- Department of Psychiatry and Behavioral Neurobiology, University of Alabama at Birmingham, Birmingham, AL, United States
It is well known that only a small proportion of the human genome code for proteins; the rest belong to the family of RNAs that do not code for protein and are known as non-coding RNAs (ncRNAs). ncRNAs are further divided into two subclasses based on size: 1) long non-coding RNAs (lncRNAs; >200 nucleotides) and 2) small RNAs (<200 nucleotides). Small RNAs contain various family members that include microRNAs (miRNAs), small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs). The roles of ncRNAs, especially lncRNAs and miRNAs, are well documented in brain development, homeostasis, stress responses, and neural plasticity. It has also been reported that ncRNAs can influence the development of psychiatric disorders including schizophrenia, major depressive disorder, and bipolar disorder. More recently, their roles are being investigated in suicidal behavior. In this article, we have comprehensively reviewed the findings of lncRNA and miRNA expression changes and their functions in various psychiatric disorders including suicidal behavior. We primarily focused on studies that have been done in postmortem human brain. In addition, we have briefly reviewed the role of other small RNAs (e.g. piwiRNA, siRNA, snRNA, and snoRNAs) and their expression changes in psychiatric illnesses.
Introduction
Non-coding RNAs (ncRNAs) are defined as RNAs that are not translated into protein. Protein-coding genes occupy only a small proportion (<3%) of the entire genome (1). However, the remaining non-protein coding genes are not a simple transcriptional noise (1, 2); instead, ~80% of them are transcriptionally active with elaborate regulatory roles. A majority of them are now considered non-coding RNA genes (3–6). Based on the size of the nucleotides, ncRNAs are divided into small ncRNAs and long ncRNAs (lncRNAs). ncRNAs that contain <200 nucleotides (nt) belong to small ncRNA family. On the other hand, those containing more >200 nt belong to long lncRNA family. Small RNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs). ncRNAs function via interactions with RNA, DNA, and protein. They regulate the transcription of mRNAs and participate in alternative splicing and epigenetic modifications such as chromatin and RNA editing (7, 8). These regulatory functions can target either neighboring transcripts (cis) or loci that are distant from their own transcription (trans). Collectively, ncRNAs constitute a unique layer of gene regulation where they function as key intermediate regulators in conveying the message from genotype to phenotype states (9).
A large number of ncRNAs are abundantly expressed in the brain (10–12). In addition, the expressions of ncRNAs are brain region- and cell type-specific (13–16). Several studies have shown the role of ncRNAs in brain evolution, development, homeostasis, stress response, and neuroplasticity (14, 17–25). Brain expressed ncRNAs can also influence the development of psychiatric disorders such as schizophrenia (SCZ), major depressive disorder (MDD), and bipolar disorder (BD) as well as neurodegenerative disorders (11, 19, 23, 26, 27). More recently, their role in suicidal behavior has been postulated (28–30). The effects of ncRNA expression changes are not restricted to the type of psychiatric disorders but, to a large extent, to different brain regions. Recent evidence suggests the inability of brain regions to act independently. Instead, they act in a coordinated manner via functional networks. In various neuropsychiatric disorders, voxel based neuroimaging has demonstrated specific roles of brain regions such as cortex (prefrontal, anterior cingulate), hippocampus, and amygdala (31). Although each of these brain regions has important roles, such as amygdala and prefrontal cortex in emotion (32) and prefrontal cortex and hippocampus in stress response (33), these brain regions crosstalk to each other via complex gene networks which could be mediated vis ncRNAs.
Earlier published reviews have primarily focused on miRNAs in psychiatric illnesses (34–36). This review comprehensively summarizes the relationship between various ncRNAs. Although the emphasis is given to lncRNAs and miRNAs, a range of other ncRNAs are also included to provide a glimpse into where the field stands and what should be the course of direction for future research. The review is restricted to studies in human postmortem brain, given that the role of peripheral ncRNAs is not very well established. We have discussed a few peripheral blood cell studies which are relevant to suicidal behavior. The criteria for selecting literature search are as follows: Electronic search was done using PubMed with search terms ncRNAs (microRNA, small interfering RNA, piwi-interacting RNA, small nucleolar RNA, and small nuclear RNA) AND psychiatric disorders (schizophrenia, major depressive disorder, and bipolar disorder). Each combination was performed separately. Additionally, postmortem research for non-suicide studies, and both postmortem brain and peripheral tissue research for suicide studies, were included in the search criteria.
Long Non-Coding RNAs
Long Non-Coding RNAs in Brain Functions
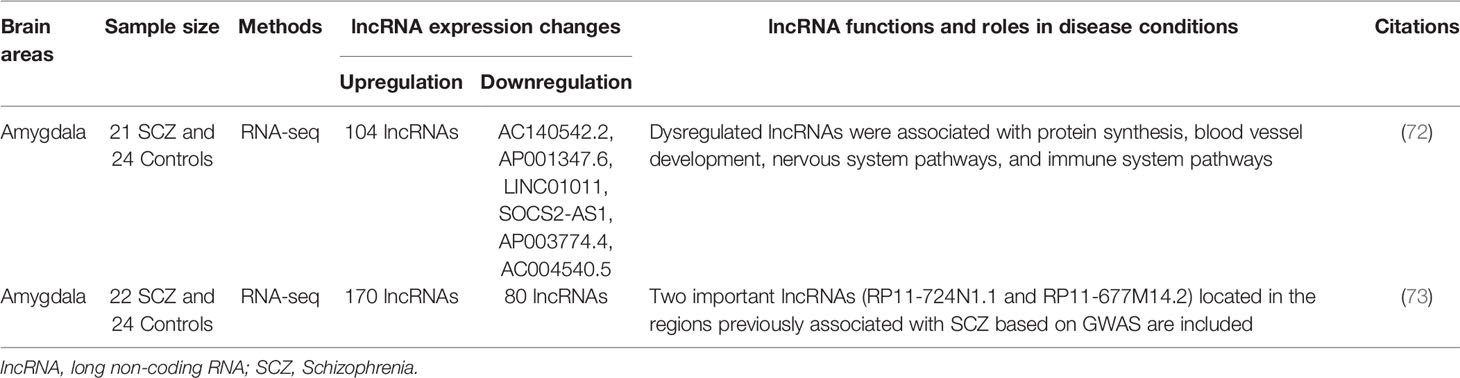
Long non-coding RNAs (lncRNAs) are defined as RNAs having >200 nucleotides with low (in the form of short peptide) or no protein coding potential (37). LncRNAs are transcribed by RNA Polymerase II and are processed to mature RNA by the same method as protein-coding mRNAs (38). LncRNAs’ lengths span ~100 kilo base-pairs (bp) and are shorter than mRNAs but have longer exons (39–41). They were originally considered to be the products of heterochronic genes (4) that control the temporal dimension of development; however, there are limitations to fully understand the function of lncRNAs from this point of view. Interestingly, the genomic structure of lncRNAs is strongly related to their function (42). In the early nineties, the X-inactive specific transcripts (Xist) and H19 were first discovered as non-protein coding RNAs by exploring cDNA libraries (43, 44). Since then, over 100,000 lncRNA genes have been found and their number is still increasing (45). Recently, several lncRNA functions have been revealed. For example, they participate in chromatin modifications (46, 47); act as ‘sponges’ that prevent miRNA functions (48); act as scaffolds that provide docking sites for proteins (49); serve as activators and suppressors of mRNA transcription (50); and act as regulators of splicing patterns (50). The functions of lncRNAs are depicted in Figure 1.
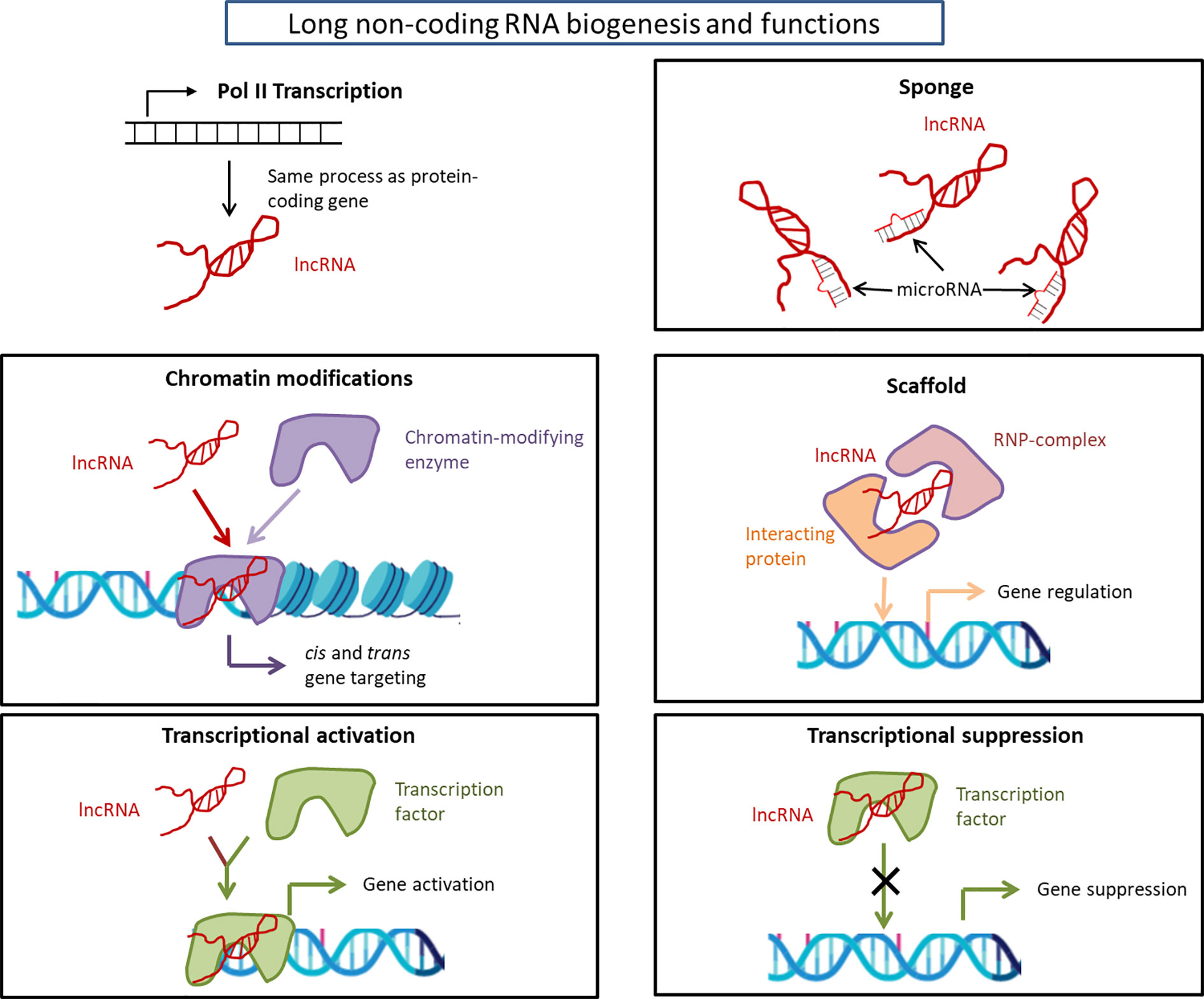
Figure 1 Long non-coding RNA biogenesis and functions. Long non-coding RNAs are transcribed by RNA polymerase II and are processed to mature RNA by the same method as protein-coding mRNAs as detailed in the text. The considerable functions of lncRNAs are: chromatin modification, transcriptional activation/suppression, sponge, and scaffold. The functions of lncRNAs depend on the type of lncRNAs. lncRNA, long non-coding RNA; RNP, ribonucleoprotein.
