- 1Proteomic Research Service, San Cecilio University Hospital, Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA), Granada, Spain
- 2Department of Genetics, University of Cambridge, Cambridge, United Kingdom
- 3Clinical Management Unit of Digestive Disease, San Cecilio University Hospital, Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA), Granada, Spain
- 4Genomic Research Service, San Cecilio University Hospital, Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA), Granada, Spain
- 5Department of Biochemistry, Cell and Molecular Biology of Plants, Estación Experimental del Zaidín, Spanish National Research Council (CSIC), Granada, Spain
- 6The UWA Institute of Agriculture and School of Agriculture and Environment, The University of Western Australia, Perth, WA, Australia
- 7Genomic Medicine Department, GENYO, Centre for Genomics and Oncological Research, Pfizer/University of Granada, Andalusian Regional Government, PTS Granada, Granada, Spain
A major consequence of the world industrialized lifestyle is the increasing period of unnatural light in environments during the day and artificial lighting at night. This major change disrupts endogenous homeostasis with external circadian cues, which has been associated to higher risk of diseases affecting human health, mainly cancer among others. Circadian disruption promotes tumor development and accelerate its fast progression. The dysregulation mechanisms of circadian genes is greatly affected by the genetic variability of these genes. To date, several core circadian genes, also called circadian clock genes, have been identified, comprising the following: ARNTL, CLOCK, CRY1, CRY2, CSNK1E, NPAS2, NR1D1, NR1D2, PER1, PER2, PER3, RORA, and TIMELESS. The polymorphic variants of these circadian genes might contribute to an individual's risk to cancer. In this short review, we focused on clock circadian clock-related genes, major contributors of the susceptibility to endocrine-dependent cancers through affecting circadian clock, most likely affecting hormonal regulation. We examined polymorphisms affecting breast, prostate and ovarian carcinogenesis, in addition to pancreatic and thyroid cancer. Further study of the genetic composition in circadian clock-controlled tumors will be of great importance by establishing the foundation to discover novel genetic biomarkers for cancer prevention, prognosis and target therapies.
Introduction
Circadian clocks have been defined as endogenous oscillators that synchronize daily both physiological and behavioral rhythms with local time cues (1, 2). In mammals, this evolutionary adaptation provides a survival advantage to anticipate environmental changes and allowing them to modify their daily rhythms in the most efficient way to meet these environmental changes (1, 3, 4).
Consequences of modern lifestyle as sleep deprivation, shift work time schedule, altered mealtime or excessive artificial light exposure at night promote metabolic imbalances that can alter the circadian system. Interestingly, dysfunction of the molecular clock is linked to uncontrolled cell proliferation in human cancer (5–7). Changes in circadian clock increase risk of different cancers in endocrine tissues which require daily proliferation to carry out their activity. Thus, a considerable part of endocrine signals are clock-controlled (8). The dysregulation mechanisms of the circadian clock genes is greatly affected by polymorphic variants of circadian clock-related genes, which can raise human cancer risk through different physiological systems (9).
This mini-review focused on the role of single polymorphisms of circadian clock genes that similarly to environmental factors might increase the risk of developing human endocrine cancers.
Circadian Clock Mechanism, Clock Genes, and Cancer
The circadian clock regulates both physiology and behavior according to the daily cycle of light and dark. In mammals, it is hierarchically organized and integrates the master clock, which is located in the suprachiasmatic nucleus (SCN) within the hypothalamus, and the peripheral clocks as well, ubiquitously found virtually in all peripheral tissues and cells (10). SCN clock is constantly coupled to environmental cues, mainly photoperiod, through the photic signals from the retina (11), daily rhythms in temperature, diet and social phenomena through a complex downstream neurohumoral pathways. Oscillators located in brain nuclei and peripheral tissues are also connected by SCN clock (12).
Circadian rhythms are controlled by circadian pathway genes. The molecular circadian clock is originated by a transcriptional/translational loop of circadian clock genes with autoregulatory feedback. The primary loop involves the genes CLOCK, BMAL1 (also known as ARNTL1), PER1–3, and CRY1-2. During the day, the complex integrated by CLOCK and BMAL1 stimulates the expression of negative regulators period genes (PER1–3) and cryptochrome genes (CRY1-2). Heterodimers constituted by PER and CRY operate as co-repressors, binding to the CLOCK-BMAL1 complex and inhibiting CRY and PER gene transcription induced by CLOCK-BMAL1. Furthermore, in the dark phase, CRY and PER expression decrease to the CRY-PER repressor complex. This leads to a new cycle of the transcription activation of the CLOCK-BMAL1 complex, which completes the basic auto regulatory loop (8, 13). Otherwise, different modulators display fine tuning of output signals in molecular clock.
