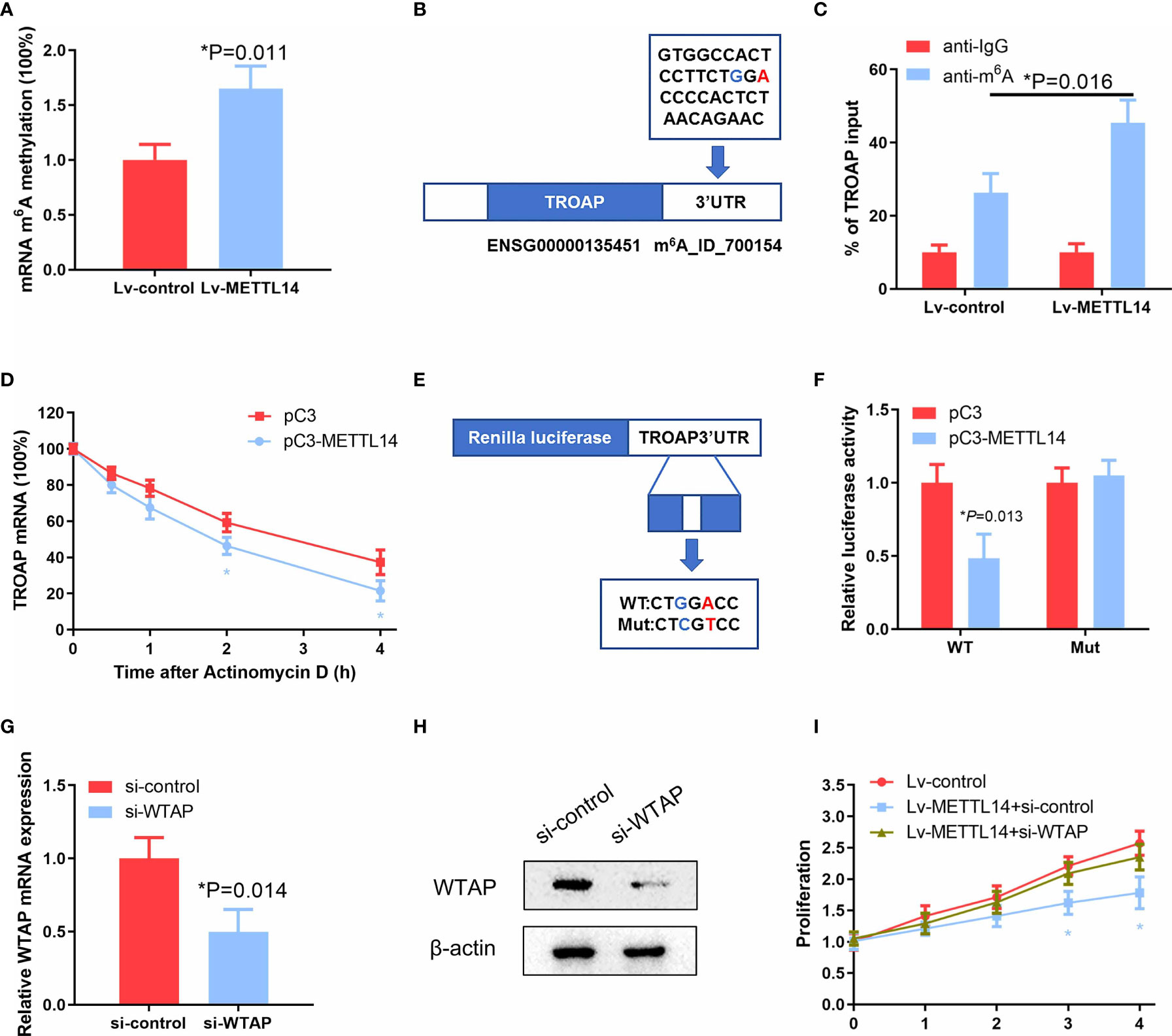
Figure 6 METTL14 destabilize TROAP mRNA through the m6A mRNA methylation. (A) Detection of mRNA m6A methylation in SKOV-3 cells transfected with the METTL14 expression plasmid (*P = 0.011). (B) Predicted m6A RNA methylation site using the m6AVar prediction website. (C) Enrichment of m6A-modified TROAP in METTL14-overexpressing SKOV-3 cells. Input percentage is shown (*P = 0.016). (D) Changes in the m6A-modified TROAP levels with METTL14 overexpression. SKOV-3 cells were transfected with pC3 or pC3-METTL14 plasmids for 36 h. The stabilization of mRNA was initiated by treatment with an RNA polymerase II inhibitor [actinomycin D (5μg/ml)]. Cells were collected at the indicated timepoints. At 0 h, the expression rates were normalized to that of GAPDH mRNA. Data are shown as the average of three or more different studies (*P < 0.05). (E) Representative cloning scheme for the wild-type and mutant TROAP 3′ UTR renilla luciferase vectors. (F) Co-transfection of these luciferase constructs with pC3 or pC3-METTL14 expression vectors into SKOV-3 cells. Data are shown as the average of three or more independent experiments (*P = 0.013). (G, H) Analyses of WTAP expression levels by qRT-PCR and western blot in SKOV-3 cells transfected with si-control or si-WTAP (*P = 0.014). (I) Cell proliferation analysis of stable METTL14-overexpressing SKOV-3 cells transfected with si-control or si-WTAP (*P < 0.05). Graphical data represented as the mean ± SD.
