- 1GMP & T Cell Therapy Unit, German Cancer Research Center (DKFZ), Heidelberg, Germany
- 2Faculty of Biosciences, University Heidelberg, Heidelberg, Germany
The NT5E (CD73) molecule represents an ecto-5′-nucleotidase expressed on the cell surface of various cell types. Hydrolyzing extracellular adenosine monophosphate into adenosine and inorganic phosphate, NT5E performs numerous homeostatic functions in healthy organs and tissues. Importantly, NT5E can act as inhibitory immune checkpoint molecule, since free adenosine generated by NT5E inhibits cellular immune responses, thereby promoting immune escape of tumor cells. MicroRNAs (miRNAs) are small non-coding RNA molecules regulating gene expression on posttranscriptional level through binding to mRNAs, resulting in translational repression or degradation of the targeted mRNA molecule. In tumor cells, miRNA expression patterns are often altered which in turn might affect NT5E surface expression and eventually influence the efficacy of antitumor immune responses. This review describes the diverse roles of NT5E, summarizes current knowledge about transcription factors controlling NT5E expression, and highlights the significance of miRNAs involved in the posttranscriptional regulation of NT5E expression.
Functions of NT5E in Healthy Tissue and Tumors
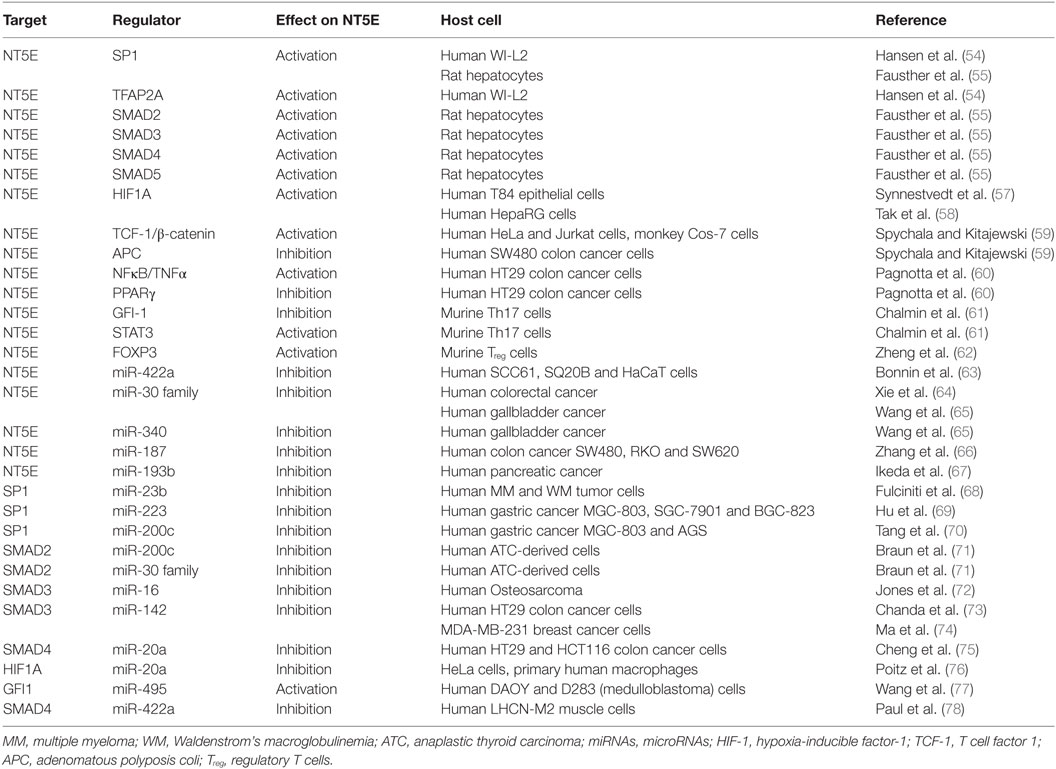
The membrane bound NT5E (CD73) is an ecto-5′-nucleotidase (NT5E) hydrolyzing extracellular adenosine monophosphate (AMP) into adenosine and inorganic phosphate (1) (Figure 1A). The enzyme consists of a homodimer inserted into the cellular membrane by glycophosphatidylinositol anchors. Besides hydrolyzing AMP to adenosine, NT5E has nucleosidase activity as shown for nicotinamide adenine dinucleotide and nicotinamide mononucleotide (2, 3). NT5E works in concert with ectonucleoside triphosphate diphosphohydrolase-1 (ENTPD1), which is also referred to as CD39, representing another ectonucleotidase acting upstream of NT5E catalyzing the hydrolysis of adenosine triphosphate (ATP) into AMP through two reversible reaction steps, whereas the final NT5E-mediated reaction from AMP to adenosine is largely irreversible (4) (Figure 1A). The two C-terminal domains of the NT5E molecule mediate noncovalent homodimer association and harbor the substrate binding sites (2). The molecular structure of NT5E can exhibit open or closed conformation and transition between these two stages occurs during substrate cleavage involving conformational changes enabled by the flexible α-helix connecting the C-terminal domains with the Zn2+ binding N-terminal domains (3), the latter being N-glycosylated at four distinct asparagine residues either by mannose saccharide chains or by a mixture of complex glycans and high mannose (2). Besides expression of the full-length molecule NT5EL (NT5E-201, 574 aa), a spliced version lacking exon 7 designated NT5ES (NT5E-203, 525 aa) can be detected in various human tissues and was found intracellularly overexpressed in human hepatocellular carcinoma cell lines (5). As depletion of amino acids 404–453 encoded by exon 7 prevents homo dimerization, NT5ES shows impaired substrate binding resulting in abrogated 5′-nucleotidase activity and lack of surface expression. Importantly, overexpression of NT5ES was shown to cause proteasome-mediated degradation of intracellular NT5EL, without affecting expression levels of native NT5EL dimers. Thus, altered splicing patterns commonly observed in many tumors (6–8) might contribute to aberrant NT5E expression levels in cancer cells.