Currently, several core circadian genes, also known as circadian clock genes, have been identified in humans (13): ARNTL (aryl hydrocarbon receptor nuclear translocator like, also identify in brain and muscle as Arnt-like protein-1, BMAL1) (14, 15), CLOCK (clock circadian regulator) (16), CRY1 (cryptochrome circadian clock 1), CRY2 (cryptochrome circadian clock 2) (17), PER1 (period circadian clock 1), PER2 (period circadian clock 2), PER3 (period circadian clock 3) (18–20), CSNK1E (casein kinase I epsilon) (6, 21), NPAS2 (neuronal PAS domain protein 2) (22, 23), NR1D1 (nuclear receptor subfamily 1 group D member 1 also called Rev-Erb alpha) (24, 25), NR1D2 (nuclear receptor subfamily 1 group D member 2 also referred to Rev-Erb beta) (26), RORA (RAR related orphan receptor A) (27) and TIMELESS (timeless circadian clock) (28, 29).
In addition, there is a large amount of circadian genes from other clock-related pathways.
Ground-breaking works in the field of molecular cancer epidemiology have unveiled a large quantity of polymorphisms affecting the clock genes (Table 1). Studies on the relationship between clock genes polymorphisms and cancer susceptibility have established that variants of ARNTL, CLOCK, CK1ε, CRY1-2, NPAS2, and PER1-3 are frequently associated to human reproductive tissues and pancreatic cancers (1).
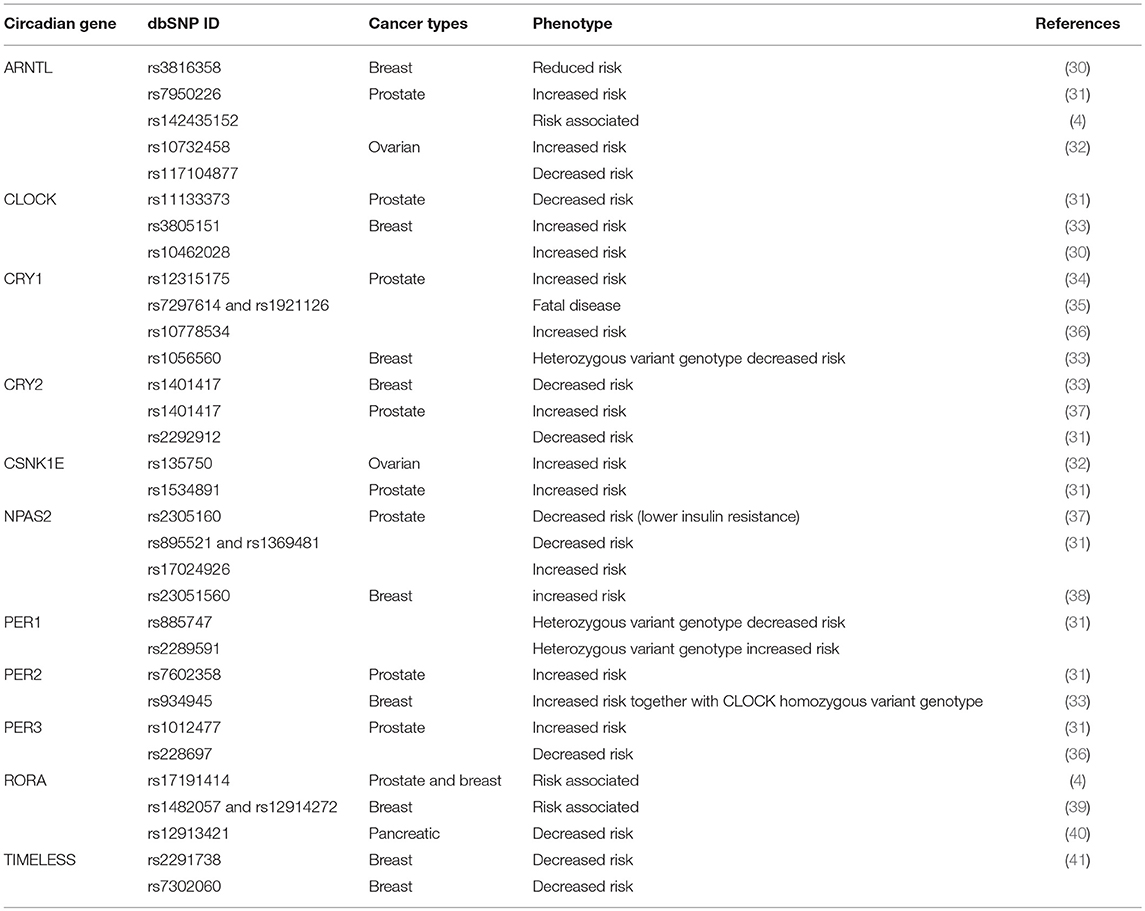
Table 1. Genetic association between circadian clock polymorphisms and different endocrine-related cancers.
Breast Cancer
Epidemiological studies have concluded an association between shift work and breast cancer risk (42–44). However, currently available experimental/epidemiological data are characterized by a great heterogeneity and some findings are in disagreement with previous observations about relationship between shift work and breast cancer risk (45).
