- 1Department of Medical Microbiology and Hygiene, University Medical Center of the Johannes Gutenberg University, Mainz, Germany
- 2Department of Membrane Biochemistry, Life & Medical Sciences Institute, University of Bonn, Bonn, Germany
Tetraspanins (Tspans) are a family of four-span transmembrane proteins, known as plasma membrane “master organizers.” They form Tspan-enriched microdomains (TEMs or TERMs) through lateral association with one another and other membrane proteins. If multiple microdomains associate with each other, larger platforms can form. For infection, viruses interact with multiple cell surface components, including receptors, activating proteases, and signaling molecules. It appears that Tspans, such as CD151, CD82, CD81, CD63, CD9, Tspan9, and Tspan7, coordinate these associations by concentrating the interacting partners into Tspan platforms. In addition to mediating viral attachment and entry, these platforms may also be involved in intracellular trafficking of internalized viruses and assist in defining virus assembly and exit sites. In conclusion, Tspans play a role in viral infection at different stages of the virus replication cycle. The present review highlights recently published data on this topic, with a focus on events at the plasma membrane. In light of these findings, we propose a model for how Tspan interactions may organize cofactors for viral infection into distinct molecular platforms.
Introduction
The contents of the cell are protected from the extracellular surroundings by the plasma membrane: a lipid bilayer densely populated with protein (1, 2). These proteins are specifically distributed throughout the membrane, a phenomenon associated with lipid microdomains, rafts, phases, or clusters. Local enrichments can be explained by spontaneous self-organization driven by thermodynamic principles (3). Conversely, the composition and architecture of membrane proteins is also actively remodeled in order to control specific functions.
Viruses are genetic entities that can form particles of sizes up to 200 nm and require multiple steps to overcome the cell barrier during entry and egress. To gain access into the cell, viruses employ different host receptors, proteases, and signaling molecules. After internalization via endocytosis, non-enveloped viruses escape the membranous organelle system in order to deliver viral genetic information into the cytoplasm or nucleus (4–6). Entry of enveloped viruses occurs through fusion of the viral and cellular membrane at the plasma membrane or in intracellular compartments (7). Each viral entry mechanism involves its own set of unique interactions between the virus and the cellular membrane system. Members of the tetraspanin (Tspan) protein family are localized to membranes and as such, associate directly and indirectly, with multiple steps of viral infection.
Tetraspanins
Tetraspanins are a family of small transmembrane proteins (8) that function in cell migration, signal-transduction, intracellular trafficking, and are used by several pathogens for infection (9–11). Of the 33 human Tspans, CD151, CD82, CD81, CD63, CD9, Tspan9, and Tspan7 have been associated with viral infections (12–16).
Structure
Structurally, Tspans consist of four transmembrane segments, a small extracellular domain, and a large extracellular loop (LEL). Intracellular domains, including the N- and C-terminal tails, are relatively small and contain palmitoylated cysteines. Homology is highly conserved between isoforms with the exception of a small variable domain located within the LEL (17), which may contribute to differences in functionality between isoforms (18, 19).
To date, structural models are only available for CD81. The first model is based on the LEL crystal structure to which the α-helical transmembrane segments were attached in a theoretical conformation. The transmembrane region was predicted to form a four-stranded coiled-coil with two helices extending vertically into the bulkier LEL (20), resulting into a mushroom-shaped structure. The second model, derived from lipidic cubic phase crystallization of the entire protein, describes an arrangement with two major differences. First, instead of assembling into one bundle the transmembrane segments form two coiled-coils resulting in a cholesterol-binding pocket. Second, two kinks exist between the helical transmembrane segments and the LEL, causing the LEL to fold back toward the membrane (21). When cholesterol is released, the kinks straighten, and the LEL adopts an orientation similar to the proposed first model (21).