In terms of functions in the brain, lncRNAs are strongly related to brain development (51). In this capacity, lncRNAs participate in spatiotemporal regulation of proliferation and differentiation of pluripotent stem cells (21, 52). High-resolution and high-throughput technologies, such as microarray and RNA-sequencing, have helped in examining the number and pattern of lncRNA expression in the brain. Microarray detects the expression of a large number of RNAs simultaneously, whereas RNA-seq detects both known and novel transcripts and quantifies a large dynamic range of expression levels with absolute rather than relative values. Ramos et al. (53) reported that more than 3,600 lncRNAs are expressed among three different brain regions: subventricular zone, olfactory bulb, and dentate gyrus. Clustering analysis revealed that lncRNAs have a tissue-specific expression pattern (39, 54). A study, using microarray analysis, investigated differential lncRNA expression in the temporal cortex ranging from infancy to adulthood (0.92–43.5 years old) and concluded that the expression pattern of lncRNA changes with age (55). At functional levels, several lncRNAs are involved in regulating neuronal development. This occurs through complex interactions between lncRNAs and other factors in transitioning neural stem cell to progenitors and eventually to differentiated neurons. Some of the lncRNAs that promote the differentiation of neural stem cells include Brn1b, RMST, and TUNA (56–58). EVF2, the first found central nervous system-specific lncRNA, regulates the differentiation of GABAergic neurons (18). The GABA system has been shown to be consistently involved in several psychiatric disorders such as SCZ (59–61), MDD (62), and BD (63). Another lncRNA, Pnky, promotes neural proliferation (64, 65). A recent study has shown that antisense lncRNAs also regulate key proteins related to psychiatric disorders. An inhibition of brain-derived neurotrophic factor (BDNF) antisense RNA (BDNF-AS) causes two- to sevenfold increase in BDNF protein level, which leads to neuronal outgrowth, differentiation, survival, and proliferation (66). An interaction between BDNF and neuropsychiatric disorders is well documented (67, 68). In addition, the mechanism of action of antidepressants (69) and electroconvulsive therapy (70) also involve changes in the levels of BDNF, both centrally and peripherally. Not only do lncRNAs impact BDNF, but changes in the expression of a large number of lncRNAs have been shown following 1 h BDNF treatment in cultured neuronal cells (71). This shows that under neuronal environment, a feed forward and feedback loop might exist between neurotrophin signaling and lncRNA expression with the potential to develop effective therapeutic strategy in psychiatric conditions.
Long Non-Coding RNAs in Psychiatric Disorders
Examining the role of ncRNAs in psychiatric illnesses is an emerging field. Two recent studies used amygdala from SCZ subjects to decipher the role of lncRNAs in this disorder (Table 1). Tian et al. (72) examined lncRNA expression by RNA-seq using 21 SCZ and 24 healthy control subjects. They found alterations in the expression of 345 lncRNAs (104 up- and six downregulated) in SCZ subjects. Subsequently, they divided 21 SCZ subjects into nine undifferentiated, seven disorganized, and five paranoid. Individually, they found 155 up- and 17 downregulated lncRNAs in the undifferentiated SCZ group, 39 up- and six downregulated lncRNAs in the disorganized SCZ subjects, and no significant alteration in paranoid SCZ subjects when compared with control subjects. They performed gene set enrichment analysis to reveal the gene ontology terms associated with dysregulated lncRNAs (all SCZ vs control subjects) and found that dysregulated lncRNAs were associated with protein synthesis, blood vessel development, nervous system pathways, and immune system pathways. In another amygdala study, Liu et al. (73) used RNA-seq for detecting dysregulation of lncRNAs in 22 SCZ subjects and 24 healthy controls. Transcriptome data identified 250 lncRNAs that showed significant expression differences between SCZ and control subjects. The study further focused on two specific lncRNAs (upregulated RP11-724N1.1 and downregulated RP11-677M14.2), which were shown to be associated with SCZ based on previous genome-wide association study (GWAS). Additional analysis by dividing SCZ subjects into three subtypes revealed the dysregulation of RP11-724N1.1 in the undifferentiated subtype and RP11-677M14.2 in the paranoid subtype. RP11-724N1.1 overlaps the CNNM2 gene locus, which has a key role in the neurodevelopmental process (74). On the other hand, RP11-677M14.2 overlaps the neurogranin (NRGN) gene locus. Several SNPs of the NRGN gene have been associated with onset (75–77), intellectual disability (75), and anterior cingulate cortex (ACC) volume (78) in SCZ subjects. These amygdala studies suggest that not only are lncRNAs involved in SCZ pathophysiology, but they can also differentiate subtypes of SCZ.
lncRNAs and Suicidal Behavior
There are four studies that show a relationship between suicidal behavior and lncRNA expression changes. Of them, three studies explored lncRNA expression in the brain (one in ACC and two in the dorsolateral prefrontal cortex [dlPFC]), and one in the blood (Table 2). Zhou et al. (79) compared lncRNA expression levels in rostral ACC between 26 MDD suicide and 24 healthy control subjects by RNA-seq. After considering nominal p values (p < 0.05), without multiple testing, 364 out of 2,670 lncRNAs were differentially regulated; of them, 60% (217 lncRNAs) were downregulated. After genome-wide multiple testing, 15 upregulated and eight downregulated lncRNAs reached statistical significance. Potential cis-targets for significantly dysregulated 23 lncRNAs based on RNA-seq data showed that six lncRNAs (RP11-326I11.3 vs IRF2; RP11-273G15.2 vs LY6E; CTD-2647L4.4 vs HMOBX1; CTC-487M23.5 vs REEP5; RP1-269M15.3 vs PTPRT; RP11-96D1.10 vs NFATC3) were significantly correlated with one antisense or overlapping gene. All of them were found to be nominally differentially expressed except REEP5. With an additional qPCR approach, the authors validated three out of five genes (IRF2, LY6E, and HMBOX1). These genes are functionally related to interferon signaling, which plays a key role in CNS homeostasis and in psychiatric disorders (81).
Punzi et al. (80) revealed a correlation between violent suicide and lncRNA LOC28758 expression in dlPFC of SCZ subjects that included 77 non-suicides, 13 non-violent suicides, and 16 violent suicides. LOC28758 is located in the MARCKS gene, which was also found to be significantly highly expressed in violent suicides than non-suicides and non-violent suicides. In a recent study, the authors also examined lncRNA expression changes in dlPFC by RNA-seq (28). LINC01268 expression was significantly higher in suicide subjects compared to non-suicide subjects. Note that both suicide and non-suicide groups included three psychiatric disorders (SCZ, MDD, and BD). Subsequently, they divided the suicide groups into violent suicide and non-violent suicide (poisoning or asphyxia = nonviolent; others = violent). The violent suicide group had higher expression of LINC01268 than the non-suicide group, but the non-violent group did not show any change. Additionally, they explored quantitative trait locus (eQTL) of LINC01268 by genotyping 23 SNPs located within 100 kb up and downstream of the gene coordinates. Of those, only rs7747961 could regulate the LINC01268 expression (C-carriers > non-C-carriers). Based on the Brown–Goodwin questionnaire score, they revealed that LINC01268 expression was higher in the aggressive group than in the non-aggressive group. Lastly, weighted correlation network analysis (WGCNA) was performed with RNA-seq data to verify the relationship between LINC01268 and the co-expression gene set. WGCNA is a systems biology method for detecting the modules correlated with clinical traits by a soft-threshold algorithm. A module including 224 genes and LINC01268 was created. It was found that among 224 genes, P2RY13, a purinergic receptor present in both the peripheral immune system and in the brain, had a strong correlation with LINC01268.
One peripheral blood mononuclear cell (PBMC) study was conducted to identify change in lncRNAs in MDD suicide subjects (63 MDD no-suicidal ideation, 57 MDD with suicidal ideation, and 63 control subjects) by qPCR method (30). qPCR method identifies amplified fragments during the PCR process, but only a smaller number of genes can be measured at once compared to RNA-seq or microarray. Six lncRNAs (TCONS_00019174, NST00000566208, NONHSAG045500, ENST00000517573, NONHSAT034045, and NONHSAT142707), previously confirmed to be downregulated in MDD subjects, were selected for this study. All six lncRNAs were downregulated in MDD with suicidal ideation, compared with MDD no suicidal ideation and control groups. The authors concluded that six lncRNAs may possibly serve as biomarker for suicidal ideation among MDD patients. Another study showed that decreased expression of TCONS_00019174 was associated with depression and expression change was rescued by antidepressant treatment (82). Additionally, an in vitro experiment showed that TCONS_00019174 was negatively correlated with phosphorylated-GSK3β (p-GSK3β) protein and β-catenin. Further, the overexpression of NONHSAG045500 inhibited the expression of serotonin transporter (SERT), while siRNA interference of NONHSAG045500 induced the upregulation of SERT (83). These studies indicate that the peripheral expression change of lncRNAs can have functional relevance and may be worth pursuing in the brain.
microRNAs
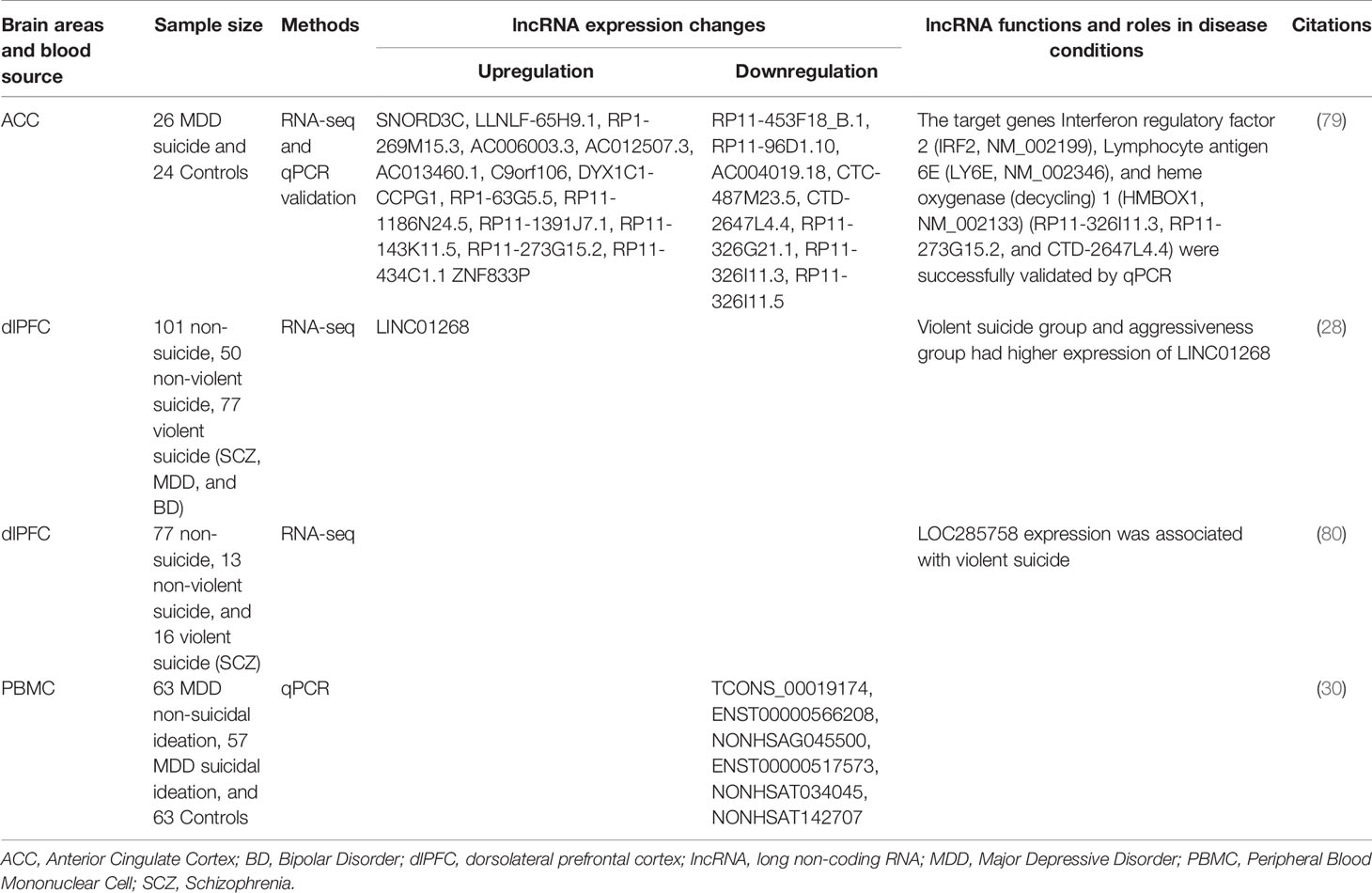
MicroRNAs (miRNAs) are the best studied ncRNAs in terms of functionality and relevance to psychiatric illnesses. Averaging 22 nucleotides in length, miRNAs are synthesized via several enzymatic processes. Initially, primary miRNA (pri-miRNA) is transcribed from an encoded gene. Next, RNase III enzyme Drosha generates precursor miRNA (pre-miRNA) by removing the flanking segments and -11 bp stem region of pri-miRNA. Drosha requires DiGeorge syndrome critical region 8 (DGCR8) to complete this process. pre-miRNAs are then translocated to cytoplasm with the help of Exportin-5 (XPO5). In the cytoplasm, dicer, a double-stranded RNA endoribonuclease, converts pre-miRNA into double-stranded mature miRNA. Dicer requires TAR RNA-binding protein (TRBP) as a cofactor (84–88). Finally, one strand of miRNA/miRNA* duplex loads into Argonaute homologue protein (Ago) to make RNA-induced silencing complex (RISC). RISC/miRNA complexes mainly work as suppressors of mRNA expression (Figure 2).