Recently, METTL3 and METTL14 have been identified to interact with WTAP, which acts as a scaffold by binding to the methyltransferases to form a complex that mediates m6A RNA methylation (18). Because the loss of WTAP significantly blocks the m6A RNA modification process, we knocked down WTAP with siRNA in METTL14-overexpressing cells (Figures 6G, H) to evaluate the effects on ovarian cancer cell proliferation. Intriguingly, we observed that WTAP knockdown almost completely abolished the proliferation inhibition levels resulting from METTL14 overexpression in SKOV-3 and A2780 cells (Figure 6I and Supplementary Figure S4), suggesting that there may be other mechanisms involved. These results suggest that METTL14 regulates TROAP expression via an m6A RNA methylation-dependent mechanism.
METTL14 Suppresses Growth of Ovarian Cancer Xenografts In Vivo
To confirm the function of METTL14 in ovarian cancer in vivo, we used the SKOV-3 cells with stable overexpression of METTL14 or control for establishing xenograft model. At 36 days after tumor cells inoculation, nude mice were sacrificed. The data indicated that METTL14 overexpression repressed the tumor volumes and weight (Figures 7A–C). The mRNA and protein expression of METTL14 were negatively correlated with TROAP in the tumor tissues dissected from the nude mice (Figures 7D, E). In addition, histological analysis of xenograft tumors revealed that the tissues from METTL14-overexpression group mice had more METTL14 and less TROAP than from the control group (Figure 7F). Therefore, our results indicated that METTL14 inhibited the proliferation ability of human ovarian cancer cells in vitro and in vivo.