Tspan-Enriched Microdomains
Tetraspanins are referred to as master organizers of the plasma membrane, largely due to the fact that they form functional units termed Tspan-enriched microdomains (TEMs or TERMs). Biochemical immunoprecipitation experiments employing detergents of varying strengths revealed two major categories of Tspan interactions: (1) robust interactions between Tspans and non-Tspan binding partners, and (2) weak interactions between Tspan family members (22). Within the second category, certain assemblies of homo–Tspan interactions are preferred over hetero-dimerization/oligomerization (23), and the specificity of oligomerization is mediated by a small segment within the LEL referred to as δ-loop (24, 25). Consistent with these biochemical findings, electron microscopy shows that CD63 and CD9 form distinct clusters (26). Using a more systematic approach, super-resolution light microscopy confirms that single Tspan family members cluster within TEMs (27). Together, these data demonstrate that Tspan isoforms segregate into individual nanoclusters within larger Tspan domains.
In immuno-electron microscopy, Tspan microdomains are highly variable in shape and size with an average surface area of 0.2 µm2 (26). When assuming a spherical shape, this corresponds to a diameter of ≈500 nm. In contrast, super-resolution light microscopy detects spherically shaped structures with a diameter in the range of 100–150 nm (27, 28). These two methods result in surface area coverage calculations that differ by more than one order of magnitude. This substantial variability is likely due to the description of multiple nanoclusters within TEMs via electron microscopy, whereas super-resolution light microscopy identifies individual nanoclusters due to a higher epitope labeling density.
At present, the sequence of events for TEM biogenesis is unknown, though we can build a model on the following observations. First, different Tspans can associate with each other, but Tspans of one type preferentially homo-oligomerize. Second, Tspans form very tight complexes with non-Tspan partners such as integrins (29, 30) and signaling receptors (31). Finally, TEMs are stabilized by weak nonspecific interactions mediated by the aforementioned palmitate residues (32, 33) and glycolipids that promote Tspan assemblies (24, 34). These different interaction modalities likely produce small TEMs (Figure 1A). Viral surfaces contain abundant identical binding sites that may crosslink small TEMs to large Tspan trafficking platforms (Figure 1B). Evidence for virus-induced large Tspan assemblies has been documented by a number of microscopic studies discussed below.