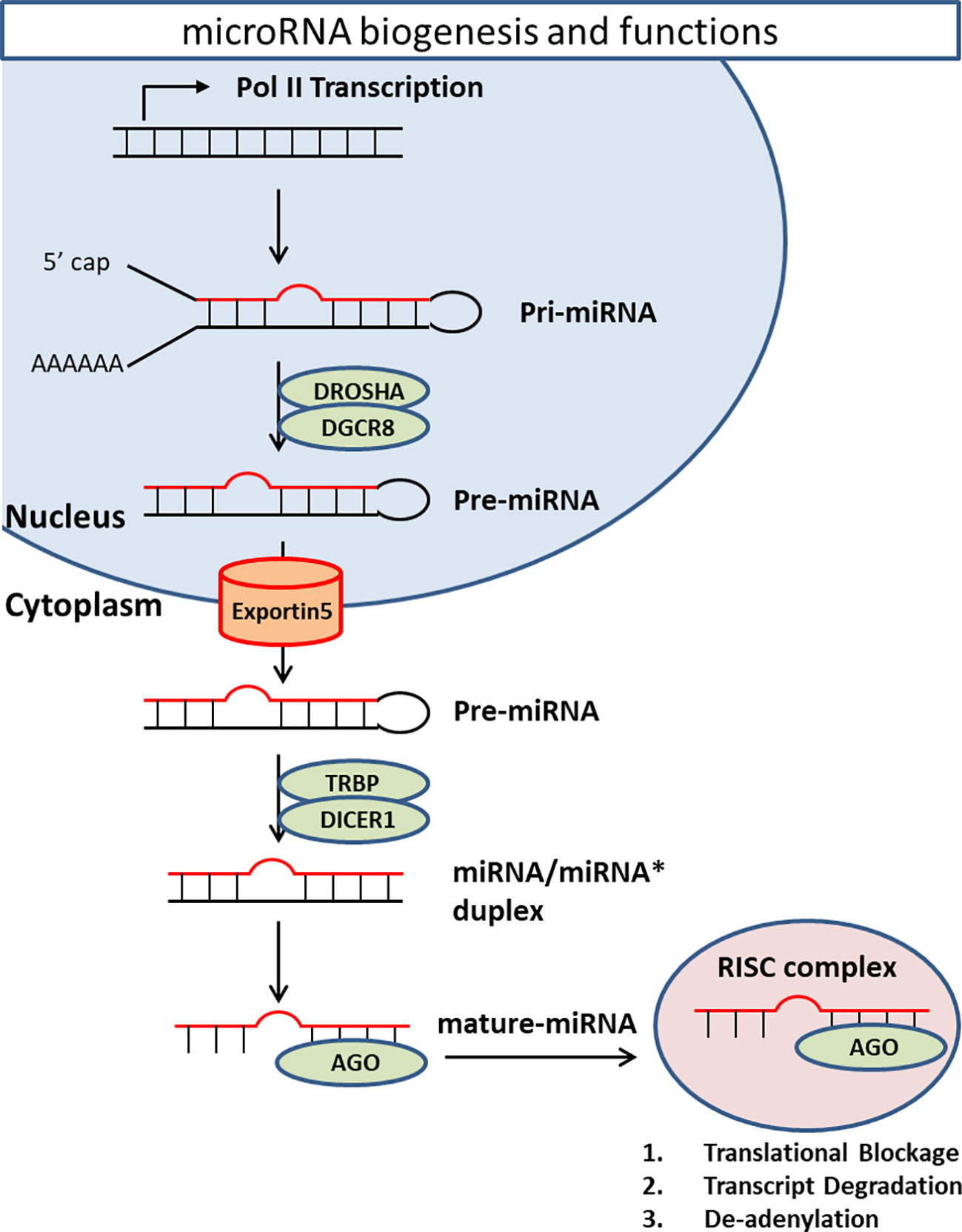
Figure 2 MiRNA biogenesis and functions. Primary miRNA is transcribed from encoded gene. RNase III enzyme Drosha generates precursor miRNA by removing the flanking segments and -11 bp stem region of pri-miRNA. Drosha requires DiGeorge syndrome critical region 8 (DGCR8) to complete this process. pre-miRNAs are then transported from nucleus to cytoplasm by a transporter Exportin-5 (XPO5) in a Ran-GTP-dependent manner. In the cytoplasm, Dicer converts pre-miRNA into double-strand mature miRNA. Dicer requires TAR RNA-binding protein (TRBP) as a cofactor. Finally, one strand of miRNA/miRNA* duplex loads onto Argonaute homolog protein (Ago) to make RNA-induced silencing complex (RISC). RISC/miRNA complex mainly works as suppressor of mRNA expression through translational blockage, transcript degradation, and deadenylation.
Several miRNAs play a key role in modulating synaptic functions and neural structures. Overexpression of miR-125b can disrupt spine structure, whereas sponging of endogenous miR-125b can induce the prolongation of dendritic protrusions (89). Introduction of miR-132 in hippocampal neurons can induce dendrite morphogenesis, which is caused by the regulation of GTPase-activating protein (p250GAP) (90, 91). At the behavioral level, transgenic miR-132 mice show altered cognitive functions through repression of MeCP2 translation, which is implicated in Rett Syndrom and mental retardation (92). The role of miRNAs in various psychiatric illnesses has been studied extensively. A summary of the human postmortem brain miRNA findings in psychiatric illnesses is provided in Table 3. Below, we discuss miRNA studies in human brain pertaining to SCZ, MDD, and BD as well as suicidal behavior.
miRNAs and Schizophrenia
MiR-137 is a well-known miRNA in schizophrenia studies (110). The first report of the involvement of miR-137 came from a GWAS study which linked a risk variant rs1625579 within miR-137 to SCZ (111). A previous study had reported that target genes for miR-137 are associated with activation in the dlPFC (112). Guella et al. (102) subsequently examined miR-137 and its target genes in the dlPFC of SCZ and BD subjects. MiR-137 expression in the dlPFC was not different between diagnoses. Interestingly, when the genotype of rs1625579 was considered, significantly lower miR-137 expression was found along with higher expression of TCF4, a target gene of miR-137, in the homozygous TT subjects compared to TG and GG subjects within the control group. The miR-137 expression was region specific with amygdala and hippocampus having the highest levels. These results suggest that miR-137 and associated TCF4 gene may be risk factors for SCZ. Using miRNA-seq, one amygdala study showed seven upregulated and 11 downregulated miRNAs (73); of them, miR-1307 was located on a locus previously found to be associated with SCZ risk allele rs11191419 in GWAS (113).
Using custom microarray, Perkins et al. (93) studied miRNA expression in BA9 of subjects with SCZ or schizoaffective disorder and compared them with psychiatrically unaffected individuals. The study found 15 downregulated and one upregulated miRNA in these subjects. They further examined sequences in the pri-miRNA motifs of those dysregulated miRNAs and found that several miRNAs shared the same sequence. For example, miR26b, miR-30a, miR-30b, and miR-7-1 had UGAGNCUU upstream sequences, whereas miRNAs miR-9-1, miR-9-2, miR-9-3, miR-7-3, and miR-30e had GUCNCUUC upstream sequences in their corresponding pre-miRNAs. This suggests the possibility that the processing of pri-miRNA to pre-miRNA might be affected, which could eventually be related to miRNA expression changes. Moreau et al. (94) reported that 19% of miRNAs in BA9 were altered based on diagnostic classification among SCZ, BD, and control subjects. Interestingly, all of the downregulated miRNAs in SCZ were downregulated in BD subjects, supporting the similarity of the genetic background between SCZ and BD. Banigan et al. (95) explored miRNA expressions in exosomes of BA9. Based on the microarray data, they validated the upregulation of miR-497. Several studies have shown that exosomes in the CNS may play a role in neuronal communications, nerve regeneration, synaptic plasticity, and immune response (114, 115).
Beveridge et al. (96) detected 25 upregulated miRNAs in BA9 of SCZ subjects; of them, 10 miRNAs (let-7d, miR-128a, miR-16, miR-181b, miR-181a, miR-20a, miR-219, miR-27a, miR-29c, and miR-7) were validated with qPCR. They also revealed that the biogenesis of miR-181b was perturbed in SCZ possibly due to the altered expression of pri- and pre-miR-181b and miRNA biogenesis-related enzymes DGCR8, DROSHA, and DICER1. Kim et al. (97) detected 7 upregulated miRNAs in another prefrontal cortical area BA46 of SCZ subjects. Of them, miR-132 was highly expressed in the forebrain and was regulated by cAMP response element-binding (CREB) and extracellular signal regulated kinase (ERK) signaling. Both CREB and ERK are targets of antipsychotic drugs (91, 116). On the contrary, Miller et al. (101) reported downregulation of miR-132 in dlPFC of SCZ subjects. The authors found that NMDA antagonist, when administered to adult mice, resulted in lower expression of miR-132 in the prefrontal cortex. Since miR-132 is highly expressed in the postnatal period, a period of synaptic pruning thought to be related to the mechanism of neurodevelopmental susceptibility to SCZ through NMDAR signaling (91), it was concluded that dysregulation in miR-132 and associated target genes may contribute to the neurodevelopmental and morphological abnormalities shown in SCZ.
Santarelli et al. (98) investigated miRNA expression in BA 46 of subjects with SCZ and schizoaffective disorder. They found differential regulation of 28 miRNA in the SCZ group compared to controls. Of them, six miRNAs (miR-17, miR-107, miR-134, miR-328, miR-382, and miR-652) were validated by qPCR. They also found increased expressions of miRNA biogenesis enzymes DGCR8, DROSHA, and DICER1 and concluded that miRNA changes may occur due to disruption in mRNA biogenesis. In another study, Zhu et al. (99) found lower expression of miR-346 in BA46 of SCZ subjects. Interestingly, miR-346 is positioned in intron 2 of the glutamate receptor ionotropic delta 1 (GRID1) gene, which was found to be downregulated in SCZ subjects. GRID1 is an important gene related to SCZ susceptibility (117). Recently, temporal dynamics of miRNA expression in dlPFC as well and their dysregulation was studied in SCZ subjects (100). It was found that miRNAs that were enriched in infants were upregulated, and those enriched in prepuberty were downregulated in SCZ. The targets of these miRNAs included genes encoding for synaptic proteins and were found to be altered in SCZ subjects. In addition, a significant upregulation in the expression of miR-3162 and miR-936 was noted. These results suggest the miRNAs may participate in the development of dlPFC. In addition, miRNAs that are enriched in infancy and prepuberty may be critical in the deployment of SCZ pathology.