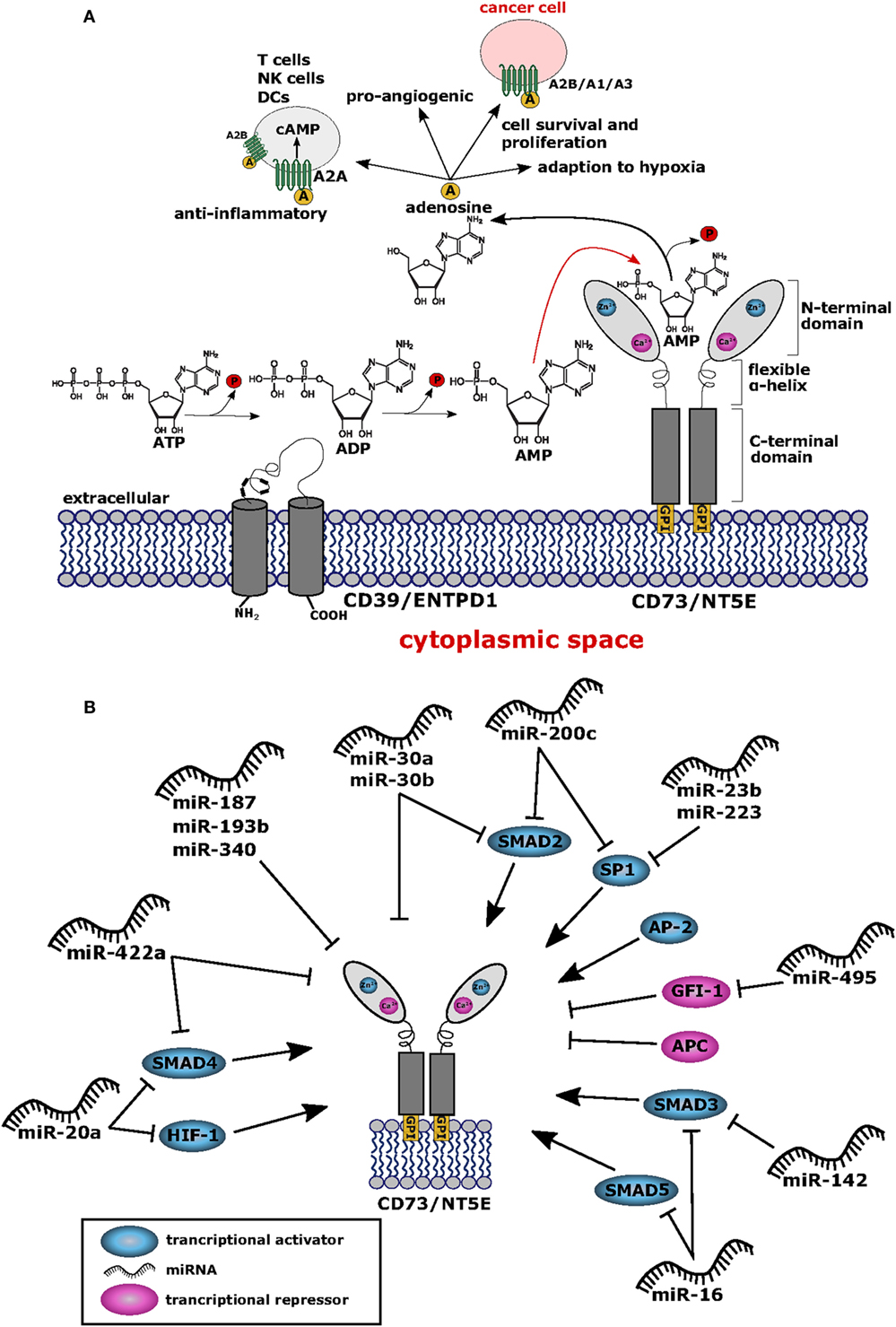
Figure 1. (A) Structure and function of CD73/NT5E. The membrane bound ecto-5′-nucleotidase NT5E hydrolyzes extracellular adenosine monophosphate (AMP) into adenosine and inorganic phosphate (P). Upstream of NT5E, adenosine triphosphate (ATP) is hydrolyzed via two reaction steps into AMP by the enzyme ectonucleoside triphosphate diphosphohydrolase-1 (ENTPD1) (CD39). Adenosine thus produced exerts anti-inflammatory effects by binding to the adenosine A2A receptor (ADORA2A) expressed by T cells, natural killer (NK) cells, and dendritic cells (DCs) resulting in cAMP mediated blocking of their effector functions. To some extent, the A2B receptor (ADORA2B) is also expressed on DCs and macrophages which are suppressed by adenosine. Thus, cancer cells can evade the immune system by upregulating NT5E protein levels. Furthermore, adenosine binds to the A2B receptor expressed by cancer cells leading to tumor cell survival and proliferation. Cancer cells also express the adenosine A1 receptor (ADORA1) and A3 receptor (ADORA3) and binding of adenosine to these receptors leads to tumor cell migration and proliferation via signaling through Gαi proteins. Adenosine is also involved in the adaption to hypoxia and shows pro-angiogenic potential. All adenosine receptors are depicted as stylized green transmembrane proteins. Adenosine is also symbolized as yellow circles marked with “A”. (B) Network of transcription factors and microRNAs (miRNAs) regulating NT5E expression. This network summarizes the current knowledge on regulation of NT5E on transcriptional (TFs) and posttranscriptional level by TFs and miRNAs, respectively. Transcriptional activators are depicted in blue and transcriptional repressors are highlighted in magenta. miRNAs targeting NT5E directly are shown, as well as miRNAs with indirect impact on NT5E expression through targeting of transcriptional regulators.
Considering healthy tissue, expression of NT5E is detectable in epithelial cells of the respiratory tract, smooth muscle cells, and cardiac myocytes and other tissues, as can be deduced from the bioGPS mRNA expression data base (9) using the data set of Primary Cell Atlas (10). Under physiological conditions, NT5E has been described as a regulator of epithelial ion transport, thereby preserving mucosal hydration (11). Moreover, NT5E can act as gate keeper on endothelial cells as free adenosine facilitates “resealing” of gaps between vascular endothelial cells left behind by transmigrating neutrophils (12). Furthermore, adenosine generated through NT5E was described to restrict inflammatory immune responses through a negative feedback loop on adenosine receptor expressing neutrophils (13). NT5E has been found expressed on regulatory T cells (Treg) and at even higher levels on anergic CD4+ T cells, thereby preserving self tolerance in healthy individuals and protecting the fetus from maternal immune attack during pregnancy (14, 15).