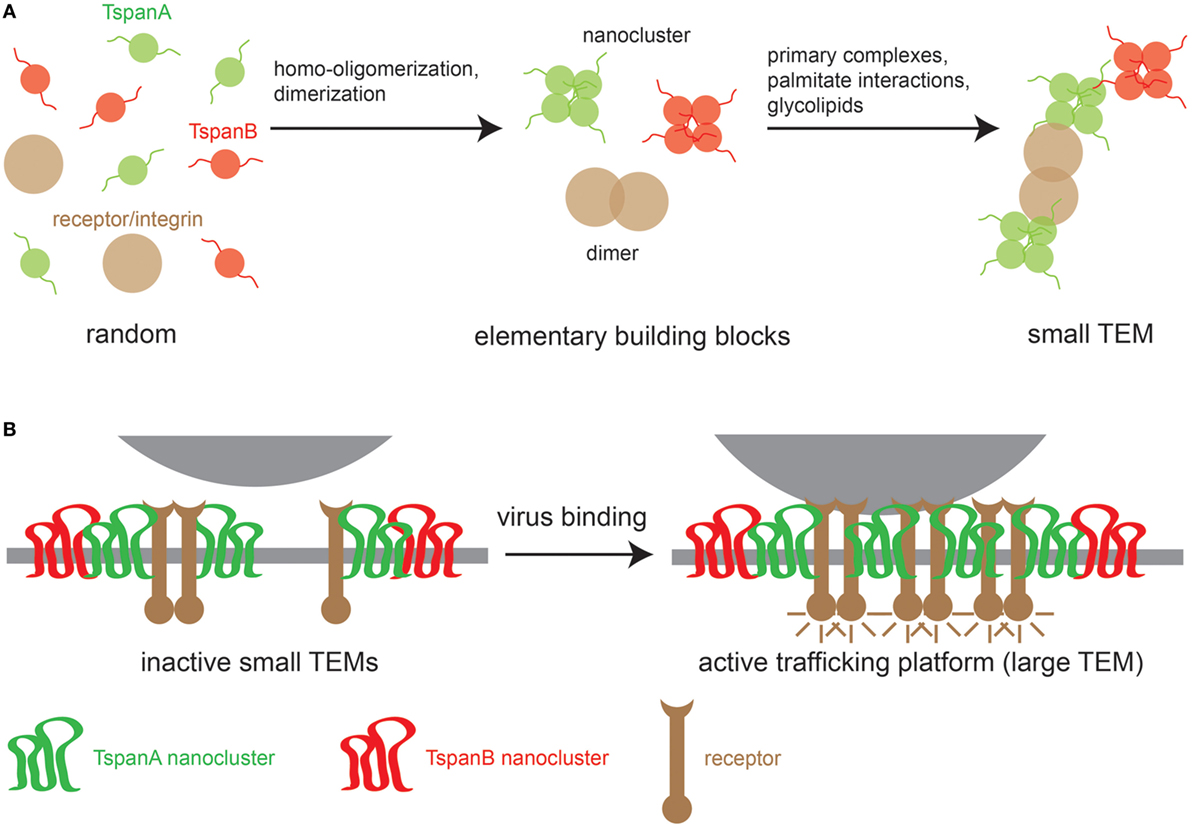
Figure 1. Model of TEM building. (A) Left, random distribution of three TEM components: two tetraspanins (TspanA and TspanB) and a primary interaction partner of TspanA [e.g., an integrin or a growth factor receptor (GFR)]. Middle, TEM building blocks; tetraspanins (Tspans) are arranged in homomeric nanoclusters and integrins/GFRs form dimers. Right, building blocks are connected via strong specific interactions between TspanA and the receptor/integrin dimer and weak nonspecific interactions between different Tspan nanoclusters. (B) When a virus encounters the cellular membrane, virus surface molecules crosslink small TEMs to form larger TEMs, thereby concentrating proteins leading to the activation of intracellular signaling cascades that trigger the uptake of the Tspan trafficking platform.
Roles of Tspans in Virus Infection
Tetraspanins are essential for specific steps in viral entry and exit (12, 13, 15). As described above, contacts between viruses and proteins on the cell surface can lead to large Tspan cluster networks or trafficking platforms (Figure 1B). Similarly, viral envelope proteins accumulate in TEMs during morphogenesis and induce large assemblies of Tspans and viral transmembrane proteins to facilitate efficient budding (Figure 2). These platforms enable the coordination of factors required for viral endocytosis, penetration, trafficking, and release. Here, we summarize and discuss the role of CD151, CD82, CD81, CD63, CD9, Tspan9, and Tspan7 in the life cycle of Tspan-facilitated viruses [for a detailed discussion on the role of Tspans in human immunodeficiency virus (HIV) infection see (Suarez et al.; Tspans, another piece in the HIV-1 replication puzzle) in this issue].
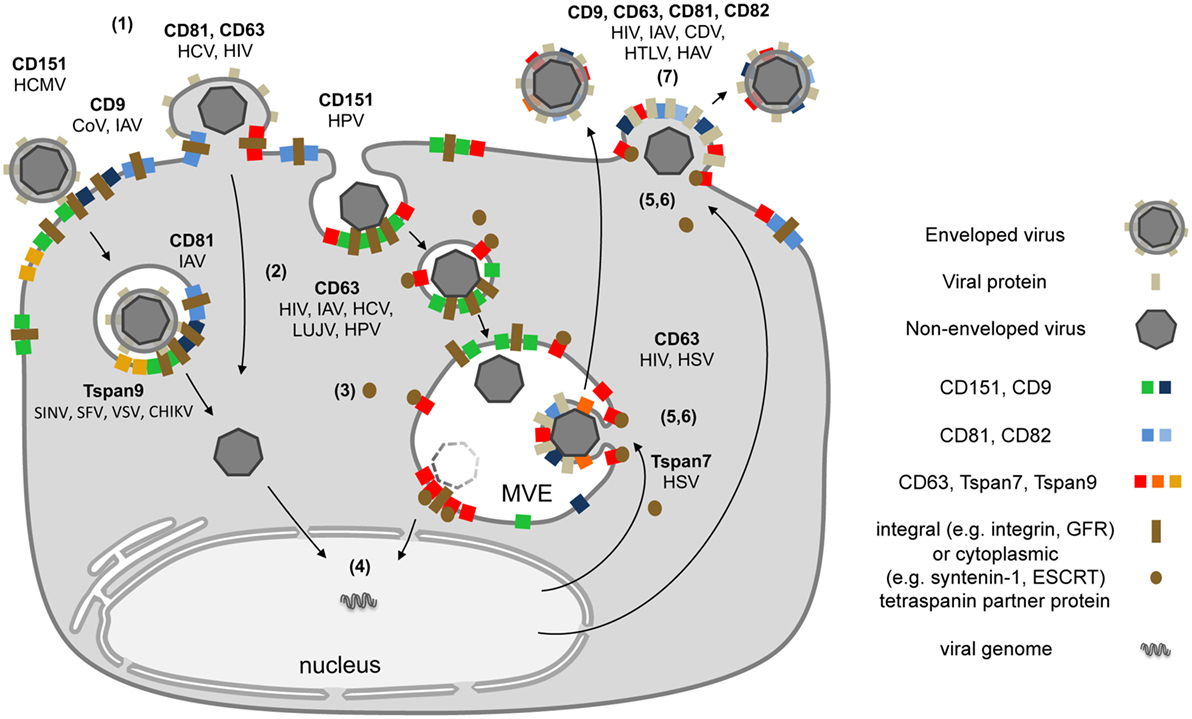
Figure 2. Schematic model depicting the subcellular localization of tetraspanins (Tspans) during virus infection. (1) Interactions between viral particles and entry receptors at Tspan clusters trigger the formation of larger cluster networks. (2) Tspan assemblies promote viral internalization by endocytosis and/or fusion. (3) Endocytosis is followed by intracellular trafficking of virus particles in transport vesicles. During this stage, Tspans mediate and organize interactions with cytoplasmic trafficking molecules. (4) These steps lead to delivery of viral genomes into the cytoplasm or the nucleus and successful infection. (5) Morphogenesis of enveloped viruses on Tspan-enriched microdomains. (6) Integration of viral proteins into Tspan clusters induces spatial enrichment of Tspans and viral proteins. (7) The resulting high concentration of virus envelope components enables efficient budding and release.