Two other reports which used superior temporal gyrus (STG) also linked miRNA abnormalities to SCZ (96, 103). Both studies showed upregulation of miR-181b in SCZ subjects. Using reporter assay, one study revealed that miR-181b could regulate VSNL1 and GRIA2 genes (103). Expression changes in GRIA2 have been reported in BA46 (118) and the medial temporal lobe (119, 120) of SCZ subjects.
miRNAs and Major Depressive Disorder
A number of studies have linked miRNA changes to major depressive disorder (MDD) pathophysiology. Azevedo et al. (104) analyzed select 29 miRNAs by qPCR in ACC of subjects with MDD, BD, and control subjects. They found that miR-132, miR-133a, and miR-212 were differentially regulated in BD, miR-184 in MDD, and miR-34a in both MDD and BD. None of the miRNAs survived significance after multiple corrections. In silico analysis identified NCOA1, NCOA2, and PDE4B as a target of miR-34a, while NCOA2 and PDE4B were targeted by miR-184. Further qPCR analyses showed that NCOA1 had an inverse relationship with miR-34a in BD, while NCOR2 had a positive relationship with MDD. It is pertinent to mention that both NCOA1 and NCOR2 regulate the GR-mediated transcription of corticotropin-releasing hormone, a risk gene for stress-related psychiatric illnesses (121).
Lopez et al. (105) studied miRNA expression in the ventrolateral prefrontal cortex (vmPFC) of depressed individuals and found that primate-specific and human brain enriched miR-1202 was the most differentially regulated miRNA in MDD subjects. They found that miR-1202 regulated the expression of gene encoding metabotropic glutamate receptor-4 (GRM4). A recent report suggests that GRM4 3′ UTR variant (rs2229901) is associated with MDD risk (122). Interestingly, the antidepressant treatment induced miR-1202 expression and downregulated GRM4 expression (105). These results suggest that miR-1202 may be associated with MDD and may serve as target for new drug development.
It has been reported that miR-124-3p is highly expressed specifically in the brain (123), and has neuron-specific expression (124–126). Our group reported a significant upregulation in miR-124-3p expression in BA46 of depressed individuals (106). In silico prediction suggested that AMPA/NMDA receptors are major targets of miR-124-3p. Of them, expression changes for GRIA2 in dlPFC (127), GRIA3 in dentate gyrus (128) and PFC (129), and GRIA4 in PFC (129, 130) have been found in depressed individuals.
miRNAs and Bipolar Disorder
Compared to MDD, postmortem brain studies of miRNAs in bipolar disorder are limited. Azevedo et al. (104) reported downregulation of 4 miRNAs (miR-34a, miR-132, miR-133a, and miR-212) in ACC of BD subjects. miR-34a was correlated with PDE4B expression in BD. PDE4B expression changes have been found in several psychiatric disorders including SCZ (131, 132), MDD (133), and BD (134). Another ACC (BA24) study conducted by Choi et al. (107) explored expression changes in five miRNAs (miR-29c, miR-31, miR-15b, miR-497, miR-219, and miR-149) in glial exosomes. They found that miR-149 was significantly upregulated in BD subjects. Since miR-149 inhibits glial proliferation, an increase in miR-149 may be associated with previously reported reduced glial cell numbers in BA24 of patients diagnosed with BD (135).
Banigan et al. (95) explored exosomal miRNA expression in BA9 of patients diagnosed with BD and found that miR-29c expression was significantly increased. miR-29c regulates canonical Wnt signaling (136). Wnt signaling is linked to BD pathogenesis such that lithium impairs GSK-3β expression, a component of Wnt signaling system (137). Kim et al. (97) investigated the expression of 667 miRNAs in the PFC of individuals with SCZ and BD using Taqman Low Density Array system. They found downregulation of seven miRNAs in SCZ and 15 miRNAs in BD. These miRNAs targeted genes within the networks overrepresented for neurodevelopment, behavior, and SCZ and BD development (97).
Genetic similarities between SCZ and BD have been well documented (138). The overlapping miRNA changes between SCZ and BD may explain the genetic similarity. In the study by Kim et al. (97), miR-154* showed downregulation in PFC of both SCZ and BD subjects. Kohen et al. (108) investigated transcriptomic changes in the dentate gyrus (DG) granule cells of hippocampus in subjects with SCZ, MDD, BD, and nonpsychiatric controls. One miRNA that stood out was miR-182. MiR-182 is involved in several biological processes such as immune response (139), DNA repair (140), and the regeneration of peripheral nerves after injury (141, 142), which play key roles in BD pathogenesis. Carriers of different genotypes among subjects with SCZ and MDD had a loss of DG miR-182 signaling. Interestingly, carriers of high (C/C) vs low-expressing genotype (C/T or T/T) of rs76481776 in both controls and BD subjects, which is located in miR-182, had different levels of miR-182 target gene expression. This suggests that miR-182 may play a critical role in shaping the DG transcriptome based on genotype. Bavamian et al. (109) studied miRNA expression changes in cerebellum and reported an increased expression of miR-34a in BD subjects. In addition, they showed an upregulation of miR-34a in induced neurons derived from BD human fibroblast collected from another cohort. Subsequently, they validated the BD risk genes ankyrin-3 (ANK3) and voltage-dependent L-type calcium channel subunit (CACNB3) as direct targets of miR-34a with luciferase reporter assay. Functionally, enhanced miR-34a expression reduced neuronal differentiation and altered neuronal morphology as well as expression of synaptic proteins. On the other hand, reduced miR-34a expression improved dendritic elaboration.
The studies mentioned above suggest that BD patients have a distinct miRNA expression profile; however, there is a significant overlap with SCZ. This is quite relevant. In the future, miRNA profiling may be useful in identifying commonality and distinction between these disorders, both functionally and at pathophysiological levels.
miRNAs and Suicide
MiRNA studies related to suicide are summarized in Table 4. Using Taqman Low Density Array system, our group was the first to examine miRNA expression changes in BA9 of 18 MDD suicides and 17 nonpsychiatric control subjects (143). We found a global downregulation of miRNAs in MDD subjects. Altogether, we found significant downregulation in 21 miRNAs. Additional 29 miRNAs were also found to be downregulated by >30%; however, they did not reach statistical significance. When the regulation of these downregulated miRNAs was studied in detail, it was observed that many of them were encoded at the same or nearby chromosomal loci. In addition, they shared 5′-seed sequences. This suggests that miRNAs that were downregulated in MDD subjects had overlapping mRNA targets. Several instances were noted where miRNAs were found to be targeting the same genes. Some of the validated target genes included transcription factors, transmembrane and signaling proteins, growth factors, epigenetic modifiers, and apoptotic regulatory proteins. Predicted targets included ubiquitin ligases, CAMK2G, splicing factor NOVA1, and the GABA-A receptor subunit GABRA4. DNMT3b was another protein that was found to be targeted and further validated. DNMT3b is a key gene in epigenetic modifications and has been shown to be involved in MDD pathophysiology (152).
Gorinski et al. (144) examined whether 5-HT1A receptor (5HT1AR) palmitoylation is a mechanism involved in MDD and suicidality. They found that miR-30e expression was increased, while ZDHHC21 expression, which they identified as a major palmitoyl acyltransferase involved in palmitoylation of 5-HT1AR, was reduced in PFC of MDD suicide subjects, suggesting that 5-HT1AR palmitoylation may be key to MDD pathogenesis and treatment. Another mechanistic study by Torres-Berrio et al. (147) examined whether the expression of Netrin-1 guidance cue receptor DCC (deleted in colorectal cancer) gene was associated with resiliency or susceptibility to PFC dysfunctions in MDD suicide via miRNAs. They found a significant correlation between the decreased expression of miR-218 in BA44 of MDD suicide subjects and the elevated expression of DCC. By conducting an animal study, they concluded that miR-218 was associated with susceptibility versus resiliency to stress-related disorders through regulation of DCC expression.
Our group assessed miRNA expression in the synaptosome of BA10 from a cohort of SCZ, MDD, and BD subjects (145). Altogether, eight out of 14 BD subjects, seven out of 15 MDD subjects, and three out of 14 SCZ subjects had died by suicide. The result of comparison between all suicides (across diagnostic categories) and all non-suicide groups showed two upregulated miRNAs (miR-376a and miR-625) and six downregulated miRNAs (miR-152, miR-181a, miR-330-3p, miR-34a, and miR-133b). Based on significance values, miR-152 was found to be strongly associated with suicide. Distinct set of miRNAs were found in all three disorders; however, several miRNAs showed overlapping changes in SCZ and BD subjects. Discrete changes were also noted in MDD and suicide groups. Interestingly, downregulated miRNAs in SCZ group were enriched within the synapse. Follow-up studies in purified synaptosomes by deep sequencing in SCZ group suggested that there was a significant loss of small RNA expression in synaptosomes only for certain sequence lengths within the miRNA range. In addition, a large number of miRNAs (n = 73) were significantly downregulated and only one miRNA was upregulated. Remarkably, greater fold change of miRNAs in SCZ was associated with lower synaptic enrichment ratio in control subjects and vice-versa. Overall, synaptic miRNAs were generally downregulated in SCZ; however, highly synaptically enriched miRNAs showed greater down-regulation. It was concluded that there may be a deficit in synthesis, transport, or processing of miRNAs in SCZ; however, this process may be more selective for those miRNAs that are localized predominantly in the synaptic compartment.
Maussion et al. (146) examined miRNA expression changes in BA10 from 38 suicide subjects across SCZ, MDD, BD, and control subjects. miR-185* and miR-491-3p were found to be upregulated in suicide subjects compared to control subjects. TrkB-T1, one of the transcript variants of TrkB, expression was significantly lowered in suicide subjects and correlated with miR-185* expression. This was specific only for TrkB-T1, but not for TrkB-T2 or TrkB-FL. TrkB has been shown to be involved in suicidal behavior as has been reported by us previously (67). In an effort to reveal the regulation of polyamine system and suicidal behavior, Lopez et al. (153) examined the expression of SAT1 and SMOX in BA44 and found them to be lower in MDD suicide subjects. They selected 10 miRNAs which were predicted to regulate SAT1 and SMOX genes. Of those, four miRNAs (miR-34c-5p, miR-320c, miR-139-5p, and miR-320c) were significantly upregulated and correlated with the polyamine stress response genes (miR-34c-5p and miR-320c with SAT1; miR-139-5p and miR-320c with SMOX). They concluded that polyamine stress response genes including SAT1 and SMOX may be linked to alterations in miRNA networks that are responsive to stress in MDD suicide subjects. In another study, Lopez et al. (150) noted upregulation of miR-425-3p, miR-146-5p, miR-24-3p, and miR-425-3p in ventrolateral prefrontal cortex (vPFC) of MDD suicide subjects. Resulting from in silico prediction and in vitro experiments, they concluded that all four miRNAs were related to the MAPK/Wnt signaling pathways. These signaling systems have been implicated in suicidal behavior (154).
In an effort to examine the regulation of pro-inflammatory cytokine genes in suicidal behavior, we examined the relationship between TNF-α and its regulatory miRNAs (29). We found an increased expression of miR-19a-3p in BA46 of MDD individuals who had died by suicide. The increased expression of miR-19a-3p was also found in PBMC of MDD subjects who had suicidal ideation. Mechanistically, we found that RNA-binding protein HuR helped in stabilizing TNF-α transcript. This occurred apparently by sequestering its 3′ untranslated region from miR-19a-3p-mediated inhibition. The study suggests that miR-19a-3p may be involved in cytokine dysregulation in suicidal individuals. Recently, we also explored miRNA changes in the locus coeruleus (LC) of MDD suicide subjects (148). A total of 10 upregulated and three downregulated miRNAs were detected in these subjects. Based on target gene prediction using the upregulated miRNAs, we narrowed our study to those genes with a strong neuropsychiatric background. We focused on RELN, GSK-3β, MAOA, CHRM1, PLCB1, and GRIK1 and found reduced expression levels for RELN, GSK-3β, and MAOA.