Interestingly, qualitative differences in NT5E function have been described depending on the NT5E expressing cell type (16, 17). Comparing epithelial cells and lymphocytes, both expressing NT5E, lymphocyte NT5E was found susceptible to phosphatidylinositol phospholipase to greater extent compared to NT5E expressed by epithelial cells (16). In the same study, antibody binding to NT5E triggered shedding from the surface of lymphocytes, but not in the case of epithelial cells. A similar observation was reported by others who proposed NT5E shedding from the surface of B16F10 cells as explanation for absent cell surface staining on murine B16F10 melanoma cells despite detection of intracellular NT5E expression (18). The study by Airas et al. (16) demonstrated also signal transduction activity of NT5E expressed on lymphocytes, whereas NT5E expressing epithelial cells lacked this function. Signal transduction by NT5E appears unexpected since this molecule lacks intracellular signaling domains (Figure 1A); however, it has been suggested that NT5E might associate with src protein kinases, thereby facilitating cellular signal transduction as proposed by Wang and colleagues (19, 20). Alternatively, NT5E might also mediate signal transduction directly (19).
In pathophysiological situations, NT5E activity was found relevant for the generation of cardio-protective adenosine in the ischemic myocardium (21) or for adaption to hypoxia (see below). Importantly, NT5E is involved in tumor development. Thus, NT5E has been described to sustain tumor angiogenesis in murine tumor models of breast cancer and prostate cancer (20, 22) as well as in xenograft models of humans breast cancer (23). Likewise, NT5E expression promoted invasion and metastasis of murine and human melanoma cells (24) and of human breast cancer cells (25).
Notably, NT5E plays a significant role as immune-inhibitory checkpoint molecule (26). Thus, infiltration of tumors by NT5E expressing regulatory immune cells such as Treg (27), MDSCs (28), or dendritic cells (DCs) (29) results in accumulation of immunosuppressive adenosine that can activate cAMP signaling in T cells expressing A2A adenosine receptors (ADORA2A). Moreover, adenosine receptors were found to be expressed on DCs, macrophages, MDSCs, and natural killer (NK) cells, implying that adenosine can repress the function of these immune cells (30, 31). Recently, an interesting phenomenon was described showing that Treg undergoing apoptosis within the metabolically abnormal tumor microenvironment release substantial amounts of ATP, that is degraded by the nucleotidases of the faded Treg resulting in accumulating adenosine levels (32). Adenosine can then trigger immune suppressive downstream effects among T cells like inhibition of chemotaxis, proliferation, activation, and effector function (33, 34). In light of its immune suppressive function and due to its expression by various tumor entities, such as melanoma (35–37), triple-negative breast cancer (34, 38), colorectal cancer (CRC) (39), and non-small cell lung cancer (40), NT5E has been considered as target checkpoint molecule for novel tumor immunotherapy approaches (41, 42). Indeed, injection of blocking NT5E-specific ab into tumor bearing mice resulted in reduced outgrowth of NT5E expressing tumors as shown for various tumor entities (43–45). Noteworthy, tumor cells can express adenosine A1 receptor (ADORA1) and ADORA3 receptors coupled to Gαi proteins, fostering tumor cell proliferation and migration (46). Moreover, therapeutic targeting of NT5E using specific inhibitors or blocking antibodies, respectively, has been proposed (22, 34, 38, 47–50) and is presently tested in a phase I clinical trial (NCT02503774).
In addition to its enzymatic function, NT5E can act as a receptor molecule shown to mediate cell–cell adhesion between lymphocytes and endothelial cells (51). Moreover, it was demonstrated that NT5E was can interact with extracellular matrix components (ECM) (34, 52, 53). This interaction occurred independently from enzymatic activity of NT5E, as blocking of ectonucleotidase function by concanavalin A did not affect interaction with ECM components like fibronectin, tenascin C, or collagen 1. In fact, NT5E turned out to mediate cell adhesion and migration via interaction with tenascin C (53).
Thus, NT5E appears to support tumor growth at multiple levels, i.e., by suppression of antitumoral immune responses via supply of adenosine and through facilitated dissemination of malignant cells from the primary tumor.
Transcriptional Regulation of NT5E Expression
The promoter region of NT5E contains binding sites for the transcription factors SP1, AP-2, and SMAD proteins as well as cAMP-responsive elements (54, 55) (Figure 1B; Table 1). Chromatin immunoprecipitation showed that transcription factors SMAD2, SMAD3, SMAD4, SMAD5, and SP1 bind to the rat NT5E promoter, with SMAD5 and SP1 being most efficient (55). As rat and human NT5E transcripts share 89% identity (56), it appears possible that human NT5E expression might be regulated by SMAD transcription factors as well.
Interestingly, hypoxia-inducible factor-1 (HIF-1) can directly bind to the NT5E promoter thereby activating NT5E expression (57), which is in line with the functional role described for NT5E in hypoxia adaptation (79). Thus, hypoxia resulting from uncontrolled tumor cell proliferation (80) might induce HIF mediated upregulation of NT5E expression on tumor cells. Another biochemical cascade often altered in tumors is the β-catenin-dependent Wnt signaling pathway (81). The promoter core sequence of NT5E is flanked upstream by a regulatory region containing consensus motifs for T cell factor 1 (TCF-1), representing a component of Wnt/β-catenin signaling pathway. In fact, expression of β-catenin could drastically enhance expression of NT5E. This upregulation was found to be dependent on the presence of TCF-1. Interestingly, the authors could also show that the antagonist of β-catenin, adenomatous polyposis coli protein, inhibits NT5E expression (59). The activation of NT5E expression by β-catenin was also confirmed by Pagnotta et al. who furthermore identified NFκB/TNFα as positive transcriptional regulators of NT5E (60). Seeking biomarkers for CRC, the authors applied a translational pathology approach and identified NT5E among others as a prognostic marker. In line with these findings, NT5E levels were found significantly upregulated in tumor specimens compared to normal colonic mucosa samples. Spranger et al. found that active β-catenin signaling was negatively associated with T cell infiltration in human melanoma samples. This was confirmed in authochtonous tumor models with inducible β-catenin expression, where absence of T cells was observed selectively in β-catenin expressing tumors (82). It is tempting to speculate that this immune suppressive effect on T cell infiltration might result from enhanced NT5E expression induced through β-catenin signaling. Of note, besides activating mechanisms on NT5E expression via NFκB/TNFα signaling pathways, also negative effects on NT5E expression through of PPARγ have been described (60).