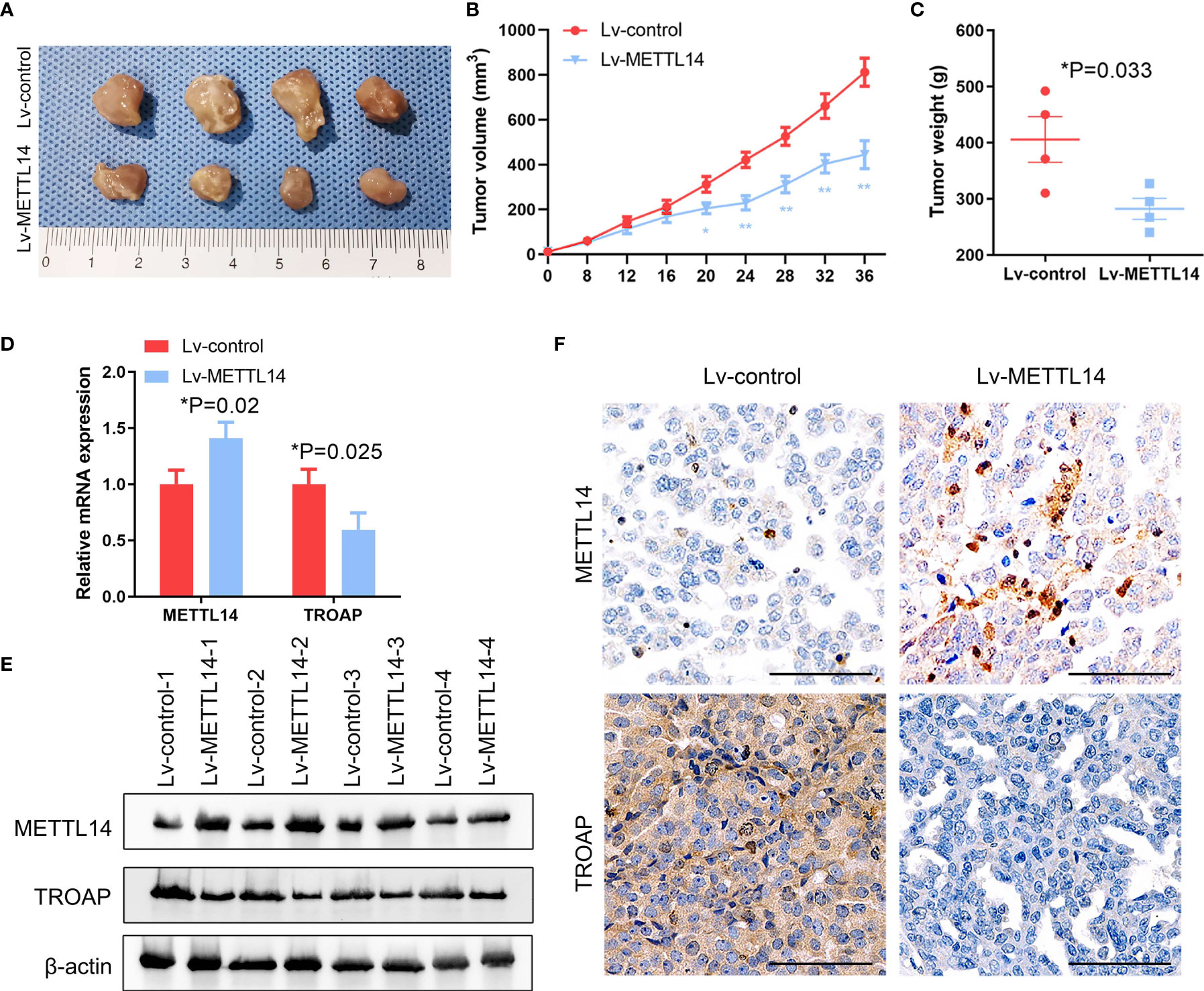
Figure 7 METTL14 inhibits ovarian cancer growth in vivo. (A) Control or METTL14–overexpressing lentivirus infected SKOV-3 cells were injected into nude mice, and tumor was evaluated after 36 days. (B, C) Tumor volume (B) and weight (C) at the end of the treatment period (**P < 0.01, *P < 0.05, *P = 0.033). (D, E) METTL14 and TROAP mRNA (D) and protein (E) expression levels assessed by real-time PCR and western blot analysis in tumors (*P = 0.02, *P = 0.025). (F) Xenograft tumor sections were subjected to IHC for METTL14 and TROAP. Magnification, 600 ×. Data are shown as mean ± SD from three independent experiments.
Discussion
In this study, we assessed and found considerable CNVs in the genes of the 10 selected regulators of m6A mRNA methylation in ovarian cancer tissues. Among these regulators, METTL14, which is an m6A mRNA methylation writer, also exhibited reduced expression levels and subsequent low levels of m6A methylation in ovarian cancer. Further investigation revealed that METTL14 reduced the stability of TROAP mRNA, which consequently arrested ovarian tumor cells at the G1 phase of the cell cycle and inhibited their proliferation. Our current data suggest a novel therapeutic target for the treatment of ovarian cancer (Figure 8).