Tspan Platforms in Virus Endocytosis and Fusion
Studies investigating different viral systems show common mechanisms for how viruses infiltrate their target cells via Tspan platforms. Several microscopic studies confirm that Tspans are enriched at viral entry sites of human papillomaviruses (HPVs) (35, 36), hepatitis C virus (HCV) (37–42), coronavirus (CoV), influenza A virus (IAV) (43–45), and HIV (46, 47), and required for penetration of human cytomegalovirus (HCMV) (48) and alphaviruses (16, 49). These viruses use specific Tspans both as receptors and by compartmentalizing host entry factors.
CD151 in Early Steps of HPV and Cytomegalovirus Entry
Host cell entry of non-enveloped DNA tumor virus HPV16 relies on a fine interplay between the virion and the host cell. HPV16 binding to primary attachment sites triggers cell signaling events and rearrangement of the viral capsid, membrane proteins, and the actin cytoskeleton (15, 50–52). These processes lead to the formation of a virus entry complex and virus uptake via a CD151-dependent and clathrin-independent endocytosis pathway (14, 15, 53). In epithelial cells, surface-bound HPV16 particles colocalize with locally enriched CD151 and CD63 on the plasma membrane during invagination and in endosomes (14, 35). Cellular depletion of CD151 and CD63 leads to significant reduction of infections by different oncogenic HPV types, suggesting that these Tspans play a more general role in HPV entry (35, 36, 54, 55). On T-cells, HPV particles are able to trigger the clustering of CD81 which results in the assembly of larger cluster networks required for particle uptake (56). Furthermore, detailed analyses using CD151 mutants revealed that palmitoylation, the δ-loop of the LEL and the C-teminus of CD151 are critical for HPV16 endocytosis (14, 36). These findings indicate that integration of the virus/receptor-complex into larger TEMs and association with cytoplasmic factors (e.g., actin) are required for this process. HPV endocytosis may also involve interactions between multiple receptors and the viral surface, crosslinking smaller TEMs to larger entry platforms (Figure 1B). HPV16 receptor-complex components include integrins (36, 57–59), growth factor receptors (GFRs) (60), the annexin A2 heterotetramer (61, 62), and other Tspans (35, 36, 56). CD151 directly interacts with integrins and GFRs (14, 22, 63, 64), and, therefore, positions these HPV receptors within TEMs (11, 65). Through this spatial arrangement of functional proteins, CD151 may control enzymatic activities and signaling pathways required for coordinated assembly of the viral entry platform and endocytosis.