In murine Th17 cells differentiated with IL-6 and TGF-β in vitro, IL-6 was found to activate Stat3, while TGF-β suppressed the transcription factor Gfi-1. As shown by the authors Stat3 sustained, whereas Gfi-1 repressed expression of ENTPD1 and NT5E through specific promoter binding, thus demonstrating transcriptional regulation of these exonucleotidases in Th17 cells through IL-6 and TGF-β (61).
A genome-wide analysis to identify forkhead box transcription factor (Foxp3) target genes in mouse led to the identification of Nt5e as one target gene of Foxp3 in mature Treg cells (62). Foxp3 is a specific transcription factor expressed in murine and human Treg cells and in recently activated human T cells (83). Thus, regulation of Nt5e by Foxp3 appears cell type specific and does not necessarily apply the same way to cancer cells. Noteworthy, high expression levels of FOXP3 in ovarian cancer has been identified as a prognostic marker for poor survival of patients (84).
MicroRNAs (miRNAs) Regulating NT5E Expression
MicroRNAs are small non-coding RNA molecules that bind to the 3′ untranslated region (3′-UTR) of target mRNAs, thereby blocking translation or inducing degradation of the targeted mRNA molecule, respectively, depending on the degree of complementary among the interacting nucleotide sequences (85). In cancer cells, aberrant miRNA expression patterns resulting in impaired regulation of target mRNA expression is commonly observed. Tumor cell-derived miRNAs have therefore gained relevance as biomarkers and as prognostic factors as described (86–88). Of note, the 3′-UTR of NT5E comprises 1,774 nucleotides (89) (NM_001204813.1), exceeding the average size of a human 3′-UTR (90) approximately threefold. Thus, regulation of NT5E expression by miRNAs appears to be particularly restrictive. To date, only a few miRNAs have been described that directly regulate NT5E expression (Table 1). Bonnin and co-workers reported the regulation of NT5E by miR-422a in head and neck squamous cell carcinoma (HNSCC) patients. The authors found a significant negative correlation between expression levels of miR-422a levels and NT5E mRNA. Blocking of endogenous miR-422a by specific antagomiRs resulted in increased NT5E protein levels with enhanced enzymatic activity. Reduced levels of miR-422a correlated with shorter relapse free survival times in HNSCC, potentially due to overexpression of NT5E (63).
Recently, miR-30a was found to target NT5E in CRC (64). In this study, transfection with miR-30a reduced NT5E expression on the mRNA and on protein level and direct interaction of miR-30a with the NT5E 3′-UTR was demonstrated via luciferase reporter assays. Similarly, direct regulation of NT5E was described through miR-30a-5p in non-small cell lung cancer (91). Enhanced expression of NT5E was accompanied by reduced miR-30a-5p expression, whereas miR-30a-5p overexpression resulted in downregulated NT5E expression on mRNA and protein levels. At the same time, proliferation, cell migration, and invasion of these cells were significantly reduced. These effects were mimicked by silencing NT5E expression using shRNA directed against NT5E.
Interestingly, the miR-30 family shares the same seed sequence (92). Thus, other miRNAs from this family might also regulate NT5E. Indeed, direct regulation of NT5E by miR-30b was shown by Wang et al. in gall bladder carcinoma (GBC). Including miR-340 in their study, the authors found that overexpression of miR-30b or miR-340 reduced GBC cell proliferation, migration, and invasion. For both miRNAs, direct interaction with the NT5E 3′-UTR could be verified and NT5E overexpression partially reverted these miRNA-mediated effects in GBC cells (65). In CRC, miR-187 levels were found strongly downregulated compared to adjacent normal tissue leading to the establishment of miR-187 expression levels as prognostic marker for CRC patients. In fact, transfection of miR-187 reduced cell proliferation and migration in vitro and decelerated tumor growth of CRC lines in vivo. In the same study, direct targeting of NT5E by miR-187 was demonstrated (66). Studies focused on miRNAs involved in the MAPK pathway of human pancreatic cancer cell lines revealed miR-193b as a direct binder of the NT5E 3′-UTR. However, in this study, binding specificity using a mutated reporter plasmid was not controlled and effects of miR-193 overexpression on NT5E expression on mRNA and protein level were not analyzed (67).
Considering the extraordinary size of the NT5E 3′-UTR region, the restricted number of validated miRNAs identified so far that directly target NT5E mRNA most likely represent just the tip of the iceberg. Further studies are needed to broaden the spectrum of known miRNAs that directly regulate NT5E surface expression, thereby potentially affecting the tumor cells’ vulnerability toward immune attack.
On the other hand, miRNAs can also function via indirect circuits, for example, by targeting transcription factors of NT5E, opening an alternative route for miRNA-mediated regulation of NT5E expression. In fact, miR-23b was found to directly suppress expression of transcription factor SP1 in multiple myeloma cells (68), and in gastric cancer, an inhibiting effect of miR-223 on epithelial to mesenchymal transition via direct posttranscriptional silencing of SP1 was reported (69). Other authors described miR-200b and miR-200c as direct inhibitors of SP1 transcription within this tumor entity (70). Whether the miRNA-mediated inhibition of SP1 expression resulted also in downstream reduction of NT5E expression levels was not investigated in these studies.