Maheu et al. (149) examined glial cell line-derived neurotrophic factor in MDD suicide subjects and found an isoform-specific decrease in GDNF family receptor alpha 1 (GFRA1) mRNA, which was associated with lower GFRα1a protein levels in basolateral amygdala. They found upregulation in several miRNAs that may target GFRA1 gene. One of them was miR-511. Under the condition of miR-511 overexpression in neural progenitor cells (NPC), they observed that protein expression of GFRα1 was significantly decreased and concluded that GFRα1 is regulated by miR-511 and may be associated with suicidality.
In terms of blood studies associated with suicidality, only one study is available. Sun et al. (151) reported downregulation of miR-34b-5p and miR-369-3p in MDD subjects with suicidal ideation compared to MDD without suicidal ideation. These changes were consistent with our previous postmortem study conducted in BA9 of MDD suicide subjects (143).
Piwi-Interacting RNAS
Piwi-interacting RNAs (piRNA) are the large small non-coding RNAs (26–32 nucleotide), which are preferentially expressed in nuclei and cytoplasm (155). Piwi proteins are recognized as one domain of argonaute (Ago) protein and bind to piRNA (156–159). The functions of Piwi/piRNA are unclear; however, they are highly expressed in germlines such as testes (156) and ovaries (160). Several studies show their role in epigenetic regulation of transposable elements in germlines (161–163). The existence of piRNA in mouse hippocampal neuron was reported by Lee et al. (164). In an Aplysia study, Rajasethupathy et al. (24) found that neuronal piRNA has a role in regulating memory-related synaptic plasticity. They found that the Piwi/piRNA complex may enable serotonin-dependent methylation of a CREB2 promoter CpG island in neurons, leading to the enhancement of long-term synaptic facilitation, learning-related synaptic plasticity and memory storage in Aplysia. More recently, Nandi et al. (165) reported that piRNA of Mili, the mouse ortholog of Piwi, exhibited behavioral deficits such as hyperactivity and less anxiety (165).
There is limited direct evidence of piRNA in psychiatric disorders. One study identified 37 piRNAs in ACC of SCZ subjects and one piRNA was correlated with antipsychotic medication (166). However, the authors did not mention the expression change between SCZ and control subjects because most of piRNAs were expressed only in a small number of samples.
Small Interfering RNAS
Small interfering RNAs (siRNA) are short (~20-24 nucleotide) double-stranded RNAs (dsRNAs) and interfere in the translation of proteins. Dicer enzyme cleaves long dsRNA and small hairpin RNAs into siRNAs (167, 168). SiRNAs have a role in mRNA cleavage by guiding RNA-induced silencing complex (RISC) to its complementary target mRNAs. Initially, siRNAs are loaded into Ago2 as duplexes. Subsequently, passenger strand of siRNAs is cleaved by Ago2; then, guide strand from siRNA duplex is liberated and produces active RISC, which has the capability to cleave target mRNAs (169, 170). Mainly, siRNAs are used for suppressing target genes as a scientific experimental method. To our knowledge, there is no report about siRNA expression changes in psychiatric disorders.
Small Nuclear RNAS
Small nuclear RNAs (snRNA), averaging about 150 nucleotides in length, are one of the components of spliceosomes. Five snRNAs (U1, U2, U4, U5, and U6) have been identified. snRNAs form ribonucleoprotein complexes (snRNPs) that can target and bind to specific sequences on pre-mRNAs (171). Spliceosomes have key roles in nuclear pre-mRNA splicing that remove the pre-mRNA regions (intron) that do not code for functional molecules. Initially, U1 snRNA binds to its associated proteins at the 5′ splice end of the hnRNA (172). Subsequently, U2 snRNP is recruited to the spliceosome binding site for making complex A. Bai et al. (173) reported that unique U1 snRNP pathology is involved in abnormal RNA splicing in Alzheimer’s disease (AD) that leads to the alteration in amyloid precursor protein (APP) expression and Aβ levels. Their group also showed U1 snRNP pathologic changes in postmortem brain of early onset AD (174). In addition, the small nuclear ribonucleoprotein U1-70K is aggregated in AD brain (175) and might contribute to neuronal toxicity in conjunction with N-terminal truncation (N40K) fragment derived from cleavage (176). In a mouse study, it is reported that dysfunction of U2 snRNA causes neurodegeneration through distortion of pre-mRNA splicing (177). A positron emission tomography study showed that amyloid accumulation is found in the brain of late-life MDD patients (178). At this stage, amyloid deposition may occur during the preclinical phase of dementia in patients who have depressive symptoms (179, 180), although MDD itself is a risk factor for the onset of AD (181). It is possible that Aβ changes through snRNAs may occur in late-life MDD patients even though the relationship between MDD and Aβ is unclear. One study, using ACC, found that 149 snRNAs were expressed in both SCZ and control subjects, and 35 snRNAs were expressed specifically for SCZ subjects (166). However, their biological impact on SCZ pathogenesis is unclear.
Small Nucleolar RNAS
Small nucleolar RNAs (snoRNAs) are generally responsible for guiding modifications in rRNAs, tRNAs, and snRNAs. snoRNAs are divided into two main classes: 1) box C/D and 2) box H/ACA. Box C/D snoRNAs, averaging 60–90 nucleotides, have a role in catalyzing 2′-O-ribose methylation. Box H/ACA snoRNA, averaging 120–140 nucleotides, guide pseudouridylation (182, 183). In 2002, brain specific C/D box snoRNAs HBII-52 (SNORD115) and HBII 85 (SNORD116) and H/ACA box snoRNA HBI-36 were detected (184). Prader–Willi syndrome (PWS) is a neurodevelopmental disorder caused by imprinted gene clusters at human chromosome region 15q11q13. It was reported that SNORD115 and SNORD116 are partially responsible for the development of PWS (185, 186). Moreover, HBI-52 and HBI-36 snoRNAs are thought to regulate 5-HT2c mRNA expression (184). 5-HT2c has a role in MDD pathogenesis. Indeed, several antidepressants work as an antagonist to 5-HT2c (187–189). In terms of schizophrenia, it is considered that 5HT-2c relates to SCZ pathogenesis through the dopaminergic pathway (190, 191) and by causing side effects to antipsychotic treatment (192). Actually, clozapine, a drug used for refractory schizophrenia, has high affinity to 5-HT2c (192). SnoRNAs may influence psychiatric disorders directly or through 5-HT2c receptors. Epigenetic changes have also been found in SNORD115 and SNORD116 in monozygotic twin study of SCZ. Hypermethylation in 12 regions on SNORD115 and five regions on SNORD116 regions were found in affected individuals (193).
In PFC synaptosomes from SCZ subjects, we noted ~50% decrease in a set of sequences that were derived from a C/D box snoRNA and SNORD85 (145). A 27-mer sequence TTCACTGATGAGAGCATTGTTCTGAGC was the most abundant one that incorporated a C/D box and terminated four bases before the 3′ end of the host snoRNA. Our study, for the first time, showed that not only is this novel class of small ncRNAs expressed in humans, and more so in synaptosomes, but they may be significantly altered in a psychiatric disorders. Another study also showed expression change in snoRNAs in SCZ (166). A total of 343 snoRNAs were expressed in both SCZ and control subjects and six snoRNAs were expressed specifically in SCZ subjects. However, none of the snoRNAs were significantly changed in SCZ compared to controls.
Limitations
There are a several limitations to ncRNA studies in neuropsychiatric disorders: 1) Several studies show changes in expression of expression lncRNAs and miRNAs; however, they are inconsistent even when studied in the same brain region with the same diagnosis. This could be attributed to several factors. For example, confounding variables such as PMI, brain pH, freeze–thaw cycle, age, and sex vary between studies. In addition, most of the studies use whole tissue. As mentioned earlier, the expression of lncRNAs and miRNAs are brain region and cell type specific. This cellular heterogeneity may affect ncRNA expressions. Thus, studying their expression patterns in neurons and glial cells (oligodendrocytes, astrocytes, and microglia) may delineate these discrepancies. Even within neurons, specific neuronal population needs to be studied. In this regard, single cell study will be helpful. 2) In recent years, many studies have focused on miRNAs and to a certain extent lncRNAs; however, the role of siRNAs, piRNAs, snoRNAs, and snRNAs are understudied. Given their important roles (e.g. epigenetic regulation of transposable elements; mRNA cleavage by guiding RISC), future research will be needed to explore their functional significance in psychiatric disorders. 3) Correct identification of new snRNAs and snoRNAs and their epigenetic roles needs to be studied (194, 195). New technologies like nanopore sequencing and structure prediction algorithms are emerging that can be helpful in this regard. 4) Although new members of ncRNA are being discovered regularly, the functions of these novel ncRNAs are not well understood. More in-vivo functional studies followed by high-throughput sequencing after chemical cross-linking or photoactivatable cross-linking and immunoprecipitation (PAR-CLIP) will be effective in probing the functional roles of novel ncRNAs at the cellular level (196). These assays can be also performed in human postmortem brain samples, but by minimizing the loss of tissue integrity and maximizing the likeliness of capturing the interaction at the molecular level. 5) Most of the studies have shown the temporal changes in ncRNA expression. Although it is very difficult to measure ncRNA expressions at different time points in postmortem samples, it is relatively easy to measure them in peripheral tissue samples. Using longitudinal studies, it will be possible to explore the diagnosis/therapeutic/disease state biomarkers for psychiatric illnesses.
Conclusions
ncRNAs are gaining traction for their role in various psychiatric illnesses. A schematic diagram of the non-coding RNAs’ impact on psychiatric illnesses and suicidality is depicted in Figure 3. Both long-non-coding RNAs and miRNAs have been extensively studied in MDD, SCZ, and BD. As can be seen in Tables 1–4, a large number of miRNAs and lncRNAs have been implicated in SCZ, MDD, and BD. There were several miRNAs that showed overlapping changes between SCZ and BD, suggesting that these two disorders may share common susceptibility factors as has been demonstrated in several genetic studies.