Likewise, entry of the enveloped HCMV depends on CD151 and CD151 partner proteins (e.g., integrins, GFRs) and additional Tspans, such as CD9 (15, 48, 66). CD151 is functionally involved post-binding during viral penetration (48). HCMV membrane fusion occurs after clathrin-independent endocytosis in many cell lines (67–69). Together, these studies suggest that CD151-mediated endocytosis might be a prerequisite for efficient HCMV and HPV infection. Mechanistically, virus-receptor, virus-Tspan, and Tspan-Tspan interactions play a vital role in organizing large Tspan platforms, which facilitate coordinated or simultaneous interactions between virus and host to induce membrane invagination by a mechanism yet to be determined [for detailed review see Ref. (15)].
CD81 and CD9 in HCV, Corona-, and Influenza-Virus Entry
Similar to HPV and HCMV, HCV entry into hepatocytes is a multistep process involving attachment to cell surface heparan sulfate proteoglycans, conformational changes, and transfer of viral particles to secondary receptors (38, 40, 70, 71). These secondary HCV binding molecules also include integrins, the epidermal GFR (EGFR), the ephrin receptor A2 and Tspans as well as claudin-1 (CLDN1), occludin, the scavenger receptor type B class I (SR-BI), and the serum response factor binding protein 1 (SRFBP1) (72–74). Tspan CD81 plays a multifunctional role in HCV entry. CD81 acts as a virus receptor by directly interacting with the HCV glycoprotein E2 (41, 42). CD81 modulates Tspan interactions after HCV binding (75) by triggering EGFR signaling pathways, which enables Tspan/receptor complex-assembly (76, 77) and promotes CD81-EGFR or CD81-CLDN1 complex formation (74, 76). These events are prerequisite for the endocytosis of CD81-HCV clusters and viral glycoprotein-dependent membrane fusion. Proteomic approaches confirmed complex formation of CLDN1, SR-BI, and SRFBP1 with CD81, and demonstrate the functional requirement of integrin β1 (ITGB1) and SRFBP1 for HCV infection and the physical interaction of the Tspan coreceptor-complex with the signaling molecule HRas (73, 77). The rat sarcoma/mitogen-activated protein kinase signaling pathways and EGFR or EphA2 activity trigger lateral diffusion of CD81 for assembly of the viral entry complex consisting of CD81- CLDN1, HRas, and ITGB1 (74, 77). Because GFRs support the uptake of multiple viruses (78), it is probable that activation of their downstream signaling cascades could trigger Tspan receptor clustering accompanied by cytoskeletal network rearrangement required for the entry of other virus families.
Influenza A-viruses and CoVs are enveloped RNA viruses (79). Tspan microdomains, especially CD81 and CD9 enriched microdomains, are preferred IAV and CoV entry sites as they are required for fusion of viral and host cell membranes in pathogenic infections by both viruses (43–45, 80, 81). IAV and CoV use a variety of coreceptors for this glycoprotein-catalyzed process (82). IAV is routed to CD81-positive endosomes and CD81 is functionally required for the fusion of the viral and the endosomal membrane (45). Here, CD81 may help organize the endosomal membrane and cofactors assisting influenza viral fusion. CoV membrane fusion is mediated by the viral spike glycoprotein (S) and depends on multiple events including proteolytic processing and conformational change of the S protein. Experiments utilizing Tspan knockout cell lines and mice revealed that infection by the human CoV strain 229E requires the Tspan CD9 (43, 44). Pulldown and proximity ligation assays uncovered the four known CoV receptors and a fusion-activating protease within CD9 microdomains. These studies also demonstrated that even in the absence of the virus, CD9 is responsible for the local accumulation of the identified entry receptors. Together, this evidence suggests that CoV uses pre-existing clusters of receptors, proteases, and Tspans for entry. Whether these viruses induce local accumulation of the pre-formed nanoclusters to enable efficient priming of the viral spike proteins during viral egress requires further investigation.