In a study by Braun et al., the authors focused on the identification of miRNAs affecting the invasive potential of anaplastic thyroid carcinoma and found miR-200c and miR-30a–e to target SMAD2 (71), representing another transcriptional activator of NT5E. Similarly, miR-16 might indirectly downregulate NT5E expression, as this miRNA was shown to inhibit expression of the transcription factors SMAD3 and, to lesser extent, SMAD5 in human osteosarcoma lines (72). Furthermore, SMAD3 is also targeted by miR-142-5p as shown in human rotavirus infected cells as well as in human breast and lung cancer cell lines (73, 74).
Further examples of miRNA-targeted transcription factors of NT5E are SMAD4 and HIF1A downregulated by miR-20a-5p (75), and Poitz et al. showed the direct downregulation of HIF1A by miR-20a (76). As mentioned above, miR-422a was shown to directly regulate NT5E, however, SMAD4 has also been described as a direct target of miR-422a (78), suggesting that miR-422a has the capacity to decrease NT5E levels both directly and indirectly. One example for a miRNA that could indirectly lead to an upregulation of NT5E levels is miR-495, which was shown to target one of NT5E’s transcriptional repressor GFI1 in medulloblastoma cells (77).
Conclusion
NT5E (CD73) has emerged as a novel target for tumor immunotherapy approaches, since functional inhibition of NT5E reversed its immunosuppressive effects resulting in tumor immune attack and eradication of cancer cells by cytotoxic CD8+ T cells and NK cells. Knowledge about NT5E regulation on the transcriptional and posttranscriptional level might provide a deeper understanding how cancer cells acquire aberrant NT5E expression to facilitate immune escape. We suggest a complex regulatory network of activatory and inhibitory transcription factors acting in conjunction with miRNAs to control NT5E expression. Interestingly, certain regulators such as miR-422a exert their effect on NT5E expression both directly as well as indirectly, i.e., through binding to the 3′-UTRs of NT5E mRNA and SMAD4 mRNA, the latter representing a transcriptional activator of NT5E. Even though the 3′-UTR region of NT5E is of extraordinary size, only few miRNAs have been described so far that regulate NT5E expression. Identification of further miRNAs targeting NT5E will help to unravel the complex regulation of NT5E expression in cancer cells.
Author Contributions
TK, WO, and SE wrote this paper. TK generated the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer IS and handling Editor declared their shared affiliation.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
1. Robson SC, Sevigny J, Zimmermann H. The E-NTPDase family of ectonucleotidases: structure function relationships and pathophysiological significance. Purinergic Signal (2006) 2(2):409–30. doi:10.1007/s11302-006-9003-5
2. Strater N. Ecto-5’-nucleotidase: structure function relationships. Purinergic Signal (2006) 2(2):343–50. doi:10.1007/s11302-006-9000-8
3. Knapp K, Zebisch M, Pippel J, El-Tayeb A, Muller CE, Strater N. Crystal structure of the human ecto-5’-nucleotidase (CD73): insights into the regulation of purinergic signaling. Structure (2012) 20(12):2161–73. doi:10.1016/j.str.2012.10.001
4. Antonioli L, Pacher P, Vizi ES, Hasko G. CD39 and CD73 in immunity and inflammation. Trends Mol Med (2013) 19(6):355–67. doi:10.1016/j.molmed.2013.03.005
5. Snider NT, Altshuler PJ, Wan S, Welling TH, Cavalcoli J, Omary MB. Alternative splicing of human NT5E in cirrhosis and hepatocellular carcinoma produces a negative regulator of ecto-5’-nucleotidase (CD73). Mol Biol Cell (2014) 25(25):4024–33. doi:10.1091/mbc.E14-06-1167
6. Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene (2014) 33(46):5311–8. doi:10.1038/onc.2013.533
7. Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep (2017) 20(9):2215–26. doi:10.1016/j.celrep.2017.08.012
8. Singh B, Eyras E. The role of alternative splicing in cancer. Transcription (2017) 8(2):91–8. doi:10.1080/21541264.2016.1268245
9. Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res (2016) 44(D1):D313–6. doi:10.1093/nar/gkv1104
10. Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics (2013) 14:632. doi:10.1186/1471-2164-14-632
11. Strohmeier GR, Lencer WI, Patapoff TW, Thompson LF, Carlson SL, Moe SJ, et al. Surface expression, polarization, and functional significance of CD73 in human intestinal epithelia. J Clin Invest (1997) 99(11):2588–601. doi:10.1172/JCI119447
12. Lennon PF, Taylor CT, Stahl GL, Colgan SP. Neutrophil-derived 5’-adenosine monophosphate promotes endothelial barrier function via CD73-mediated conversion to adenosine and endothelial A2B receptor activation. J Exp Med (1998) 188(8):1433–43. doi:10.1084/jem.188.8.1433
13. Eltzschig HK, Ibla JC, Furuta GT, Leonard MO, Jacobson KA, Enjyoji K, et al. Coordinated adenine nucleotide phosphohydrolysis and nucleoside signaling in posthypoxic endothelium: role of ectonucleotidases and adenosine A2B receptors. J Exp Med (2003) 198(5):783–96. doi:10.1084/jem.20030891
14. Kalekar LA, Schmiel SE, Nandiwada SL, Lam WY, Barsness LO, Zhang N, et al. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat Immunol (2016) 17(3):304–14. doi:10.1038/ni.3331
15. Kalekar LA, Mueller DL. Relationship between CD4 regulatory T cells and anergy in vivo. J Immunol (2017) 198(7):2527–33. doi:10.4049/jimmunol.1602031
16. Airas L, Niemelä J, Salmi M, Puurunen T, Smith DJ, Jalkanen S. Differential regulation and function of CD73, a glycosyl-phosphatidylinositol-linked 70-kD adhesion molecule, on lymphocytes and endothelial cells. J Cell Biol (1997) 136(2):421–31. doi:10.1083/jcb.136.2.421
17. Yegutkin GG, Auvinen K, Rantakari P, Hollmén M, Karikoski M, Grénman R, et al. Ecto-5′-nucleotidase/CD73 enhances endothelial barrier function and sprouting in blood but not lymphatic vasculature. Eur J Immunol (2015) 45(2):562–73. doi:10.1002/eji.201444856
18. Koszalka P, Pryszlak A, Golunska M, Kolasa J, Stasilojc G, Skladanowski AC, et al. Inhibition of CD73 stimulates the migration and invasion of B16F10 melanoma cells in vitro, but results in impaired angiogenesis and reduced melanoma growth in vivo. Oncol Rep (2014) 31(2):819–27. doi:10.3892/or.2013.2883
19. Zhi X, Wang Y, Yu J, Yu J, Zhang L, Yin L, et al. Potential prognostic biomarker CD73 regulates epidermal growth factor receptor expression in human breast cancer. IUBMB Life (2012) 64(11):911–20. doi:10.1002/iub.1086
20. Wang L, Tang S, Wang Y, Xu S, Yu J, Zhi X, et al. Ecto-5′-nucleotidase (CD73) promotes tumor angiogenesis. Clin Exp Metastasis (2013) 30(5):671–80. doi:10.1007/s10585-013-9571-z
21. Kitakaze M, Minamino T, Node K, Komamura K, Hori M. Activation of ecto-5’-nucleotidase and cardioprotection by ischemic preconditioning. Basic Res Cardiol (1996) 91(1):23–6.