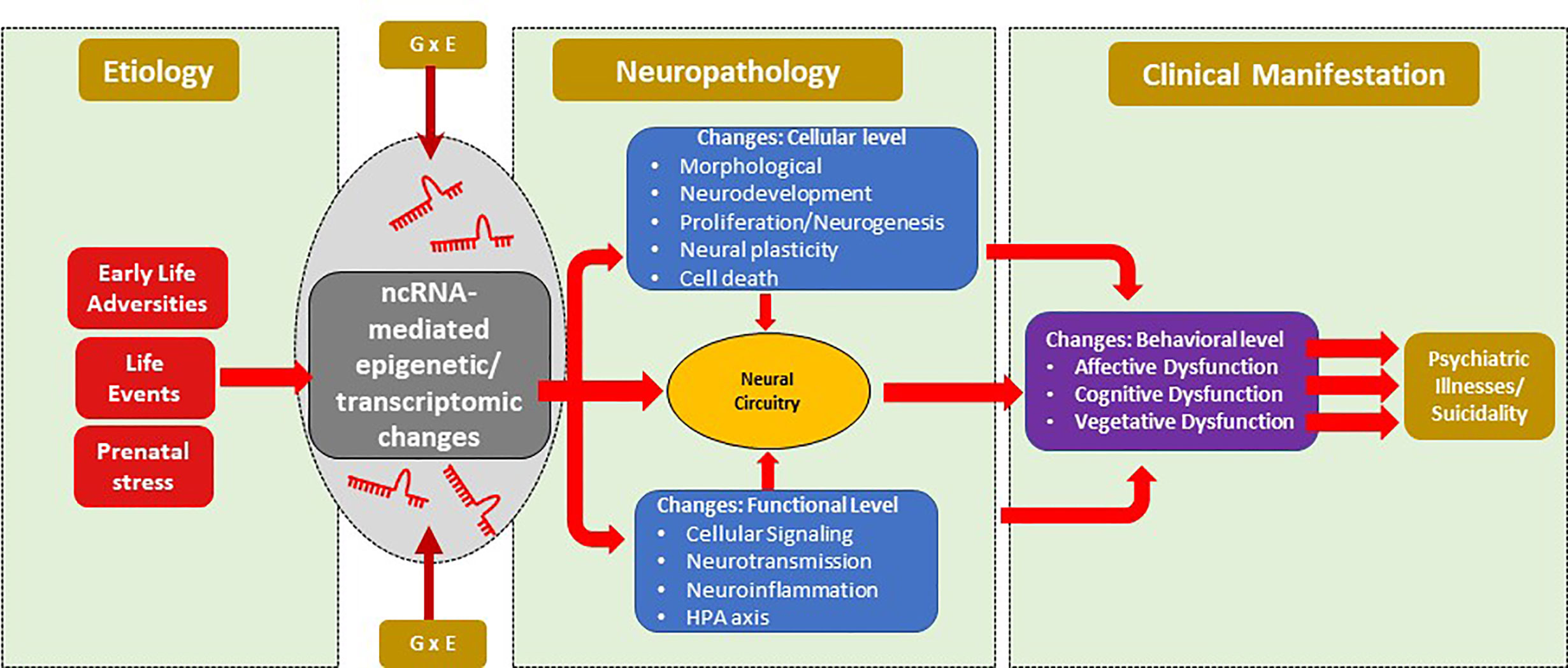
Figure 3 Schematic diagram of the non-coding RNAs’ impact on psychiatric illnesses and suicidality. Risk factors for mental illnesses include early life adversity, current or recurrent life events that along with gene environment interaction can lead to epigenetic modifications mediated by ncRNAs. These modifications can give rise to neuropathology mediated by changes at cellular and/or molecular levels, which can subsequently alter neural circuitry. Phenotypic changes can arise from circuitry changes that can mediate the development of psychiatric illnesses such as major depression, bipolar disorder and schizophrenia. Suicidal behavior could be a manifestation of psychiatric illnesses or may be independent of psychiatric illnesses. ncRNAs, non-coding RNA.
The population-based studies have explored the expression changes of lncRNA and miRNA among psychiatric patients who have suicidal ideation. A couple of postmortem brain studies have also explored suicide associated ncRNAs. Interestingly, both lncRNAs and miRNAs were specifically changed in psychiatric patients who died by suicide even though the sample size and number of studies are quite small. The results are encouraging and need further exploration.
In terms of other small RNAs, the studies are quite limited; nevertheless, they appear to be promising. piRNAs and snoRNAs show interesting results in SCZ patients that may shed light on their role in regulating other RNAs and their role in this psychiatric illness. These studies need to be expanded in order to understand their precise role in regulatory mechanisms that can be involved in the pathogenesis of psychiatric disorders.
Author Contributions
YD conceptualized, outlined, and edited the paper. YY reviewed literature and co-wrote the paper.
Funding
This work was supported by funding from the National Institutes of Health (R01MH082802; R01MH101890; R01MH100616; R01MH107183; R01MH118884) to YD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489:101–8. doi: 10.1038/nature11233
2. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (2005) 309:1559–63. doi: 10.1126/science.1112014
3. Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet (2014) 15:423–37. doi: 10.1038/nrg3722
4. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
5. Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature (2017) 543:199–204. doi: 10.1038/nature21374
6. Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489:57–74. doi: 10.1038/nature11247
7. Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev (2007) 87:799–823. doi: 10.1152/physrev.00036.2006
8. Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol (2010) 17:1030–4. doi: 10.1038/nsmb.1841
9. Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell (2011) 147:370–81. doi: 10.1016/j.cell.2011.09.041
10. Kadakkuzha BM, Liu XA, Mccrate J, Shankar G, Rizzo V, Afinogenova A, et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci (2015) 9:63. doi: 10.3389/fncel.2015.00063
11. Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol (2012) 11:189–200. doi: 10.1016/S1474-4422(11)70286-1
12. Webb A, Papp AC, Curtis A, Newman LC, Pietrzak M, Seweryn M, et al. RNA sequencing of transcriptomes in human brain regions: protein-coding and non-coding RNAs, isoforms and alleles. BMC Genomics (2015) 16:990. doi: 10.1186/s12864-015-2207-8
13. Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci (2008) 28:11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008
14. Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci (2010) 11:14. doi: 10.1186/1471-2202-11-14
15. Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA (2010) 16:1516–29. doi: 10.1261/rna.1833310
16. Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PloS Genet (2009) 5:e1000617. doi: 10.1371/journal.pgen.1000617
17. Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J (2010) 29:3082–93. doi: 10.1038/emboj.2010.199
18. Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci (2009) 12:1020–7. doi: 10.1038/nn.2371
19. Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron (2015) 88:861–77. doi: 10.1016/j.neuron.2015.09.045
20. Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neuroscientist (2008) 14:434–45. doi: 10.1177/1073858408319187
21. Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J (2012) 31:522–33. doi: 10.1038/emboj.2011.459
22. Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci USA (2012) 109:16939–44. doi: 10.1073/pnas.1202956109
23. Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics (2013) 10:632–46. doi: 10.1007/s13311-013-0199-0
24. Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell (2012) 149:693–707. doi: 10.1016/j.cell.2012.02.057
25. Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, et al. A Primate lncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron (2016) 90:1174–88. doi: 10.1016/j.neuron.2016.05.005
26. Kocerha J, Dwivedi Y, Brennand KJ. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Mol Psychiatry (2015) 20:677–84. doi: 10.1038/mp.2015.30
27. Sartor GC, St Laurent G, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Front Genet (2012) 3:106. doi: 10.3389/fgene.2012.00106
28. Punzi G, Ursini G, Viscanti G, Radulescu E, Shin JH, Quarto T, et al. Association of a Noncoding RNA Postmortem With Suicide by Violent Means and In Vivo With Aggressive Phenotypes. Biol Psychiatry (2019) 85:417–24. doi: 10.1016/j.biopsych.2018.11.002
29. Wang Q, Roy B, Turecki G, Shelton RC, Dwivedi Y. Role of Complex Epigenetic Switching in Tumor Necrosis Factor-alpha Upregulation in the Prefrontal Cortex of Suicide Subjects. Am J Psychiatry (2018) 175:262–74. doi: 10.1176/appi.ajp.2017.16070759
30. Cui X, Niu W, Kong L, He M, Jiang K, Chen S, et al. Long noncoding RNA expression in peripheral blood mononuclear cells and suicide risk in Chinese patients with major depressive disorder. Brain Behav (2017) 7:e00711. doi: 10.1002/brb3.711
31. Downar J, Blumberger DM, Daskalakis ZJ. The Neural Crossroads of Psychiatric Illness: An Emerging Target for Brain Stimulation. Trends Cognit Sci (2016) 20:107–20. doi: 10.1016/j.tics.2015.10.007
32. Resibois M, Verduyn P, Delaveau P, Rotge JY, Kuppens P, Van Mechelen I, et al. The neural basis of emotions varies over time: different regions go with onset- and offset-bound processes underlying emotion intensity. Soc Cognit Affect Neurosci (2017) 12:1261–71. doi: 10.1093/scan/nsx051
33. Godoy LD, Rossignoli MT, Delfino-Pereira P, Garcia-Cairasco N, De Lima Umeoka EH. A Comprehensive Overview on Stress Neurobiology: Basic Concepts and Clinical Implications. Front Behav Neurosci (2018) 12:127. doi: 10.3389/fnbeh.2018.00127
34. Luoni A, Riva MA. MicroRNAs and psychiatric disorders: From aetiology to treatment. Pharmacol Ther (2016) 167:13–27. doi: 10.1016/j.pharmthera.2016.07.006
35. O’connor RM, Gururajan A, Dinan TG, Kenny PJ, Cryan JF. All Roads Lead to the miRNome: miRNAs Have a Central Role in the Molecular Pathophysiology of Psychiatric Disorders. Trends Pharmacol Sci (2016) 37:1029–44. doi: 10.1016/j.tips.2016.10.004
36. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci (2014) 16:43–61.
37. Hartford CCR, Lal A. When Long Noncoding Becomes Protein Coding. Mol Cell Biol (2020) 40:e00528–19. doi: 10.1128/MCB.00528-19
38. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17:47–62. doi: 10.1038/nrg.2015.10
39. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev (2011) 25:1915–27. doi: 10.1101/gad.17446611
40. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458:223–7. doi: 10.1038/nature07672
41. Lyle R, Watanabe D, Te Vruchte D, Lerchner W, Smrzka OW, Wutz A, et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet (2000) 25:19–21. doi: 10.1038/75546
42. Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol (2015) 22:5–7. doi: 10.1038/nsmb.2942
43. Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature (1991) 351:153–5. doi: 10.1038/351153a0
44. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature (1991) 349:38–44. doi: 10.1038/349038a0
45. Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res (2016) 44:D203–8. doi: 10.1093/nar/gkv1252
46. Bohmdorfer G, Wierzbicki AT. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol (2015) 25:623–32. doi: 10.1016/j.tcb.2015.07.002
47. Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol (2015) 12:1094–8. doi: 10.1080/15476286.2015.1063770
48. Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun (2016) 7:10982. doi: 10.1038/ncomms10982
49. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature (2012) 482:339–46. doi: 10.1038/nature10887
50. Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature (2011) 470:284–8. doi: 10.1038/nature09701
51. Shi C, Zhang L, Qin C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res Bull (2017) 132:160–9. doi: 10.1016/j.brainresbull.2017.03.010
52. Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, et al. A transcriptomic atlas of mouse neocortical layers. Neuron (2011) 71:605–16. doi: 10.1016/j.neuron.2011.06.039
53. Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, et al. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell (2013) 12:616–28. doi: 10.1016/j.stem.2013.03.003
54. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi: 10.1101/gr.132159.111
55. Lipovich L, Tarca AL, Cai J, Jia H, Chugani HT, Sterner KN, et al. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb Cortex (2014) 24:1451–9. doi: 10.1093/cercor/bhs414
56. Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell (2014) 53:1005–19. doi: 10.1016/j.molcel.2014.01.021
57. Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet (2013) 29:461–8. doi: 10.1016/j.tig.2013.03.002
58. Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife (2013) 2:e01749. doi: 10.7554/eLife.01749
59. Gonzalez-Burgos G, Cho RY, Lewis DA. Alterations in cortical network oscillations and parvalbumin neurons in schizophrenia. Biol Psychiatry (2015) 77:1031–40. doi: 10.1016/j.biopsych.2015.03.010
60. Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci (2005) 6:312–24. doi: 10.1038/nrn1648
61. Nakazawa K, Zsiros V, Jiang Z, Nakao K, Kolata S, Zhang S, et al. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology (2012) 62:1574–83. doi: 10.1016/j.neuropharm.2011.01.022
62. Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry (2011) 16:383–406. doi: 10.1038/mp.2010.120
63. Brambilla P, Perez J, Barale F, Schettini G, Soares JC. GABAergic dysfunction in mood disorders. Mol Psychiatry (2003) 8:721–737, 715. doi: 10.1038/sj.mp.4001362
64. Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PloS One (2011) 6:e23356. doi: 10.1371/journal.pone.0023356
65. Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell (2015) 16:439–47. doi: 10.1016/j.stem.2015.02.007
66. Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol (2012) 30:453–9. doi: 10.1038/nbt.2158
67. Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry (2003) 60:804–15. doi: 10.1001/archpsyc.60.8.804
68. Molendijk ML, Spinhoven P, Polak M, Bus BA, Penninx BW, Elzinga BM. Serum BDNF concentrations as peripheral manifestations of depression: evidence from a systematic review and meta-analyses on 179 associations (N=9484). Mol Psychiatry (2014) 19:791–800. doi: 10.1038/mp.2013.105