A recent study in breast cancer showed that circadian genes CRY2 and PER1-3 were down-regulated, while CLOCK and TIMELESS were over-expressed (46). This study confirms previous results where PER1, PER2, and PER3 exhibited changes associated with the tumor suppressor activity (47, 48). In breast epithelial cells, an altered estrogen receptor signaling has been related to breast cancer and two circadian clock genes, PER2 and ARNTL, both required for breast epithelial acinar morphogenesis in vitro (49).
Therefore, polymorphism studies in circadian clock-related genes and breast cancer, most of them have focused on the evaluation of the polymorphisms in core circadian genes, melatonin biosynthesis and signaling pathways. In this line, polymorphisms affecting CLOCK (rs3805151), CRY1 (rs1056560), CRY2 (rs1401417), and PER2 (rs934945) (33) have been associated to breast cancer risk in a Chinese population. Concerning CLOCK gene, it was found that carrier subjects CT and CT+TT genotypes exhibited an increased risk of breast cancer in comparison to CC carriers. However, the GT genotype of the CRY1 gene was the genetic variant with the lower risk of suffering cancer, as well as for those subjects carrying CC genotype of the CRY2 gene having an even lower risk. Finally, simultaneous presence of CLOCK CC and PER2 AA genotypes resulted in a higher risk of developing breast cancer (33).
An alternative study supporting the involvement of the CLOCK gene in the development of breast cancer came from a Bonn (Germany) population of shift workers. This case-control study analyzed the associations between breast cancer and polymorphisms in circadian clock genes (ARTNL, CLOCK, CRY2, NPAS, and PER2) and in genes of the melatonin pathway as well (AANAT and MTNR1B). In these workers with the rs8150 polymorphism from AANAT gene, as well as for the rs10462028 polymorphism from the CLOCK gene, was observed an increased frequency of breast cancer, while a lower risk was observed for the rs3816358 from ARNTL gene (30). Interaction analysis was performed in two-ways, detecting interactions between shift work and CLOCK gene. Therefore, interactions were found for shift work and MTNR1B, ARNTL, and NPAS2 genes (50).
Additionally, a combined analysis of three genome wide associations studies (GWAS) was performed, including a total 285,984 SNPs (Single Nucleotid Polymorphisms) in an European population of 2.702 women with invasive breast cancer, in comparison to 5,726 subject controls. The most important association to breast cancer was found for genes implicated in the circadian clock (51). Monsees et al. conducted a study on 1,825 women finding common variants across 15 circadian system genes that were tested for association with breast cancer risk. The interaction study was designed in a subset of 1,318 women, showing interactions between genotype and rotating shift-work. NPAS2 gene was strongly associated to the risk of developing breast cancer, and the rs23051560 variant was related with potential effects. The estimated risk value for the minor allele (A) was lower among women with <2 years of shift-work compared to women with more than 2 years of shift-work (38).
In Europe, among a French population analyzed, 577 SNPs were found in 23 circadian clock genes associated with breast cancer risk in more than 1,000 cases and control subjects. RORA SNPs rs1482057 and rs12914272148 were also associated with breast cancer (39). Furthermore, a significant association has been shown between two SNPs in the TIMELESS gene (rs2291738 and rs7302060) (33, 41). In this study, C allele and CC genotype from rs7302060 polymorphism were associated with lower breast cancer risk. In rs2291738 polymorphism, GG genotype was related to decreased breast cancer risk.
On the other hand, limitations of the selected studies include the stratification of the populations according to different conditions among the different studies, such as shift work duration, menopausal status and expression levels of hormonal receptor in tumor tissues. Moreover, the polymorphism associations to breast cancer risk remain controversial, since some of these results were not able to be replicated in other populations or significance of associations is lost after adjusting for confounding factors. Furthermore, the described investigations included other limitations which will be further discussed in more detail.
Prostate Cancer
Prostate cancer exhibits the highest cancer prevalence in men, being the second cause of cancer-related deaths (52). Normal prostate cancer development is dependent on androgens levels. Circadian clock genes regulate androgen production (53), affecting prostate cancer evolution (54). On the other hand, a balanced regulation of the circadian clock genes might modulate and even suppress tumor growth by controlling DNA replication, repair mechanisms and cell proliferation (55). Although a limited number of epidemiologic studies have been realized, several circadian genes have been implicated in prostate cancer regulation: ARNTL, CLOCK, CRY1-2, CSNK1e, MTNR1A and MTNR1B, NPAS2, NR1D1, PER1-3, RORA, RORB, and TIMELESS (4, 31, 56, 57).