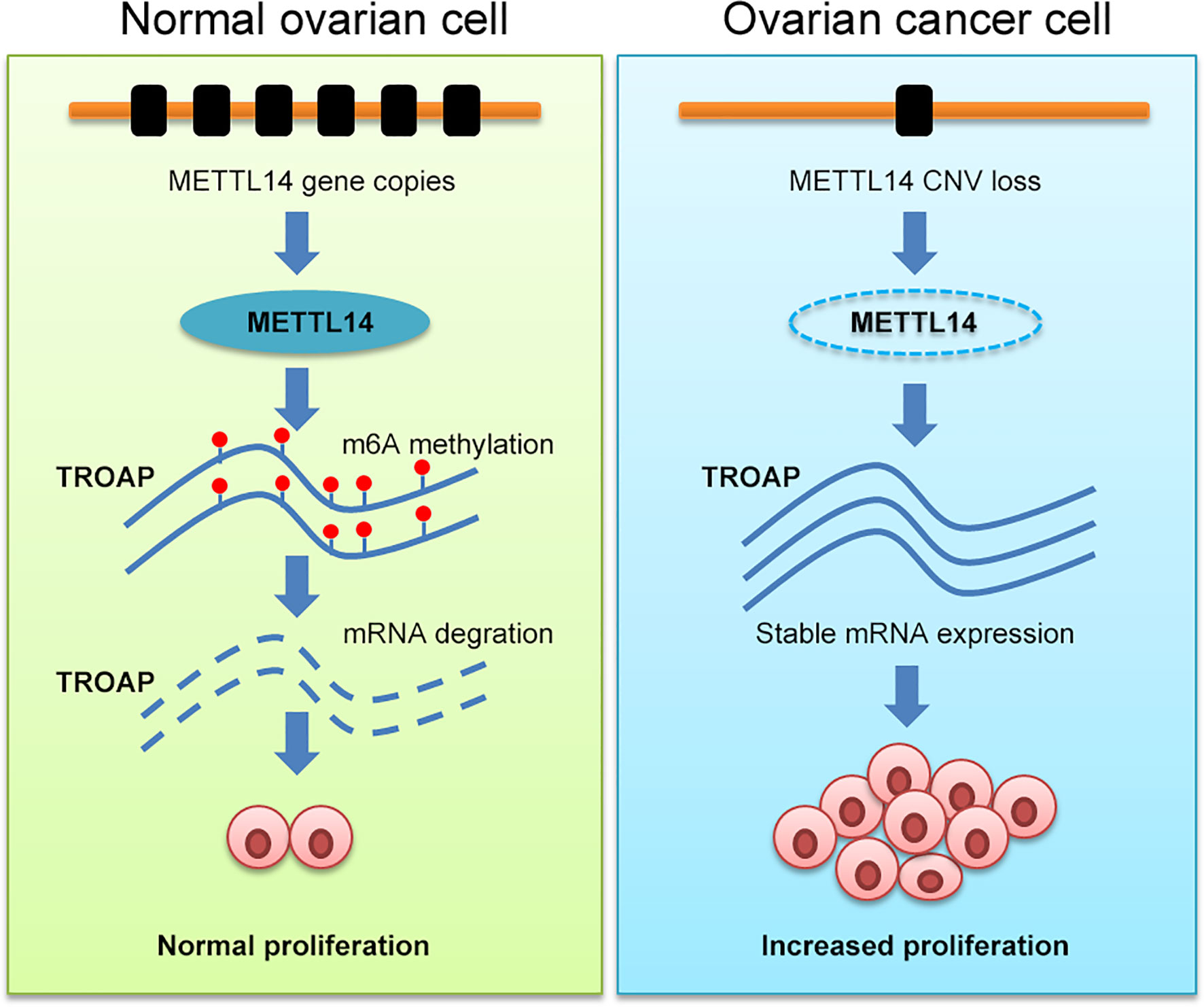
Figure 8 Schematic illustration of the regulatory role of METTL14 via its m6A RNA methylation function on TROAP expression and ovarian cancer cell proliferation.
Indeed, a previous study has reported that m6A modification improves the transcript stability of long non-coding RNA rhophillin Rho GTPase-binding protein 1-antisense RNA 1 (RHPN1-AS1) and reduces its degradation. Additionally, RHPN1-AS1 overexpression is associated with tumorigenesis and metastasis of ovarian cancer (11). However, the underlying mechanism of increased m6A RNA methylation remains unclear (12). Another recent study has shown that alterations in m6A modifications contribute to oncogenesis and at the same time represent an emerging class of therapeutic targets (3). Additionally, Li et al. have demonstrated that approximately 70% of all endometrial tumors exhibit a reduction in m6A methylation levels compared with that of normal endometrial tissues and that the downregulation or mutation of either METTL3 or METTL14 may be responsible for this reduction (6). In the current study, we assessed 10 regulators of m6A methylation for gene mutations and CNVs and found that reduced m6A modification levels were associated with the poor overall survival of ovarian cancer patients. Indeed, previous studies have shown that the induction of ALKBH5 expression occurs in ovarian cancer tissues and is associated with tumor cell proliferation and invasion in vitro (27). Elevated ALKBH5 expression has also been shown to contribute to the m6A modification of frizzled class receptor 10 (FZD10) mRNA and to the resistance of ovarian cancer cells to treatment with a poly-ADP ribose polymerase inhibitor, which may also be associated with altered FTO levels (28). Moreover, METTL3 expression is upregulated in ovarian cancer and is associated with advanced tumor grades and stages, lymph node and distant metastases, the International Federation of Gynecology and Obstetrics stage, and overall patient survival. Likewise, stable METTL3 overexpression significantly enhances tumor cell proliferation and invasion as well as tumor formation in nude mice (29). In addition, WTAP has been evaluated as a prognostic marker of high-grade serous ovarian cancer and is associated with tumor metastasis (30). The expression of YTHDF1, which is an m6A modification reader, is also associated with ovarian cancer progression (31). Similar to these findings, our current data showed that METTL14 had a significant reduction in CNVs in ovarian cancer tissues and that METTL14 overexpression inhibited ovarian cancer cell proliferation in vitro, further confirming the TCGA and GEO data. However, there are currently no previous reports on the role of METTL14 in ovarian cancer; thus, further studies are warranted. METTL14 is a critical part of the m6A methyltransferase complex and is responsible for binding to the RNA (32). Furthermore, it has been well documented that various methyltransferases play an important role in the growth and metastasis of multiple types of cancer, including renal clear cell carcinoma (8), hepatocellular carcinoma (5), and acute myeloid leukemia (33). Although increased METTL14 expression has been shown to suppress the progression of certain types of cancer, it leads to increased proliferation of acute myeloid cells.