CD63 and Tspan9 as Regulator of Virus Trafficking and Fusion in Infections by HIV, IAV, HPV, and Lujo Virus (LUJV)
Tetraspanins not only organize plasma membrane molecules but also regulate the trafficking of cellular proteins and the transport of endocytosed viruses (11, 15, 83). Many viruses, including HIV, IAV, HPV, and LUJV, localize to CD63-positive endosomes during entry (35, 46, 55, 81, 84). CD63 is most abundant in late endosomes or multivesicular bodies (MVBs) (85) and involved in the membrane organization and trafficking of cellular transmembrane proteins that interact with viruses such as HIV-receptor component CXCR4 (83, 86, 87). Therefore, a functional involvement of CD63 in viral fusion and transport is conceivable. Cellular depletion of CD63 or treatment of cells with CD63 antibodies leads to decreased infectivity of HIV-1, HCV, LUJV, and oncogenic HPV types presenting CD63 as a more general mediator of virus infection (46, 55, 84, 88–90). In contrast to the proviral role of CD63, it is believed that CD9 and CD81 negatively regulate HIV-1 entry by interfering with the formation of the entry receptor complex (47).
Tetraspanin CD63 forms complexes with HPV16 capsid protein L1 (55). As CD63 is involved in the transport of proteins to multiple subcellular locations, it is plausible that different adaptors are required for regulating its trafficking and sorting. For example, syntenin-1 modulates trafficking of CD63 by binding to its C-terminus (91). Consequently, ultrastructural analyses demonstrated the importance of the CD63/syntenin-1 complex for HPV trafficking to MVBs, a process that is required for capsid disassembly (55). The complexity of CD63-mediated viral trafficking is highlighted by the finding that components of the cellular endosomal sorting complex required for transport (ESCRT) are also integrated into the HPV transport platform (55, 92, 93). ESCRT proteins are able to interact with both, syntenin-1 and viral proteins like the HPV16 capsid protein L2 (55, 92–97). Therefore, both viral and cytoplasmic proteins may be targeted to CD63 platforms in a virus-modulated endosomal trafficking pathway.
In addition to its role in trafficking, CD63 facilitates membrane fusion of enveloped viruses. For example, LUJV glycoprotein-mediated membrane fusion is dependent on CD63 and low pH (84), highlighting the importance of the endo/lysosomal system in cell entry. Similarly, Tspan TSPAN9 promotes membrane penetration in early endosomes by the alphaviruses Sindbis virus, Semliki Forest virus, vesicular stomatitis virus, and chikungunya virus (16, 49). Together, CD63 and TSPAN9 may modulate the endosomal compartment to be more permissive for the fusion of viral and cellular membranes.
Tspans in Virus Exit
Morphogenesis of enveloped viruses occurs on membranes of intracellular compartments or at the plasma membrane. Like virus entry, virus exit is a multi-step process driven by viral proteins. This process includes the targeting of viral proteins to specific membrane domains, local concentration of these proteins, virus budding, and release of virus particles. During these processes, Tspans are incorporated into the enveloping membrane of virions, such as HIV, feline immunodeficiency virus, canine distemper virus (CDV), HCMV, influenza, or hepatitis A virus (HAV) (98), implicating TEMs at the site of virus budding.
Earlier reports support this hypothesis using electron and fluorescence microscopy to demonstrate that the HIV core (Gag) and envelope (Env) proteins (26, 99–101), the HTLV-1 Gag protein (102, 103), the Marburgvirus matrix protein VP40 (104, 105), and influenza proteins (45) accumulate in CD9, CD63, CD81, and/or CD82 containing TEMs.