One of the first epidemiologic studies performed on prostate cancer and their associated SNPs, Chu et al. identified five polymorphisms in five circadian genes. This case-control study was conducted in a Chinese population with 187 cases and 242 control subjects (37). These polymorphisms encompassed CRY2 rs1401417, CSNK1E rs1005473, NPAS2 rs2305160, and PER1 rs2585405. The C allele from CRY2 presented an elevated prostate cancer risk compared to GG genotype carriers. Higher risk was found in men whom also sustained elevated insulin resistance (IR) compared to these with the GG genotype and lower IR. Moreover, the A allele from NPAS2 polymorphism was associated with a reduced risk of developing prostate cancer in men with reduced IR when compared to the GG genotype carriers.
Zhu and colleagues also investigated the link between circadian gene with prostate tumors. The case-control study in a Caucasian men population included 1,308 cases and 1,266 control subjects. In this study was genotyped 41 variants in ten genes related with circadian clock (31). At least one polymorphism in nine clock circadian genes was significantly associated with prostate cancer risk. Specifically, it was found the variants rs7950226 in ARNTL, rs11133373 in CLOCK, rs12315175 in CRY1, rs2292912 in CRY2, rs1534891 in CSNK1E, rs1369481, rs895521, and rs17024926 in NPAS2, rs885747 and rs2289591 in PER1, rs7602358 in PER2 and rs1012477 in PER3. They observed that the estimate risk for variants rs885747 and rs2289591 in PER1, rs1012477 in PER3, and rs11133373 in CLOCK significantly changed depending on disease aggressiveness.
Lin et al. carried out studies in prostate cancer using two populations, from Seattle and Sweden, respectively) (36). They genotyped 937 polymorphisms corresponding to 156 genes in 1,309 men with prostate cancer in a Seattle cohort. They identified 22 variants associated to prostate cancer-specific mortality (PCSM), and validated them afterward in a Swedish cohort (2,875 patients. In the Swedish cohort, five polymorphisms out of the 22 SNPs identified in the Seattle cohort, were found to be significantly associated with PCSM, with a statistical significance variant in the CRY1 gene (rs10778534). The study also identified another variant in the Seattle cohort, rs228697 in PER3, associated with PCSM, which was not further tested in the Swedish cohort due to genotyping drawbacks.
Another study evaluating the association between mortality in prostate cancer and circadian clock-related genes was carried out by Markt et al. (35). Authors tested 96 variants in 12 circadian-related genes using 3 patient cohorts (24, 40, and 105 cases/respectively). It was also analyzed the association with lower levels of melatonin (measured by 6-sulfatoxymelatonin). This study showed no variants significantly associated with overall risk of prostate cancer, however in all cohorts was observed that variation in the CRY1 gene was associated with mortality in prostate cancer This study of individual cohorts revealed that two polymorphisms from CRY1, rs7297614, and rs1921126 were associated to increased mortality in 2 out 3 prostate cancer cohorts, and a similar association was proved for rs12315175 in the CRY1 gene in a single cohort. Finally, their analysis of the 6-sulfatoxymelatonin levels showed that patients with metabolite levels lower than the median had an increased risk of advanced disease, where polymorphisms in CSNK1E, NPAS2, PER3, and TIMELESS were associated to changes in these 6-sulfatoxymelatonin production. Future investigations should be designed including a large population compared to the one used in this study, and similar clinicopathological factors as well to ensure statistical power and allow for results comparisons.
Recently, Mocellin et al. performed an analysis using adaptive rank truncated product (ARTP)-based gene and pathway analysis to discern the relevance of the variation in circadian clock genes and cancer susceptibility (4). In this analysis using previously published dataset of prostate cancer (58), they found a highly significant association between genetic variation of circadian pathway and susceptibility to prostate cancer. This result was founded on data regarding 17 SNPs located in seven genes, with the most significant SNP rs142435152 from ARNTL gene. Their analysis of subgroups revealed that the risk of suffering aggressive prostate cancer was also highly associated with circadian pathway variation. This finding was based on 28 SNPs located in seven genes, where the most significant gene was RORA with the rs17191414 SNP.
Ovarian Cancer
Ovaries express circadian genes at high levels to regulate hormonal levels during reproductive cycles, and the disruption of this expression is associated to different risk factors for ovarian cancer (e.g., endometriosis).
An association between nightshift work and elevated risk of invasive and borderline ovarian cancers have been described in women aged over 50 years (59). A GWAS and a replication study for epithelial ovarian cancer (EOC) analyzed variants of several circadian genes (ARNTL, CRY2, CSNK1E, NPAS2, PER3, REV1, and TIMELESS) and two transcription factors (KLF10 and SENP3) (32). The study examined 3,761 EOC patients and 2,722 control subjects. A replication stage was evaluated with ~44,000 subjects with European ancestry. This study indicates that circadian clock genes could act in the development of EOC, particularly ARNTL rs10732458, CSNK1E rs135750, SENP3 (rs6608), and REV1 rs3792152 variants. SNPs in KLF10 (rs2513928, rs2511703, rs3191333, and rs2513927) were also associated with serious risk of EOC. Interestingly, the most significant association was the rs117104877 variant in ARNTL. Ablation of ARNTL in mice causes ovary tumors via reduction of p53 expression (60) and dysregulation response to anti-cancer drugs (61). In addition, lower ARNTL and CRY1 expression levels in EOC cells were found after comparison to normal ovarian tissue (62).