22. Allard B, Turcotte M, Spring K, Pommey S, Royal I, Stagg J. Anti-CD73 therapy impairs tumor angiogenesis. Int J Cancer (2014) 134(6):1466–73. doi:10.1002/ijc.28456
23. Zhou X, Zhi X, Zhou P, Chen S, Zhao F, Shao Z, et al. Effects of ecto-5’-nucleotidase on human breast cancer cell growth in vitro and in vivo. Oncol Rep (2007) 17(6):1341–6. doi:10.3892/or.17.6.1341
24. Burghoff S, Gong X, Viethen C, Jacoby C, Flogel U, Bongardt S, et al. Growth and metastasis of B16-F10 melanoma cells is not critically dependent on host CD73 expression in mice. BMC Cancer (2014) 14:898. doi:10.1186/1471-2407-14-898
25. Wang L, Zhou X, Zhou T, Ma D, Chen S, Zhi X, et al. Ecto-5′-nucleotidase _promotes invasion, migration and adhesion of human breast cancer cells. J Cancer Res Clin Oncol (2008) 134(3):365–72. doi:10.1007/s00432-007-0292-z
26. Beavis PA, Stagg J, Darcy PK, Smyth MJ. CD73: a potent suppressor of antitumor immune responses. Trends Immunol (2012) 33(5):231–7. doi:10.1016/j.it.2012.02.009
27. Alam MS, Kurtz CC, Rowlett RM, Reuter BK, Wiznerowicz E, Das S, et al. CD73 is expressed by human regulatory T helper cells and suppresses proinflammatory cytokine production and Helicobacter felis-induced gastritis in mice. J Infect Dis (2009) 199(4):494–504. doi:10.1086/596205
28. Ryzhov S, Novitskiy SV, Goldstein AE, Biktasova A, Blackburn MR, Biaggioni I, et al. Adenosinergic regulation of the expansion and immunosuppressive activity of CD11b+Gr1+ cells. J Immunol (2011) 187(11):6120–9. doi:10.4049/jimmunol.1101225
29. Berchtold S, Ogilvie ALJ, Bogdan C, Mühl-Zürbes P, Ogilvie A, Schuler G, et al. Human monocyte derived dendritic cells express functional P2X and P2Y receptors as well as ecto-nucleotidases. FEBS Lett (1999) 458(3):424–8. doi:10.1016/S0014-5793(99)01197-7
30. Allard B, Beavis PA, Darcy PK, Stagg J. Immunosuppressive activities of adenosine in cancer. Curr Opin Pharmacol (2016) 29:7–16. doi:10.1016/j.coph.2016.04.001
31. Young A, Ngiow SF, Barkauskas DS, Sult E, Hay C, Blake SJ, et al. Co-inhibition of CD73 and A2AR adenosine signaling improves anti-tumor Immune responses. Cancer Cell (2018) 30(3):391–403. doi:10.1016/j.ccell.2016.06.025
32. Maj T, Wang W, Crespo J, Zhang H, Wang W, Wei S, et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat Immunol (2017) 18:1332. doi:10.1038/ni.3868
33. Sitkovsky MV, Lukashev D, Apasov S, Kojima H, Koshiba M, Caldwell C, et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu Rev Immunol (2004) 22:657–82. doi:10.1146/annurev.immunol.22.012703.104731
34. Allard B, Turcotte M, Stagg J. Targeting CD73 and downstream adenosine receptor signaling in triple-negative breast cancer. Expert Opin Ther Targets (2014) 18(8):863–81. doi:10.1517/14728222.2014.915315
35. Sadej R, Spychala J, Skladanowski AC. Ecto-5’-nucleotidase (eN, CD73) is coexpressed with metastasis promoting antigens in human melanoma cells. Nucleosides Nucleotides Nucleic Acids (2006) 25(9–11):1119–23. doi:10.1080/15257770600894188
36. Sadej R, Spychala J, Skladanowski AC. Expression of ecto-5’-nucleotidase (eN, CD73) in cell lines from various stages of human melanoma. Melanoma Res (2006) 16(3):213–22. doi:10.1097/01.cmr.0000215030.69823.11
37. Wang H, Lee S, Nigro CL, Lattanzio L, Merlano M, Monteverde M, et al. NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br J Cancer (2012) 106(8):1446–52. doi:10.1038/bjc.2012.95
38. Buisseret L, Pommey S, Allard B, Garaud S, Bergeron M, Cousineau I, et al. Clinical significance of CD73 in triple-negative breast cancer: multiplex analysis of a phase III clinical trial. Ann Oncol (2017) mdx730. doi:10.1093/annonc/mdx730
39. Liu N, Fang XD, Vadis Q. CD73 as a novel prognostic biomarker for human colorectal cancer. J Surg Oncol (2012) 106(7):918–9. doi:10.1002/jso.23159
40. Inoue Y, Yoshimura K, Kurabe N, Kahyo T, Kawase A, Tanahashi M, et al. Prognostic impact of CD73 and A2A adenosine receptor expression in non-small-cell lung cancer. Oncotarget (2017) 8(5):8738–51. doi:10.18632/oncotarget.14434
41. Allard B, Longhi MS, Robson SC, Stagg J. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol Rev (2017) 276(1):121–44. doi:10.1111/imr.12528
42. Antonioli L, Novitskiy SV, Sachsenmeier KF, Fornai M, Blandizzi C, Haskó G. Switching off CD73: a way to boost the activity of conventional and targeted antineoplastic therapies. Drug Discov Today (2017) 22(11):1686–96. doi:10.1016/j.drudis.2017.06.005
43. Young A, Ngiow SF, Gao Y, Patch A-M, Barkauskas DS, Messaoudene M, et al. A2AR adenosine signaling suppresses natural killer cell maturation in the tumor microenvironment. Cancer Res (2018) 78(4):1003–16. doi:10.1158/0008-5472.can-17-2826
44. Stagg J, Divisekera U, McLaughlin N, Sharkey J, Pommey S, Denoyer D, et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci U S A (2010) 107(4):1547–52. doi:10.1073/pnas.0908801107