69. Bjorkholm C, Monteggia LM. BDNF - a key transducer of antidepressant effects. Neuropharmacology (2016) 102:72–9. doi: 10.1016/j.neuropharm.2015.10.034
70. Rocha RB, Dondossola ER, Grande AJ, Colonetti T, Ceretta LB, Passos IC, et al. Increased BDNF levels after electroconvulsive therapy in patients with major depressive disorder: A meta-analysis study. J Psychiatr Res (2016) 83:47–53. doi: 10.1016/j.jpsychires.2016.08.004
71. Aliperti V, Donizetti A. Long Non-coding RNA in Neurons: New Players in Early Response to BDNF Stimulation. Front Mol Neurosci (2016) 9:15. doi: 10.3389/fnmol.2016.00015
72. Tian T, Wei Z, Chang X, Liu Y, Gur RE, Sleiman PMA, et al. The Long Noncoding RNA Landscape in Amygdala Tissues from Schizophrenia Patients. EBioMedicine (2018) 34:171–81. doi: 10.1016/j.ebiom.2018.07.022
73. Liu Y, Chang X, Hahn CG, Gur RE, Sleiman PAM, Hakonarson H. Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl Psychiatry (2018) 8:44. doi: 10.1038/s41398-017-0030-5
74. Arjona FJ, De Baaij JH, Schlingmann KP, Lameris AL, Van Wijk E, Flik G, et al. CNNM2 mutations cause impaired brain development and seizures in patients with hypomagnesemia. PloS Genet (2014) 10:e1004267. doi: 10.1371/journal.pgen.1004267
75. Ohi K, Hashimoto R, Yasuda Y, Fukumoto M, Yamamori H, Umeda-Yano S, et al. Functional genetic variation at the NRGN gene and schizophrenia: evidence from a gene-based case-control study and gene expression analysis. Am J Med Genet B Neuropsychiatr Genet (2012) 159B:405–13. doi: 10.1002/ajmg.b.32043
76. Ruano D, Aulchenko YS, Macedo A, Soares MJ, Valente J, Azevedo MH, et al. Association of the gene encoding neurogranin with schizophrenia in males. J Psychiatr Res (2008) 42:125–33. doi: 10.1016/j.jpsychires.2006.10.008
77. Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature (2009) 460:744–7. doi: 10.1038/nature08186
78. Walton E, Geisler D, Hass J, Liu J, Turner J, Yendiki A, et al. The impact of genome-wide supported schizophrenia risk variants in the neurogranin gene on brain structure and function. PloS One (2013) 8:e76815. doi: 10.1371/journal.pone.0076815
79. Zhou Y, Lutz PE, Wang YC, Ragoussis J, Turecki G. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl Psychiatry (2018) 8:224. doi: 10.1038/s41398-018-0267-7
80. Punzi G, Ursini G, Shin JH, Kleinman JE, Hyde TM, Weinberger DR. Increased expression of MARCKS in post-mortem brain of violent suicide completers is related to transcription of a long, noncoding, antisense RNA. Mol Psychiatry (2014) 19:1057–9. doi: 10.1038/mp.2014.41
81. Mostafavi S, Battle A, Zhu X, Potash JB, Weissman MM, Shi J, et al. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol Psychiatry (2014) 19:1267–74. doi: 10.1038/mp.2013.161
82. Ni X, Liao Y, Li L, Zhang X, Wu Z. Therapeutic role of long non-coding RNA TCONS_00019174 in depressive disorders is dependent on Wnt/beta-catenin signaling pathway. J Integr Neurosci (2018) 17:125–32. doi: 10.31083/JIN-170052
83. Liu S, Zhou B, Wang L, Hu H, Yao C, Cai Z, et al. Therapeutic Antidepressant Potential of NONHSAG045500 in Regulating Serotonin Transporter in Major Depressive Disorder. Med Sci Monit (2018) 24:4465–73. doi: 10.12659/MSM.908543
84. Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature (2007) 447:823–8. doi: 10.1038/nature05841
85. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature (2005) 436:740–4. doi: 10.1038/nature03868
86. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell (2005) 123:631–40. doi: 10.1016/j.cell.2005.10.022
87. Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell (2006) 125:887–901. doi: 10.1016/j.cell.2006.03.043
88. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature (2003) 425:415–9. doi: 10.1038/nature01957
89. Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron (2010) 65:373–84. doi: 10.1016/j.neuron.2010.01.005
90. Impey S, Davare M, Lesiak A, Fortin D, Ando H, Varlamova O, et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci (2010) 43:146–56. doi: 10.1016/j.mcn.2009.10.005
91. Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA (2008) 105:9093–8. doi: 10.1073/pnas.0803072105
92. Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PloS One (2010) 5:e15497. doi: 10.1371/journal.pone.0015497
93. Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol (2007) 8:R27. doi: 10.1186/gb-2007-8-2-r27
94. Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry (2011) 69:188–93. doi: 10.1016/j.biopsych.2010.09.039
95. Banigan MG, Kao PF, Kozubek JA, Winslow AR, Medina J, Costa J, et al. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PloS One (2013) 8:e48814. doi: 10.1371/journal.pone.0048814
96. Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry (2010) 15:1176–89. doi: 10.1038/mp.2009.84
97. Kim AH, Reimers M, Maher B, Williamson V, Mcmichael O, Mcclay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res (2010) 124:183–91. doi: 10.1016/j.schres.2010.07.002
98. Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry (2011) 69:180–7. doi: 10.1016/j.biopsych.2010.09.030
99. Zhu Y, Kalbfleisch T, Brennan MD, Li Y. A MicroRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res (2009) 109:86–9. doi: 10.1016/j.schres.2009.01.022
100. Hu Z, Gao S, Lindberg D, Panja D, Wakabayashi Y, Li K, et al. Temporal dynamics of miRNAs in human DLPFC and its association with miRNA dysregulation in schizophrenia. Transl Psychiatry (2019) 9:196. doi: 10.1038/s41398-019-0572-9
101. Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci USA (2012) 109:3125–30. doi: 10.1073/pnas.1113793109
102. Guella I, Sequeira A, Rollins B, Morgan L, Torri F, Van Erp TG, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res (2013) 47:1215–21. doi: 10.1016/j.jpsychires.2013.05.021
103. Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet (2008) 17:1156–68. doi: 10.1093/hmg/ddn005
104. Azevedo JA, Carter BS, Meng F, Turner DL, Dai M, Schatzberg AF, et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J Psychiatr Res (2016) 82:58–67. doi: 10.1016/j.jpsychires.2016.07.012
105. Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med (2014) 20:764–8. doi: 10.1038/nm.3582
106. Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology (2017) 42:864–75. doi: 10.1038/npp.2016.175
107. Choi JL, Kao PF, Itriago E, Zhan Y, Kozubek JA, Hoss AG, et al. miR-149 and miR-29c as candidates for bipolar disorder biomarkers. Am J Med Genet B Neuropsychiatr Genet (2017) 174:315–23. doi: 10.1002/ajmg.b.32518
108. Kohen R, Dobra A, Tracy JH, Haugen E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl Psychiatry (2014) 4:e366. doi: 10.1038/tp.2014.9
109. Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry (2015) 20:573–84. doi: 10.1038/mp.2014.176
110. Sakamoto K, Crowley JJ. A comprehensive review of the genetic and biological evidence supports a role for MicroRNA-137 in the etiology of schizophrenia. Am J Med Genet B Neuropsychiatr Genet (2018) 177:242–56. doi: 10.1002/ajmg.b.32554
111. Schizophrenia Psychiatric Genome-Wide Association Study, C. Genome-wide association study identifies five new schizophrenia loci. Nat Genet (2011) 43:969–76. doi: 10.1038/ng.940
112. Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, et al. Identifying gene regulatory networks in schizophrenia. Neuroimage (2010) 53:839–47. doi: 10.1016/j.neuroimage.2010.06.036
113. Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature (2014) 511:421–7. doi: 10.1038/nature13595
114. Bahrini I, Song JH, Diez D, Hanayama R. Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia. Sci Rep (2015) 5:7989. doi: 10.1038/srep07989
115. Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci (2011) 46:409–18. doi: 10.1016/j.mcn.2010.11.004
116. Ruso-Julve F, Pombero A, Pilar-Cuellar F, Garcia-Diaz N, Garcia-Lopez R, Juncal-Ruiz M, et al. Dopaminergic control of ADAMTS2 expression through cAMP/CREB and ERK: molecular effects of antipsychotics. Transl Psychiatry (2019) 9:306. doi: 10.1038/s41398-019-0647-7
117. Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, Mcgrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet (2005) 77:918–36. doi: 10.1086/497703
118. Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res (2002) 58:11–20. doi: 10.1016/S0920-9964(01)00377-2
119. Eastwood SL, Kerwin RW, Harrison PJ. Immunoautoradiographic evidence for a loss of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate-preferring non-N-methyl-D-aspartate glutamate receptors within the medial temporal lobe in schizophrenia. Biol Psychiatry (1997) 41:636–43. doi: 10.1016/S0006-3223(96)00220-X
120. Eastwood SL, Mcdonald B, Burnet PW, Beckwith JP, Kerwin RW, Harrison PJ. Decreased expression of mRNAs encoding non-NMDA glutamate receptors GluR1 and GluR2 in medial temporal lobe neurons in schizophrenia. Brain Res Mol Brain Res (1995) 29:211–23. doi: 10.1016/0169-328X(94)00247-C
121. Van Der Laan S, Lachize SB, Vreugdenhil E, De Kloet ER, Meijer OC. Nuclear receptor coregulators differentially modulate induction and glucocorticoid receptor-mediated repression of the corticotropin-releasing hormone gene. Endocrinology (2008) 149:725–32. doi: 10.1210/en.2007-1234
122. Dadkhah T, Rahimi-Aliabadi S, Jamshidi J, Ghaedi H, Taghavi S, Shokraeian P, et al. A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J Affect Disord (2017) 208:218–22. doi: 10.1016/j.jad.2016.10.008
123. Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol (2004) 5:R13. doi: 10.1186/gb-2004-5-3-r13
124. Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci (2013) 33:5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013
125. Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell (2007) 27:435–48. doi: 10.1016/j.molcel.2007.07.015
126. Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci (2005) 21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x
127. Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry (2015) 20:1057–68. doi: 10.1038/mp.2015.91
128. Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, et al. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol (2013) 16:69–82. doi: 10.1017/S1461145712000016
129. Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of AMPA receptor trafficking and signaling molecule transcripts in the prefrontal cortex in schizophrenia. Synapse (2006) 60:585–98. doi: 10.1002/syn.20329
130. Meador-Woodruff JH, Hogg AJ Jr., Smith RE. Striatal ionotropic glutamate receptor expression in schizophrenia, bipolar disorder, and major depressive disorder. Brain Res Bull (2001) 55:631–40. doi: 10.1016/S0361-9230(01)00523-8
131. Fatemi SH, King DP, Reutiman TJ, Folsom TD, Laurence JA, Lee S, et al. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr Res (2008) 101:36–49. doi: 10.1016/j.schres.2008.01.029
132. Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science (2005) 310:1187–91. doi: 10.1126/science.1112915
133. Numata S, Iga J, Nakataki M, Tayoshi S, Taniguchi K, Sumitani S, et al. Gene expression and association analyses of the phosphodiesterase 4B (PDE4B) gene in major depressive disorder in the Japanese population. Am J Med Genet B Neuropsychiatr Genet (2009) 150B:527–34. doi: 10.1002/ajmg.b.30852
134. Padmos RC, Hillegers MH, Knijff EM, Vonk R, Bouvy A, Staal FJ, et al. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch Gen Psychiatry (2008) 65:395–407. doi: 10.1001/archpsyc.65.4.395