Studies investigating Tspan dynamics in virus budding have shown that the herpes simplex virus (HSV)-1 capsid protein VP26 physically interacts with Tspan7 (earlier known as CTMP-7) (106), and that formation of this complex supports viral egress. Moreover, influenza infection induced redistribution of CD81 on the plasma membrane into concentrated patches of viral budding sites which also contain different viral proteins (45). Likewise, CD63 coordinates sorting of specific viral proteins into extracellular vesicles, such as the major oncoprotein latent membrane protein 1 of the Epstein–Barr virus (107, 108). Comparable to influenza budding, HIV Gag insertion into the plasma membrane induces recruitment of CD81 and CD9 and the coalescence of different membrane microdomains (100, 101, 109–111). Co-immunoprecipitation experiments revealed that Gag proteins interact, directly or indirectly, with CD81 (100). The process of Gag accumulation in Tspan assemblies leads to the formation of larger membrane domains that extend over a few hundred nanometers (109, 112) and contain up to 2,500 tightly packed Gag molecules (113). In addition, multiple studies have shown that modulation of Tspan expression levels and redistribution via anti-Tspan antibody treatment in viral or cellular membranes interferes with different steps of the HIV and CDV life cycle including virus-to-cell fusion, reverse transcription, release, and virus-induced cell–cell fusion (114–121). Thus, Tspans can regulate, for example, viral release and cell–cell fusion by controlling the access of the required cellular machineries to the specific areas.
In addition to Tspans and viral proteins, HIV and HAV exit platforms accumulate cytoplasmic factors, such as components of the ESCRT machinery, which are required for the budding process (113, 122–127). This Tspan-mediated pre-assembly of viral and cellular proteins enables the formation of large budding platforms, a precondition for coordinated viral morphogenesis.
Conclusion
At present, the various interaction modalities between viral and cellular proteins preclude the development of a simple model for viral entry. Common molecular mechanisms in viral infection may be revealed by characterizing Tspan platforms in different systems, from their initial involvement at the plasma membrane to their roles in intracellular trafficking and viral egress (Figure 2). We hypothesize that active accumulation of molecules into Tspan platforms drives viral infection forward in a defined step-wise sequence.
Author Contributions
LF and TL wrote the manuscript and designed the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Julia R. Taylor for her critical reading of the manuscript.
Funding
The research in our laboratories is supported by grants from the German Research Foundation (DFG; FL 696/2-1, FL 696/3-1 | LA 1272/8-1).
Abbreviations
CD, cluster of differentiation; CDV, canine distemper virus; CLDN1, claudin; CoV, coronavirus; EBV, Epstein–Barr virus; ESCRT, endosomal sorting complexes required for transport; FIV, feline immunodeficiency virus, GFRs, growth factor receptors; HAV, hepatitis A virus; HCMV, human cytomegalovirus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HPV, human papillomavirus; HSV, herpes simplex virus; IAV, influenza A virus; ITGB1, integrin β1; LEL, large extracellular loop; LUJV, Lujo virus; MAPK, mitogen-activated protein kinase; MVB, multivesicular body; Ras, rat sarcoma; SR-BI, scavenger receptor type B class I; TEMs or TERMs, tetraspanin-enriched microdomains; TIRF, total internal reflection fluorescence microscopy; TM, trans-membrane; Tspan, tetraspanin; vDNA, viral DNA.
References
1. Sowers AE, Hackenbrock CR. Rate of lateral diffusion of intramembrane particles: measurement by electrophoretic displacement and rerandomization. Proc Natl Acad Sci U S A (1981) 78(10):6246–50. doi:10.1073/pnas.78.10.6246
2. Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, et al. Molecular anatomy of a trafficking organelle. Cell (2006) 127(4):831–46. doi:10.1016/j.cell.2006.10.030
3. Destainville N, Schmidt TH, Lang T. Where biology meets physics – a converging view on membrane microdomain dynamics. Curr Top Membr (2016) 77:27–65. doi:10.1016/bs.ctm.2015.10.004
4. Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol (2014) 6(8):a016972. doi:10.1101/cshperspect.a016972
5. Smith AE, Helenius A. How viruses enter animal cells. Science (2004) 304(5668):237–42. doi:10.1126/science.1094823
6. Marsh M, Helenius A. Virus entry: open sesame. Cell (2006) 124(4):729–40. doi:10.1016/j.cell.2006.02.007
7. White JM, Whittaker GR. Fusion of enveloped viruses in endosomes. Traffic (2016) 17(6):593–614. doi:10.1111/tra.12389
8. Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, et al. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics (2005) 86(6):674–84. doi:10.1016/j.ygeno.2005.08.004
9. Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci (2014) 127(Pt 17):3641–8. doi:10.1242/jcs.154906
10. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol (2005) 6(10):801–11. doi:10.1038/nrm1736