Nevertheless, an alternative study has evaluated polymorphism association in nine circadian clock-related genes (ARNTL, CKIE, CLOCK, CRY1-2, CSNKIE, NPAS2, and PER1-3), but however, it did fail finding any association between genes of the circadian rhythm pathway and ovarian cancer (63).
Pancreatic and Thyroid Cancer
Pancreatic cancer is a major cause of cancer mortality in western countries population. Several studies have shown disruption of circadian genes expression associated to pancreatic cancer (64–68). However, only one study was able to show an association between circadian genes and pancreatic cancer. It found association between the SNP rs12913421 in RORA gene and pancreatic cancer, but the significance disappeared after correcting for multiple comparisons (40). Nevertheless, this study was not specifically designed to evaluate association between polymorphisms in genes of the circadian rhythm pathway and pancreatic cancer. Additional studies of polymorphisms in the circadian pathway are urgently needed, with systematical studies of fine mapping and sufficient sample size.
On the other hand, the disruption of circadian clock genes has been associated to a higher risk of thyroid tumors (69, 70).
Despite this result, no studies have examined the association between polymorphism in circadian genes and thyroid cancer.
Discussion and Future Perspectives
A better understanding of molecular mechanisms in endocrine-related cancers may facilitate early diagnosis, prognosis and therapies development. Therefore, studies should be improved in several aspects. The reviewed studies about the association of circadian gene polymorphisms to cancer risk have been only conducted in European an East Asian population. Additional studies should be implemented in other ethnic groups to validate epidemiological data.
On the other hand, selection of genetic polymorphisms and experimental and statistical analysis approaches were different among cancer studies and thus, they were not comparable in most cases. These differences might consequently influence the associations found between germline variants and cancer risk.
In future studies, functional implications of the circadian pathway polymorphisms associated to endocrine cancer should be also evaluated. Polymorphisms localized in the 3′UTR could regulate gene transcription. However, the majority of the studied polymorphisms are located in intron regions, thus it is unknown how polymorphisms modulate their functions resulting in a higher or lower cancer risk.
Our review reports that the polymorphisms of some circadian genes are related to cancers of reproductive tissues, where some genes are associated and implicated in three types of tumors (ARNTL gene in prostate, breast and ovary), two types of tumors (CLOCK, CRY1, CRY2, NPAS2, and PER2 genes in breast and prostate and CSNK1E gene in prostate and ovary) or alternatively they are more specific to one type of cancer (PER1 and PER3 genes in prostate and TIMELESS gene in breast). Other circadian genes are associated and implicated by several endocrine-related cancers (RORA gen in prostate, breast and pancreas).
The current findings suggest that some genes must be involved in the predisposition to cancer development of reproductive tissues, other genes must be specific to a type of cancer, and other genes should affect tissues modulated by endocrine hormones. The effect of these genes is probably showed up at the hormone pathways level, as in the CLOCK gene. The activity of the CLOCK gen product regulates estrogenic and androgenic hormonal pathways (71, 72). This could be related to the fact that polymorphisms of this gene alter the regulation of these pathways and produce an uncontrolled proliferation of prostate, breast and ovarian tissue cells.
We reckon that a screening of polymorphisms related with the circadian clock could provide valuable information regarding predisposition of suffering a particular type of cancer, thus facilitating its prognosis. When cancer is already present, malignancy intervention strategies could be immediately applied due to earlier detection.
Concluding Remarks
Compelling evidence supports the notion that loss of circadian homeostasis promotes endocrine cancer development. Genetic component is a key factor that contributes to dysregulation of the circadian clock. Polymorphisms of circadian clock genes are associated to the risk of suffering an endocrine cancer and poor performance to of therapies with anticancer treatments, particularly, these related to reproductive tissues. The findings detailed in this review indicate an exciting research line in order to investigate clock-controlled tumor suppression, also in other organs regulated by circadian rhythms that need high levels of cell proliferation to support their functions, such as thyroid or pancreas.
Author Contributions
SM-S conceived the project. SM-S, SM, and MM wrote the paper. JL, AC-G, and JJ-L contributed to the revision of literature. All authors have read, corrected, and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SM-S would like to thank the grant awards from SEIOMM (2017) and SEEN (2018). JJ-L was funded by a Ramon y Cajal Research Program from the Spanish Ministry of Economy, Industry and Competitiveness (grant ref. number YC-2014-16536); the grant ref. BFU2016-77243-P; by the CSIC—intramural grant ref. 201540E065; and by the EU Marie Curie Research Program FP7-PEOPLE-2011-IOF, grant ref. PIOF-GA-2011-301550. All authors thank Michela Verbeni for the English language edition performed on the manuscript.