TROAP (Trophinin-associated protein, also known as TASTIN) is found to participate in the proliferation, invasion, and migration of many cancers (34). Additionally, aberrant TROAP expression occurs in different human cancers (35). Recently, Jing et al. demonstrated that TROAP knockdown in gastric cancer cells results in reduced tumor cell growth, migration and invasion (36). Emerging evidence indicates that positive proliferation related gene, such as cyclin D1, survivin, and p-AKT, are positively regulated by TROAP (24–26). In the current study, we revealed TROAP as a downstream target of METTL14. We further demonstrated that METTL14 inhibited the expression of TROAP via the m6A RNA modification in ovarian cancer cells, which also inhibited cell proliferation by suppress cyclin D1, survivin, and p-AKT. Additionally, knockdown of TROAP reversed these inhibitory effects of METTL14 on tumor cell proliferation. However, we cannot exclude the possibility of other gene signaling pathways that may be involved in this regulatory mechanism. Therefore, additional investigations, including high-throughput analyses, are required to identify other possible genes that are targeted by METTL14.
In summary, the current study delineates that METTL14 negatively regulates TROAP expression in an m6A RNA methylation-dependent manner and that its expression is reduced in ovarian cancer and promotes cancer cell proliferation. These results suggest that the m6A modification plays a critical role in ovarian cancer growth and may be a novel therapeutic target that warrants further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the First Affiliated Hospital of Fourth Military Medical University. The patients/participants provided their written informed consent to participate in this study The animal study was reviewed and approved by Animal Care Committee of the Fourth Military Medical University.
Author Contributions
WL and WB conceived and designed the study. YL, HP, PJ, JZ, and XF performed the in vitro analysis. YZ, CP, and JR analyzed the data and prepared the figures. HZ, WB, and WL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81702554 to YL, 81802661 to WB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.824258/full#supplementary-material
References
1. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-Methyladenosine Modulators Promote Breast Cancer Progression. BMC Cancer (2019) 19(1):326. doi: 10.1186/s12885-019-5538-z
2. Jiang L, Li Y, He Y, Wei D, Yan L, Wen H. Knockdown of M6a Reader IGF2BP3 Inhibited Hypoxia-Induced Cell Migration and Angiogenesis by Regulating Hypoxia Inducible Factor-1α in Stomach Cancer. Front Oncol (2021) 11:711207. doi: 10.3389/fonc.2021.711207
3. Chen XY, Zhang J, Zhu JS. The Role of M6a RNA Methylation in Human Cancer. Mol Cancer (2019) 18(1):103. doi: 10.1186/s12943-019-1033-z
4. Yao Q, He L, Gao X, Tang N, Lin L, Yu X, et al. The M6a Methyltransferase METTL14-Mediated N6-Methyladenosine Modification of PTEN mRNA Inhibits Tumor Growth and Metastasis in Stomach Adenocarcinoma. Front Oncol (2021) 11:699749. doi: 10.3389/fonc.2021.699749
5. Qu N, Bo X, Li B, Ma L, Wang F, Zheng Q, et al. Role of N6-Methyladenosine (M6a) Methylation Regulators in Hepatocellular Carcinoma. Front Oncol (2021) 11:755206. doi: 10.3389/fonc.2021.755206
6. Li Q, He W, Wan G. Methyladenosine Modification in RNAs: Classification and Roles in Gastrointestinal Cancers. Front Oncol (2021) 10:586789. doi: 10.3389/fonc.2020.586789
7. Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li Z, et al. M6a Methyltransferase METTL14-Mediated Upregulation of Cytidine Deaminase Promoting Gemcitabine Resistance in Pancreatic Cancer. Front Oncol (2021) 11:696371. doi: 10.3389/fonc.2021.696371
8. Wang J, Zhang C, He W, Gou X. Effect of M6a RNA Methylation Regulators on Malignant Progression and Prognosis in Renal Clear Cell Carcinoma. Front Oncol (2020) 10:3. doi: 10.3389/fonc.2020.00003
9. Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. Mettl14 Inhibits Bladder TIC Self-Renewal and Bladder Tumorigenesis Through N6-Methyladenosine of Notch1. Mol Cancer (2019) 18(1):168. doi: 10.1186/s12943-019-1084-1
10. Feng ZY, Gao HY, Feng TD. Immune Infiltrates of M6a RNA Methylation-Related lncRNAs and Identification of PD-L1 in Patients With Primary Head and Neck Squamous Cell Carcinoma. Front Cell Dev Biol (2021) 9:672248. doi: 10.3389/fcell.2021.672248
11. Li Q, Ren CC, Chen YN, Yang L, Zhang F, Wang BJ, et al. A Risk Score Model Incorporating Three M6a RNA Methylation Regulators and a Related Network of miRNAs-M6a Regulators-M6a Target Genes to Predict the Prognosis of Patients With Ovarian Cancer. Front Cell Dev Biol (2021) 9:703969. doi: 10.3389/fcell.2021.703969
12. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N6-Methyladenosine (M6a) RNA Modification in Gastrointestinal Tract Cancers: Roles, Mechanisms, and Applications. Mol Cancer (2019) 18(1):178. doi: 10.1186/s12943-019-1099-7
13. Kumar V, Gupta S, Chaurasia A, Sachan M. Evaluation of Diagnostic Potential of Epigenetically Deregulated MiRNAs in Epithelial Ovarian Cancer. Front Oncol (2021) 11:681872. doi: 10.3389/fonc.2021.681872
14. Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, et al. FTO-Dependent N6-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res (2020) 80(16):3200–14. doi: 10.1158/0008-5472.CAN-19-4044
15. Zheng M, Hu Y, Gou R, Nie X, Li X, Liu J, et al. Identification Three LncRNA Prognostic Signature of Ovarian Cancer Based on Genome-Wide Copy Number Variation. BioMed Pharmacother (2020) 124:109810. doi: 10.1016/j.biopha.2019.109810
16. Fan L, Lin Y, Lei H, Shu G, He L, Yan Z, et al. A Newly Defined Risk Signature, Consisting of Three M6a RNA Methylation Regulators, Predicts the Prognosis of Ovarian Cancer. Aging (Albany NY) (2020) 12(18):18453–75. doi: 10.18632/aging.103811
17. Lian Y, Fan W, Huang Y, Wang H, Wang J, Zhou L, et al. Downregulated Trophinin-Associated Protein Plays a Critical Role in Human Hepatocellular Carcinoma Through Upregulation of Tumor Cell Growth and Migration. Oncol Res (2018) 26(5):691–701. doi: 10.3727/096504017X15101398724809
18. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-Coding RNA XIST. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
19. Feng Y, Dong H, Sun B, Hu Y, Yang Y, Jia Y, et al. METTL3/METTL14 Transactivation and M6a-Dependent TGF-β1 Translation in Activated Kupffer Cells. Cell Mol Gastroenterol Hepatol (2021) 12(3):839–56. doi: 10.1016/j.jcmgh.2021.05.007
20. Hu N, Ji H. N6-Methyladenosine (M6a)-Mediated Up-Regulation of Long Noncoding RNA LINC01320 Promotes the Proliferation, Migration, and Invasion of Gastric Cancer via Mir495-5p/RAB19 Axis. Bioengineered (2021) 12(1):4081–91. doi: 10.1080/21655979.2021.1953210
21. Huang R, Yang L, Zhang Z, Liu X, Fei Y, Tong WM, et al. RNA M6a Demethylase ALKBH5 Protects Against Pancreatic Ductal Adenocarcinoma via Targeting Regulators of Iron Metabolism. Front Cell Dev Biol (2021) 9:724282. doi: 10.3389/fcell.2021.724282
22. Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The M6a Demethylase ALKBH5 Controls Trophoblast Invasion at The Maternal-Fetal Interface by Regulating The Stability of CYR61 mRNA. Theranostics (2019) 9(13):3853–65. doi: 10.7150/thno.31868
23. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M6a Enhances The Phase Separation Potential of mRNA. Nature (2019) 571(7765):424–8. doi: 10.1038/s41586-019-1374-1
24. Zhao ZQ, Wu XJ, Cheng YH, Zhou YF, Ma XM, Zhang J, et al. TROAP Regulates Cell Cycle and Promotes Tumor Progression Through Wnt/β-Catenin Signaling Pathway in Glioma Cells. CNS Neurosci Ther (2021) 27(9):1064–76. doi: 10.1111/cns.13688
25. Ye J, Chu C, Chen M, Shi Z, Gan S, Qu F, et al. TROAP Regulates Prostate Cancer Progression via The WNT3/Survivin Signalling Pathways. Oncol Rep (2019) 41(2):1169–79. doi: 10.3892/or.2018.6854
26. Li L, Wei JR, Song Y, Fang S, Du Y, Li Z, et al. TROAP Switches DYRK1 Activity to Drive Hepatocellular Carcinoma Progression. Cell Death Dis (2021) 12(1):125. doi: 10.1038/s41419-021-03422-3
27. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer Through miR-7 and BCL-2. J Exp Clin Cancer Res (2019) 38(1):163. doi: 10.1186/s13046-019-1159-2
28. Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N6-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res (2019) 79(11):2812–20. doi: 10.1158/0008-5472.CAN-18-3592
29. Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 Promotes Ovarian Carcinoma Growth and Invasion Through The Regulation of AXL Translation and Epithelial to Mesenchymal Transition. Gynecol Oncol (2018) 151(2):356–65. doi: 10.1016/j.ygyno.2018.09.015
30. Yu HL, Ma XD, Tong JF, Li JQ, Guan XJ, Yang JH. WTAP Is a Prognostic Marker of High-Grade Serous Ovarian Cancer and Regulates The Progression of Ovarian Cancer Cells. Onco Targets Ther (2019) 12:6191–201. doi: 10.2147/OTT.S205730
31. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The M6a Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048
32. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA M6a Modification. Cell Stem Cell (2018) 22(2):191–205.e9. doi: 10.1016/j.stem.2017.11.016
33. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med (2016) 374(23):2209–21. doi: 10.1056/NEJMoa1516192
34. Li K, Zhang R, Wei M, Zhao L, Wang Y, Feng X, et al. TROAP Promotes Breast Cancer Proliferation and Metastasis. BioMed Res Int (2019) 2019:6140951. doi: 10.1155/2019/6140951
35. Hu H, Xu L, Chen Y, Luo SJ, Wu YZ, Xu SH, et al. The Upregulation of Trophinin-Associated Protein (TROAP) Predicts a Poor Prognosis in Hepatocellular Carcinoma. J Cancer (2019) 10(4):957–67. doi: 10.7150/jca.26666
Keywords: Ovarian cancer, Copy number variations, m6A RNA methylation, METTL14, TROAP
Citation: Li Y, Peng H, Jiang P, Zhang J, Zhao Y, Feng X, Pang C, Ren J, Zhang H, Bai W and Liu W (2022) Downregulation of Methyltransferase-Like 14 Promotes Ovarian Cancer Cell Proliferation Through Stabilizing TROAP mRNA. Front. Oncol. 12:824258. doi: 10.3389/fonc.2022.824258
Received: 29 November 2021; Accepted: 31 January 2022;
Published: 18 February 2022.
Edited by:
Dmitry Shcherbo, Pirogov Russian National Research Medical University, RussiaCopyright © 2022 Li, Peng, Jiang, Zhang, Zhao, Feng, Pang, Ren, Zhang, Bai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Zhang, zhm@fmmu.edu.cn; Wendong Bai, bwddcgzl@fmmu.edu.cn; Wenchao Liu, liuch@fmmu.edu.cn
†These authors have contributed equally to this work
 Yize Li1†
Yize Li1† Wendong Bai
Wendong Bai