45. Beavis PA, Divisekera U, Paget C, Chow MT, John LB, Devaud C, et al. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci U S A (2013) 110(36):14711–6. doi:10.1073/pnas.1308209110
46. Ohta A. A metabolic immune checkpoint: adenosine in tumor microenvironment. Front Immunol (2016) 7(109):1–11. doi:10.3389/fimmu.2016.00109
47. Zhang B. CD73: a novel target for cancer immunotherapy. Cancer Res (2010) 70(16):6407–11. doi:10.1158/0008-5472.CAN-10-1544
48. Antonioli L, Yegutkin GG, Pacher P, Blandizzi C, Hasko G. Anti-CD73 in cancer immunotherapy: awakening new opportunities. Trends Cancer (2016) 2(2):95–109. doi:10.1016/j.trecan.2016.01.003
49. Geoghegan JC, Diedrich G, Lu X, Rosenthal K, Sachsenmeier KF, Wu H, et al. Inhibition of CD73 AMP hydrolysis by a therapeutic antibody with a dual, non-competitive mechanism of action. MAbs (2016) 8(3):454–67. doi:10.1080/19420862.2016.1143182
50. Turcotte M, Allard D, Mittal D, Bareche Y, Buisseret L, Jose V, et al. CD73 promotes resistance to HER2/ErbB2 antibody therapy. Cancer Res (2017) 77(20):5652–63. doi:10.1158/0008-5472.CAN-17-0707
51. Airas L, Hellman J, Salmi M, Bono P, Puurunen T, Smith DJ, et al. CD73 is involved in lymphocyte binding to the endothelium: characterization of lymphocyte-vascular adhesion protein 2 identifies it as CD73. J Exp Med (1995) 182(5):1603–8. doi:10.1084/jem.182.5.1603
52. Andrade CM, Lopez PL, Noronha BT, Wink MR, Borojevic R, Margis R, et al. Ecto-5’-nucleotidase/CD73 knockdown increases cell migration and mRNA level of collagen I in a hepatic stellate cell line. Cell Tissue Res (2011) 344(2):279–86. doi:10.1007/s00441-011-1140-7
53. Sadej R, Skladanowski AC. Dual, enzymatic and non-enzymatic, function of ecto-5’-nucleotidase (eN, CD73) in migration and invasion of A375 melanoma cells. Acta Biochim Pol (2012) 59(4):647–52.
54. Hansen KR, Resta R, Webb CF, Thompson LF. Isolation and characterization of the promoter of the human 5’-nucleotidase (CD73)-encoding gene. Gene (1995) 167(1–2):307–12. doi:10.1016/0378-1119(95)00574-9
55. Fausther M, Sheung N, Saiman Y, Bansal MB, Dranoff JA. Activated hepatic stellate cells upregulate transcription of ecto-5’-nucleotidase/CD73 via specific SP1 and SMAD promoter elements. Am J Physiol Gastrointest Liver Physiol (2012) 303(8):G904–14. doi:10.1152/ajpgi.00015.2012
56. Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol (2000) 7(1–2):203–14. doi:10.1089/10665270050081478
57. Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK, et al. Ecto-5’-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest (2002) 110(7):993–1002. doi:10.1172/JCI15337
58. Tak E, Jung DH, Kim SH, Park GC, Jun DY, Lee J, et al. Protective role of hypoxia-inducible factor-1alpha-dependent CD39 and CD73 in fulminant acute liver failure. Toxicol Appl Pharmacol (2017) 314:72–81. doi:10.1016/j.taap.2016.11.016
59. Spychala J, Kitajewski J. Wnt and beta-catenin signaling target the expression of ecto-5’-nucleotidase and increase extracellular adenosine generation. Exp Cell Res (2004) 296(2):99–108. doi:10.1016/j.yexcr.2003.11.001
60. Pagnotta SM, Laudanna C, Pancione M, Sabatino L, Votino C, Remo A, et al. Ensemble of gene signatures identifies novel biomarkers in colorectal cancer activated through PPARgamma and TNFalpha signaling. PLoS One (2013) 8(8):e72638. doi:10.1371/journal.pone.0072638
61. Chalmin F, Mignot G, Bruchard M, Chevriaux A, Vegran F, Hichami A, et al. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity (2012) 36(3):362–73. doi:10.1016/j.immuni.2011.12.019
62. Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature (2007) 445(7130):936–40. doi:10.1038/nature05563
63. Bonnin N, Armandy E, Carras J, Ferrandon S, Battiston-Montagne P, Aubry M, et al. miR-422a promotes loco-regional recurrence by targeting NT5E/CD73 in head and neck squamous cell carcinoma. Oncotarget (2016) 7(28):44023–38. doi:10.18632/oncotarget.9829
64. Xie M, Qin H, Luo Q, Huang Q, He X, Yang Z, et al. MicroRNA-30a regulates cell proliferation and tumor growth of colorectal cancer by targeting CD73. BMC Cancer (2017) 17(1):305. doi:10.1186/s12885-017-3291-8
65. Wang N, Xiang X, Chen K, Liu P, Zhu A. Targeting of NT5E by miR-30b and miR-340 attenuates proliferation, invasion and migration of gallbladder carcinoma. Biochimie (2017) 146:56–67. doi:10.1016/j.biochi.2017.10.027
66. Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, et al. MicroRNA-187, a downstream effector of TGFbeta pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett (2016) 373(2):203–13. doi:10.1016/j.canlet.2016.01.037
67. Ikeda Y, Tanji E, Makino N, Kawata S, Furukawa T. MicroRNAs associated with mitogen-activated protein kinase in human pancreatic cancer. Mol Cancer Res (2012) 10(2):259–69. doi:10.1158/1541-7786.MCR-11-0035