References
1. Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. (2013) 119:221–82. doi: 10.1016/B978-0-12-396971-2.00009-9
2. Padmanabhan K, Billaud M. Desynchronization of circadian clocks in cancer: a metabolic and epigenetic connection. Front Endocrinol (Lausanne) (2017) 8:136. doi: 10.3389/fendo.2017.00136
3. Do MT, Yau KW. Intrinsically photosensitive retinal ganglion cells. Physiol Rev. (2010) 90:1547–81. doi: 10.1152/physrev.00013.2010
4. Mocellin S, Tropea S, Benna C, Rossi CR. Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies. BMC Med. (2018) 16:20. doi: 10.1186/s12916-018-1010-1
5. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell (2006) 22:375–82. doi: 10.1016/j.molcel.2006.03.038
6. Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. (2008) 9:R92. doi: 10.1186/gb-2008-9-6-r92
7. Jenwitheesuk A, Nopparat C, Mukda S, Wongchitrat P, Govitrapong P. Melatonin regulates aging and neurodegeneration through energy metabolism, epigenetics, autophagy and circadian rhythm pathways. Int J Mol Sci. (2014) 15:16848–84. doi: 10.3390/ijms150916848
8. Angelousi A, Kassi E, Nasiri-Ansari N, Weickert MO, Randeva H, Kaltsas G. Clock genes alterations and endocrine disorders. Eur J Clin Invest. (2018) 48:e12927. doi: 10.1111/eci.12927
9. Valenzuela FJ, Vera J, Venegas C, Munoz S, Oyarce S, Munoz K, et al. Evidences of polymorphism associated with circadian system and risk of pathologies: a review of the literature. Int J Endocrinol. (2016) 2016:2746909. doi: 10.1155/2016/2746909
10. Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms (2003) 18:250–60. doi: 10.1177/0748730403018003007
11. Liu F, Chang HC. Physiological links of circadian clock and biological clock of aging. Protein Cell (2017) 8:477–88. doi: 10.1007/s13238-016-0366-2
12. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. (2010) 72:517–49. doi: 10.1146/annurev-physiol-021909-135821
13. Benna C, Helfrich-Forster C, Rajendran S, Monticelli H, Pilati P, Nitti D, et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget (2017) 8:23978–95. doi: 10.18632/oncotarget.15074
14. Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun. (1997) 233:258–64. doi: 10.1006/bbrc.1997.6371
15. Yu W, Ikeda M, Abe H, Honma S, Ebisawa T, Yamauchi T, et al. Characterization of three splice variants and genomic organization of the mouse BMAL1 gene. Biochem Biophys Res Commun. (1999) 260:760–7. doi: 10.1006/bbrc.1999.0970
16. King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, et al. Positional cloning of the mouse circadian clock gene. Cell (1997) 89:641–53. doi: 10.1016/S0092-8674(00)80245-7
17. Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry (1996) 35:13871–7. doi: 10.1021/bi962209o
18. Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron (1997) 19:1261–9. doi: 10.1016/S0896-6273(00)80417-1
19. Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature (1997) 389:512–6. doi: 10.1038/39086
20. Kim P, Oster H, Lehnert H, Schmid SM, Salamat N, Barclay JL, et al. Coupling the circadian clock to homeostasis: the role of Period in timing physiology. Endocr Rev. (2018) 40:66–95. doi: 10.1210/er.2018-00049
21. Zheng X, Sowcik M, Chen D, Sehgal A. Casein kinase 1 promotes synchrony of the circadian clock network. Mol Cell Biol. (2014) 34:2682–94. doi: 10.1128/MCB.01571-13
22. Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science (2001) 293:506–9. doi: 10.1126/science.1060699
23. Landgraf D, Wang LL, Diemer T, Welsh DK. NPAS2 Compensates for loss of CLOCK in peripheral circadian oscillators. PLoS Genet. (2016) 12:e1005882. doi: 10.1371/journal.pgen.1005882
24. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110:251–60. doi: 10.1016/S0092-8674(02)00825-5
25. Kim YH, Marhon SA, Zhang Y, Steger DJ, Won KJ, Lazar MA. Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science (2018) 359:1274–7. doi: 10.1126/science.aao6891
26. Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature (2012) 485:123–7. doi: 10.1038/nature11048
27. Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron (2004) 43:527–37. doi: 10.1016/j.neuron.2004.07.018
28. Gotter AL, Manganaro T, Weaver DR, Kolakowski LF Jr, Possidente B, Sriram S, et al. A time-less function for mouse timeless. Nat Neurosci. (2000) 3:755–6. doi: 10.1038/77653