135. Ongur D, Drevets WC, Price JL. Glial reduction in the subgenual prefrontal cortex in mood disorders. Proc Natl Acad Sci USA (1998) 95:13290–5. doi: 10.1073/pnas.95.22.13290
136. Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem (2009) 108:216–24. doi: 10.1002/jcb.22243
137. Ferres-Coy A, Galofre M, Pilar-Cuellar F, Vidal R, Paz V, Ruiz-Bronchal E, et al. Therapeutic antidepressant potential of a conjugated siRNA silencing the serotonin transporter after intranasal administration. Mol Psychiatry (2016) 21:328–38. doi: 10.1038/mp.2015.80
138. Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet (2009) 373:234–9. doi: 10.1016/S0140-6736(09)60072-6
139. Ceribelli A, Satoh M, Chan EK. MicroRNAs and autoimmunity. Curr Opin Immunol (2012) 24:686–91. doi: 10.1016/j.coi.2012.07.011
140. Yao E, Ventura A. A new role for miR-182 in DNA repair. Mol Cell (2011) 41:135–7. doi: 10.1016/j.molcel.2011.01.005
141. Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience (2009) 164:711–23. doi: 10.1016/j.neuroscience.2009.08.033
142. Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, et al. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res (2012) 40:10356–65. doi: 10.1093/nar/gks750
143. Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PloS One (2012) 7:e33201. doi: 10.1371/journal.pone.0033201
144. Gorinski N, Bijata M, Prasad S, Wirth A, Abdel Galil D, Zeug A, et al. Attenuated palmitoylation of serotonin receptor 5-HT1A affects receptor function and contributes to depression-like behaviors. Nat Commun (2019) 10:3924. doi: 10.1038/s41467-019-11876-5
145. Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PloS One (2014) 9:e86469. doi: 10.1371/journal.pone.0086469
146. Maussion G, Yang J, Yerko V, Barker P, Mechawar N, Ernst C, et al. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PloS One (2012) 7:e39301. doi: 10.1371/journal.pone.0039301
147. Torres-Berrio A, Lopez JP, Bagot RC, Nouel D, Dal Bo G, Cuesta S, et al. DCC Confers Susceptibility to Depression-like Behaviors in Humans and Mice and Is Regulated by miR-218. Biol Psychiatry (2017) 81:306–15. doi: 10.1016/j.biopsych.2016.08.017
148. Roy B, Wang Q, Palkovits M, Faludi G, Dwivedi Y. Altered miRNA expression network in locus coeruleus of depressed suicide subjects. Sci Rep (2017) 7:4387. doi: 10.1038/s41598-017-04300-9
149. Maheu M, Lopez JP, Crapper L, Davoli MA, Turecki G, Mechawar N. MicroRNA regulation of central glial cell line-derived neurotrophic factor (GDNF) signalling in depression. Transl Psychiatry (2015) 5:e511. doi: 10.1038/tp.2015.11
150. Lopez JP, Fiori LM, Cruceanu C, Lin R, Labonte B, Cates HM, et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat Commun (2017) 8:15497. doi: 10.1038/ncomms15497
151. Sun N, Lei L, Wang Y, Yang C, Liu Z, Li X, et al. Preliminary comparison of plasma notch-associated microRNA-34b and -34c levels in drug naive, first episode depressed patients and healthy controls. J Affect Disord (2016) 194:109–14. doi: 10.1016/j.jad.2016.01.017
152. Higuchi F, Uchida S, Yamagata H, Otsuki K, Hobara T, Abe N, et al. State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. J Psychiatr Res (2011) 45:1295–300. doi: 10.1016/j.jpsychires.2011.04.008
153. Lopez JP, Fiori LM, Gross JA, Labonte B, Yerko V, Mechawar N, et al. Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int J Neuropsychopharmacol (2014) 17:23–32. doi: 10.1017/S1461145713000941
154. Ren X, Rizavi HS, Khan MA, Dwivedi Y, Pandey GN. Altered Wnt signalling in the teenage suicide brain: focus on glycogen synthase kinase-3beta and beta-catenin. Int J Neuropsychopharmacol (2013) 16:945–55. doi: 10.1017/S1461145712001010
155. Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development (2008) 135:3–9. doi: 10.1242/dev.006486
156. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature (2006) 442:203–7. doi: 10.1038/nature04916
157. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature (2006) 442:199–202. doi: 10.1038/nature04917
158. Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev (2006) 20:1709–14. doi: 10.1101/gad.1434406
159. Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science (2006) 313:363–7. doi: 10.1126/science.1130164
160. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature (2008) 453:534–8. doi: 10.1038/nature06904
161. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science (2007) 316:744–7. doi: 10.1126/science.1142612
162. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science (2008) 322:1387–92. doi: 10.1126/science.1165171
163. Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev (2008) 22:908–17. doi: 10.1101/gad.1640708
164. Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, et al. Identification of piRNAs in the central nervous system. RNA (2011) 17:1090–9. doi: 10.1261/rna.2565011
165. Nandi S, Chandramohan D, Fioriti L, Melnick AM, Hebert JM, Mason CE, et al. Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc Natl Acad Sci USA (2016) 113:12697–702. doi: 10.1073/pnas.1609287113
166. Ragan C, Patel K, Edson J, Zhang ZH, Gratten J, Mowry B. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr Res (2017) 183:82–7. doi: 10.1016/j.schres.2016.11.024
167. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature (2001) 409:363–6. doi: 10.1038/35053110
168. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell (2004) 118:57–68. doi: 10.1016/j.cell.2004.06.017
169. Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep (2006) 7:314–20. doi: 10.1038/sj.embor.7400637
170. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell (2005) 123:607–20. doi: 10.1016/j.cell.2005.08.044
171. Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol (2011) 3. doi: 10.1101/cshperspect.a003707
172. Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol (1988) 8:3755–60. doi: 10.1128/MCB.8.9.3755
173. Bai B, Hales CM, Chen PC, Gozal Y, Dammer EB, Fritz JJ, et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc Natl Acad Sci USA (2013) 110:16562–7. doi: 10.1073/pnas.1310249110
174. Hales CM, Seyfried NT, Dammer EB, Duong D, Yi H, Gearing M, et al. U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer’s disease due to autosomal dominant genetic mutations and trisomy 21. Mol Neurodegener (2014) 9:15. doi: 10.1186/1750-1326-9-15
175. Diner I, Hales CM, Bishof I, Rabenold L, Duong DM, Yi H, et al. Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. J Biol Chem (2014) 289:35296–313. doi: 10.1074/jbc.M114.562959
176. Bai B, Chen PC, Hales CM, Wu Z, Pagala V, High AA, et al. Integrated approaches for analyzing U1-70K cleavage in Alzheimer’s disease. J Proteome Res (2014) 13:4526–34. doi: 10.1021/pr5003593
177. Jia Y, Mu JC, Ackerman SL. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell (2012) 148:296–308. doi: 10.1016/j.cell.2011.11.057
178. Wu KY, Hsiao IT, Chen CS, Chen CH, Hsieh CJ, Wai YY, et al. Increased brain amyloid deposition in patients with a lifetime history of major depression: evidenced on 18F-florbetapir (AV-45/Amyvid) positron emission tomography. Eur J Nucl Med Mol Imaging (2014) 41:714–22. doi: 10.1007/s00259-013-2627-0
179. Byers AL, Yaffe K. Depression and risk of developing dementia. Nat Rev Neurol (2011) 7:323–31. doi: 10.1038/nrneurol.2011.60
180. Ownby RL, Crocco E, Acevedo A, John V, Loewenstein D. Depression and risk for Alzheimer disease: systematic review, meta-analysis, and metaregression analysis. Arch Gen Psychiatry (2006) 63:530–8. doi: 10.1001/archpsyc.63.5.530
181. Da Silva J, Goncalves-Pereira M, Xavier M, Mukaetova-Ladinska EB. Affective disorders and risk of developing dementia: systematic review. Br J Psychiatry (2013) 202:177–86. doi: 10.1192/bjp.bp.111.101931
182. Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie (2002) 84:775–90. doi: 10.1016/S0300-9084(02)01402-5
183. Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol (2002) 14:319–27. doi: 10.1016/S0955-0674(02)00334-4
184. Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA (2000) 97:14311–6. doi: 10.1073/pnas.250426397
185. Cavaille J. Box C/D small nucleolar RNA genes and the Prader-Willi syndrome: a complex interplay. Wiley Interdiscip Rev RNA (2017) 8:e1417. doi: 10.1002/wrna.1417
186. De Los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which Is highly expressed in brain. Am J Hum Genet (2000) 67:1067–82. doi: 10.1086/303106
187. Jenck F, Bos M, Wichmann J, Stadler H, Martin JR, Moreau JL. The role of 5-HT2C receptors in affective disorders. Expert Opin Invest Drugs (1998) 7:1587–99. doi: 10.1517/13543784.7.10.1587
188. Ni YG, Miledi R. Blockage of 5HT2C serotonin receptors by fluoxetine (Prozac). Proc Natl Acad Sci USA (1997) 94:2036–40. doi: 10.1073/pnas.94.5.2036
189. Palvimaki EP, Roth BL, Majasuo H, Laakso A, Kuoppamaki M, Syvalahti E, et al. Interactions of selective serotonin reuptake inhibitors with the serotonin 5-HT2c receptor. Psychopharmacol (Berl) (1996) 126:234–40. doi: 10.1007/BF02246453
190. Alex KD, Pehek EA. Pharmacologic mechanisms of serotonergic regulation of dopamine neurotransmission. Pharmacol Ther (2007) 113:296–320. doi: 10.1016/j.pharmthera.2006.08.004
191. Millan MJ, Lejeune F, Gobert A. Reciprocal autoreceptor and heteroreceptor control of serotonergic, dopaminergic and noradrenergic transmission in the frontal cortex: relevance to the actions of antidepressant agents. J Psychopharmacol (2000) 14:114–38. doi: 10.1177/026988110001400202
192. Lett TA, Wallace TJ, Chowdhury NI, Tiwari AK, Kennedy JL, Muller DJ. Pharmacogenetics of antipsychotic-induced weight gain: review and clinical implications. Mol Psychiatry (2012) 17:242–66. doi: 10.1038/mp.2011.109
193. Castellani CA, Laufer BI, Melka MG, Diehl EJ, O’reilly RL, Singh SM. DNA methylation differences in monozygotic twin pairs discordant for schizophrenia identifies psychosis related genes and networks. BMC Med Genomics (2015) 8:17. doi: 10.1186/s12920-015-0093-1
194. Hardwick SA, Bassett SD, Kaczorowski D, Blackburn J, Barton K, Bartonicek N, et al. Targeted, High-Resolution RNA Sequencing of Non-coding Genomic Regions Associated With Neuropsychiatric Functions. Front Genet (2019) 10:309. doi: 10.3389/fgene.2019.00309
195. Fiannaca A, La Rosa M, La Paglia L, Rizzo R, Urso A. nRC: non-coding RNA Classifier based on structural features. BioData Min (2017) 10:27. doi: 10.1186/s13040-017-0148-2
196. Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res (2006) 34:635–46. doi: 10.1093/nar/gkj469
Glossary

Keywords: major depressive disorder, schizophrenia, bipolar disorder, long non-coding RNAs, microRNAs
Citation: Yoshino Y and Dwivedi Y (2020) Non-Coding RNAs in Psychiatric Disorders and Suicidal Behavior. Front. Psychiatry 11:543893. doi: 10.3389/fpsyt.2020.543893
Received: 18 March 2020; Accepted: 14 August 2020;
Published: 15 September 2020.
Edited by:
Maurizio Pompili, Sapienza University of Rome, ItalyReviewed by:
Miao Qu, Xuanwu Hospital, Capital Medical University, ChinaPeter Petschner, Semmelweis University, Hungary
Copyright © 2020 Yoshino and Dwivedi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yogesh Dwivedi, yogeshdwivedi@uabmc.edu
 Yuta Yoshino
Yuta Yoshino Yogesh Dwivedi
Yogesh Dwivedi