68. Fulciniti M, Amodio N, Bandi RL, Cagnetta A, Samur MK, Acharya C, et al. miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J (2016) 6:e380. doi:10.1038/bcj.2015.106
69. Hu J, Shan Z, Hu K, Ren F, Zhang W, Han M, et al. miRNA-223 inhibits epithelial-mesenchymal transition in gastric carcinoma cells via Sp1. Int J Oncol (2016) 49(1):325–35. doi:10.3892/ijo.2016.3533
70. Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res (2013) 19(20):5602–12. doi:10.1158/1078-0432.CCR-13-1326
71. Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene (2010) 29(29):4237–44. doi:10.1038/onc.2010.169
72. Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res (2012) 72(7):1865–77. doi:10.1158/0008-5472.CAN-11-2663
73. Chanda S, Nandi S, Chawla-Sarkar M. Rotavirus-induced miR-142-5p elicits proviral milieu by targeting non-canonical transforming growth factor beta signalling and apoptosis in cells. Cell Microbiol (2016) 18(5):733–47. doi:10.1111/cmi.12544
74. Ma Z, Liu T, Huang W, Liu H, Zhang HM, Li Q, et al. MicroRNA regulatory pathway analysis identifies miR-142-5p as a negative regulator of TGF-beta pathway via targeting SMAD3. Oncotarget (2016) 7(44):71504–13. doi:10.18632/oncotarget.12229
75. Cheng D, Zhao S, Tang H, Zhang D, Sun H, Yu F, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget (2016) 7(29):45199–213. doi:10.18632/oncotarget.9900
76. Poitz DM, Augstein A, Gradehand C, Ende G, Schmeisser A, Strasser RH. Regulation of the Hif-system by micro-RNA 17 and 20a – role during monocyte-to-macrophage differentiation. Mol Immunol (2013) 56(4):442–51. doi:10.1016/j.molimm.2013.06.014
77. Wang C, Yun Z, Zhao T, Liu X, Ma X. miR-495 is a predictive biomarker that downregulates GFI1 expression in medulloblastoma. Cell Physiol Biochem (2015) 36(4):1430–9. doi:10.1159/000430308
78. Paul R, Lee J, Donaldson AV, Connolly M, Sharif M, Natanek SA, et al. miR-422a suppresses SMAD4 protein expression and promotes resistance to muscle loss. J Cachexia Sarcopenia Muscle (2018) 9:119–28. doi:10.1002/jcsm.12236
79. Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles for ecto-5’-nucleotidase (CD73). Purinergic Signal (2006) 2(2):351–60. doi:10.1007/s11302-005-5302-5
80. Eales KL, Hollinshead KE, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis (2016) 5:e190. doi:10.1038/oncsis.2015.50
81. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene (2017) 36(11):1461–73. doi:10.1038/onc.2016.304
82. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature (2015) 523:231. doi:10.1038/nature14404
83. Lu L, Barbi J, Pan F. The regulation of immune tolerance by FOXP3. Nat Rev Immunol (2017) 17(11):703–17. doi:10.1038/nri.2017.75
84. Wolf D, Wolf AM, Rumpold H, Fiegl H, Zeimet AG, Muller-Holzner E, et al. The expression of the regulatory T cell-specific forkhead box transcription factor FoxP3 is associated with poor prognosis in ovarian cancer. Clin Cancer Res (2005) 11(23):8326–31. doi:10.1158/1078-0432.CCR-05-1244
85. Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol (2015) 25(3):137–47. doi:10.1016/j.tcb.2014.11.004
86. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med (2014) 20(8):460–9. doi:10.1016/j.molmed.2014.06.005
87. Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, et al. MicroRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep (2016) 6:25997. doi:10.1038/srep25997
88. Larrea E, Sole C, Manterola L, Goicoechea I, Armesto M, Arestin M, et al. New concepts in cancer biomarkers: circulating miRNAs in liquid biopsies. Int J Mol Sci (2016) 17(5):1–42. doi:10.3390/ijms17050627
89. Dassi E, Re A, Leo S, Tebaldi T, Pasini L, Peroni D, et al. AURA 2: empowering discovery of post-transcriptional networks. Translation (Austin) (2014) 2(1):e27738. doi:10.4161/trla.27738
90. Mazumder B, Seshadri V, Fox PL. Translational control by the 3’-UTR: the ends specify the means. Trends Biochem Sci (2003) 28(2):91–8. doi:10.1016/S0968-0004(03)00002-1
91. Zhu J, Zeng Y, Li W, Qin H, Lei Z, Shen D, et al. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol Cancer (2017) 16(1):34. doi:10.1186/s12943-017-0591-1
Keywords: checkpoint molecule, CD73, NT5E, microRNA, transcription factor, T cell, tumor, A2A receptor
Citation: Kordaß T, Osen W and Eichmüller SB (2018) Controlling the Immune Suppressor: Transcription Factors and MicroRNAs Regulating CD73/NT5E. Front. Immunol. 9:813. doi: 10.3389/fimmu.2018.00813
Received: 01 March 2018; Accepted: 04 April 2018;
Published: 18 April 2018
Edited by:
Alexandr Bazhin, Klinikum der Universität München, GermanyReviewed by:
Jörg Wischhusen, Universität Würzburg, GermanyIvan Shevchenko, Ludwig-Maximilians-Universität München, Germany
Camilla Jandus, Université de Lausanne, Switzerland
Copyright: © 2018 Kordaß, Osen and Eichmüller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan B. Eichmüller, s.eichmueller@dkfz.de
 Theresa Kordaß1,2
Theresa Kordaß1,2 Stefan B. Eichmüller
Stefan B. Eichmüller