29. Mazzoccoli G, Laukkanen MO, Vinciguerra M, Colangelo T, Colantuoni V. A timeless link between circadian patterns and disease. Trends Mol Med. (2016) 22:68–81. doi: 10.1016/j.molmed.2015.11.007
30. Rabstein S, Harth V, Justenhoven C, Pesch B, Plottner S, Heinze E, et al. Polymorphisms in circadian genes, night work and breast cancer: results from the GENICA study. Chronobiol Int. (2014) 31:1115–22. doi: 10.3109/07420528.2014.957301
31. Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. (2009) 69:9315–22. doi: 10.1158/0008-5472.CAN-09-0648
32. Jim HS, Lin HY, Tyrer JP, Lawrenson K, Dennis J, Chornokur G, et al. Common genetic variation in circadian rhythm genes and risk of epithelial ovarian cancer (EOC). J Genet Genome Res. (2015) 2:017. doi: 10.23937/2378-3648/1410017
33. Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. (2011) 127:531–40. doi: 10.1007/s10549-010-1231-2
34. Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. (2005) 14:268–70. Available online at: http://cebp.aacrjournals.org/content/14/1/268.long
35. Markt SC, Valdimarsdottir UA, Shui IM, Sigurdardottir LG, Rider JR, Tamimi RM, et al. Circadian clock genes and risk of fatal prostate cancer. Cancer Causes Control. (2015) 26:25–33. doi: 10.1007/s10552-014-0478-z
36. Lin DW, FitzGerald LM, Fu R, Kwon EM, Zheng SL, Kolb S, et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol Biomarkers Prev. (2011) 20:1928–36. doi: 10.1158/1055-9965.EPI-11-0236
37. Chu LW, Zhu Y, Yu K, Zheng T, Yu H, Zhang Y, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. (2008) 11:342–8. doi: 10.1038/sj.pcan.4501024
38. Monsees GM, Kraft P, Hankinson SE, Hunter DJ, Schernhammer ES. Circadian genes and breast cancer susceptibility in rotating shift workers. Int J Cancer (2012) 131:2547–52. doi: 10.1002/ijc.27564
39. Truong T, Liquet B, Menegaux F, Plancoulaine S, Laurent-Puig P, Mulot C, et al. Breast cancer risk, nightwork, and circadian clock gene polymorphisms. Endocr Relat Cancer (2014) 21:629–38. doi: 10.1530/ERC-14-0121
40. Cotterchio M, Lowcock E, Bider-Canfield Z, Lemire M, Greenwood C, Gallinger S, et al. Association between variants in atopy-related immunologic candidate genes and pancreatic cancer risk. PLoS ONE (2015) 10:e0125273. doi: 10.1371/journal.pone.0125273
41. Fu A, Leaderer D, Zheng T, Hoffman AE, Stevens RG, Zhu Y. Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol Carcinog. (2012) 51:923–9. doi: 10.1002/mc.20862
42. Ijaz S, Verbeek J, Seidler A, Lindbohm ML, Ojajarvi A, Orsini N, et al. Night-shift work and breast cancer–a systematic review and meta-analysis. Scand J Work Environ Health (2013) 39:431–47. doi: 10.5271/sjweh.3371
43. Blakeman V, Williams JL, Meng QJ, Streuli CH. Circadian clocks and breast cancer. Breast Cancer Res. (2016) 18:89. doi: 10.1186/s13058-016-0743-z
44. Wegrzyn LR, Tamimi RM, Rosner BA, Brown SB, Stevens RG, Eliassen AH, et al. Rotating night-shift work and the risk of breast cancer in the Nurses' health studies. Am J Epidemiol. (2017) 186:532–40. doi: 10.1093/aje/kwx140
45. Johns LE, Jones ME, Schoemaker MJ, McFadden E, Ashworth A, Swerdlow AJ. Domestic light at night and breast cancer risk: a prospective analysis of 105,000 UK women in the Generations Study. Br J Cancer (2018) 118:600–6. doi: 10.1038/bjc.2017.359
46. Lesicka M, Jablonska E, Wieczorek E, Seroczynska B, Siekierzycka A, Skokowski J, et al. Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS ONE (2018) 13:e0199622. doi: 10.1371/journal.pone.0199622
47. Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. (2004) 24:584–94. doi: 10.1128/MCB.24.2.584-594.2004
48. Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. (2009) 117:423–31. doi: 10.1007/s10549-008-0133-z
49. Rossetti S, Corlazzoli F, Gregorski A, Azmi NH, Sacchi N. Identification of an estrogen-regulated circadian mechanism necessary for breast acinar morphogenesis. Cell Cycle (2012) 11:3691–700. doi: 10.4161/cc.21946
50. Kochan DZ, Kovalchuk O. Circadian disruption and breast cancer: an epigenetic link? Oncotarget (2015) 6:16866–82. doi: 10.18632/oncotarget.4343
51. Li J, Humphreys K, Heikkinen T, Aittomaki K, Blomqvist C, Pharoah PD, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. (2011) 126:717–27. doi: 10.1007/s10549-010-1172-9
52. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. (2009) 59:225–49. doi: 10.3322/caac.20006
53. Plymate SR, Tenover JS, Bremner WJ. Circadian variation in testosterone, sex hormone-binding globulin, and calculated non-sex hormone-binding globulin bound testosterone in healthy young and elderly men. J Androl. (1989) 10:366–71. doi: 10.1002/j.1939-4640.1989.tb00120.x
54. Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer (2001) 1:34–45. doi: 10.1038/35094009
55. Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer (2003) 3:350–61. doi: 10.1038/nrc1072
56. Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. (2009) 69:7619–25. doi: 10.1158/0008-5472.CAN-08-4199
57. Kiss Z, Ghosh PM. WOMEN IN CANCER THEMATIC REVIEW: Circadian rhythmicity and the influence of ‘clock’ genes on prostate cancer. Endocr Relat Cancer (2016) 23:T123–T134. doi: 10.1530/ERC-16-0366
58. Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. (2014) 46:1103–9. doi: 10.1038/ng.3094
59. Bhatti P, Cushing-Haugen KL, Wicklund KG, Doherty JA, Rossing MA. Nightshift work and risk of ovarian cancer. Occup Environ Med. (2013) 70:231–7. doi: 10.1136/oemed-2012-101146
60. Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS ONE (2010) 5:e10995. doi: 10.1371/journal.pone.0010995
61. Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, Huang YJ, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem. (2010) 148:319–26. doi: 10.1093/jb/mvq069
62. Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. (2008) 87:1060–70. doi: 10.1080/00016340802348286
63. Gu F, Zhang H, Hyland PL, Berndt S, Gapstur SM, Wheeler W, et al. Inherited variation in circadian rhythm genes and risks of prostate cancer and three other cancer sites in combined cancer consortia. Int J Cancer (2017) 141:1794–802. doi: 10.1002/ijc.30883
64. Oda A, Katayose Y, Yabuuchi S, Yamamoto K, Mizuma M, Shirasou S, et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. (2009) 29:1201–9. Available online at: http://ar.iiarjournals.org/content/29/4/1201.long
65. Wu Y, Sato F, Yamada T, Bhawal UK, Kawamoto T, Fujimoto K, et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol. (2012) 41:1337–46. doi: 10.3892/ijo.2012.1559
66. Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg. (2013) 17:443–50. doi: 10.1007/s11605-012-2112-2
67. Tavano F, Pazienza V, Fontana A, Burbaci FP, Panebianco C, Saracino C, et al. SIRT1 and circadian gene expression in pancreatic ductal adenocarcinoma: effect of starvation. Chronobiol Int. (2015) 32:497–512. doi: 10.3109/07420528.2014.1003351
68. Jiang W, Zhao S, Jiang X, Zhang E, Hu G, Hu B, et al. The circadian clock gene Bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett. (2016) 371:314–25. doi: 10.1016/j.canlet.2015.12.002
69. Mannic T, Meyer P, Triponez F, Pusztaszeri M, Le Martelot G, Mariani O, et al. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab. (2013) 98:4446–56. doi: 10.1210/jc.2013-2568
70. Wilson C. Cancer: Thyroid circadian clock–altered in cancer? Nat Rev Endocrinol. (2013) 9:628. doi: 10.1038/nrendo.2013.182
71. Zhu Y, Zheng T, Stevens RG, Zhang Y, Boyle P. Does “clock” matter in prostate cancer? Cancer Epidemiol Biomarkers Prev. (2006) 15:3–5. doi: 10.1158/1055-9965.EPI-05-0631
Keywords: polymorphism, circadian clock genes, endocrine cancer, breast cancer, ovarian cancer, prostate cancer, pancreatic cancer, thyroid cancer
Citation: Morales-Santana S, Morell S, Leon J, Carazo-Gallego A , Jimenez-Lopez JC and Morell M (2019) An Overview of the Polymorphisms of Circadian Genes Associated With Endocrine Cancer. Front. Endocrinol. 10:104. doi: 10.3389/fendo.2019.00104
Received: 30 September 2018; Accepted: 04 February 2019;
Published: 26 February 2019.
Edited by:
Veronica Vella, Università degli Studi di Catania, ItalyReviewed by:
Barbara Belletti, Centro di Riferimento Oncologico di Aviano (IRCCS), ItalyPaola Defilippi, University of Turin, Italy
Copyright © 2019 Morales-Santana, Morell, Leon, Carazo-Gallego, Jimenez-Lopez and Morell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonia Morales-Santana, sonia.morales.exts@juntadeandalucia.es
Santiago Morell, sm2322@cam.ac.uk
†These authors have contributed equally to this work
 Sonia Morales-Santana
Sonia Morales-Santana Santiago Morell
Santiago Morell Josefa Leon
Josefa Leon Angel Carazo-Gallego
Angel Carazo-Gallego Jose C. Jimenez-Lopez
Jose C. Jimenez-Lopez María Morell7
María Morell7