11. Berditchevski F, Rubinstein E. Tetraspanins. Dordrecht; Heidelberg; New York; London: Springer (2013). 1 p.
12. van Spriel AB, Figdor CG. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect (2010) 12(2):106–12. doi:10.1016/j.micinf.2009.11.001
13. Monk PN, Partridge LJ. Tetraspanins: gateways for infection. Infect Disord Drug Targets (2012) 12(1):4–17. doi:10.2174/187152612798994957
14. Scheffer KD, Berditchevski F, Florin L. The tetraspanin CD151 in papillomavirus infection. Viruses (2014) 6(2):893–908. doi:10.3390/v6020893
15. Fast LA, Lieber D, Lang T, Florin L. Tetraspanins in infections by human cytomegalo- and papillomaviruses. Biochem Soc Trans (2017) 45(2):489–97. doi:10.1042/BST20160295
16. Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog (2013) 9(12):e1003835. doi:10.1371/journal.ppat.1003835
17. Seigneuret M, Delaguillaumie A, Lagaudrière-Gesbert C, Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem (2001) 276(43):40055–64. doi:10.1074/jbc.M105557200
18. Stipp C, Kolesnikova T, Hemler M. Functional domains in tetraspanin proteins. Trends Biochem Sci (2003) 28(2):106–12. doi:10.1016/S0968-0004(02)00014-2
19. DeSalle R, Mares R, Garcia-España A. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol Phylogenet Evol (2010) 56(1):486–91. doi:10.1016/j.ympev.2010.02.015
20. Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys J (2006) 90(1):212–27. doi:10.1529/biophysj.105.069666
21. Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, et al. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell (2016) 167(4):1041–1051.e11. doi:10.1016/j.cell.2016.09.056
22. Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci (2001) 58(9):1189–205. doi:10.1007/PL00000933
23. Kovalenko OV, Yang X, Kolesnikova TV, Hemler ME. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem J (2004) 377(Pt 2):407–17. doi:10.1042/bj20031037
24. Schmidt TH, Homsi Y, Lang T. Oligomerization of the Tetraspanin CD81 via the Flexibility of Its δ-Loop. Biophys J (2016) 110(11):2463–74. doi:10.1016/j.bpj.2016.05.003
25. Homsi Y, Lang T. The specificity of homomeric clustering of CD81 is mediated by its δ-loop. FEBS Open Bio (2017) 7(2):274–83. doi:10.1002/2211-5463.12187
26. Nydegger S, Khurana S, Krementsov D, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol (2006) 173(5):795. doi:10.1083/jcb.200508165
27. Zuidscherwoude M, Göttfert F, Dunlock VME, Figdor CG, van den Bogaart G, van Spriel AB. The tetraspanin web revisited by super-resolution microscopy. Sci Rep (2015) 5:12201. doi:10.1038/srep12201
28. Termini CM, Cotter ML, Marjon KD, Buranda T, Lidke KA, Gillette JM. The membrane scaffold CD82 regulates cell adhesion by altering α4 integrin stability and molecular density. Mol Biol Cell (2014) 25(10):1560–73. doi:10.1091/mbc.E13-11-0660
29. Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci (2001) 114(23):4143–51.
30. Bassani S, Cingolani LA. Tetraspanins: interactions and interplay with integrins. Int J Biochem Cell Biol (2012) 44(5):703–8. doi:10.1016/j.biocel.2012.01.020
31. Termini CM, Gillette JM. Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol (2017) 5:34. doi:10.3389/fcell.2017.00034
32. Yang X, Claas C, Kraeft S-K, Chen LB, Wang Z, Kreidberg JA, et al. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell (2002) 13(3):767–81. doi:10.1091/mbc.01-05-0275
33. Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett (2002) 516(1–3):139–44. doi:10.1016/S0014-5793(02)02522-X
34. Kawakami Y, Kawakami K, Steelant WFA, Ono M, Baek RC, Handa K, et al. Tetraspanin CD9 is a “proteolipid,” and its interaction with alpha 3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin-5-dependent cell motility. J Biol Chem (2002) 277(37):34349–58. doi:10.1074/jbc.M200771200
35. Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, et al. Clathrin- and caveolin-independent entry of human papillomavirus type 16 – involvement of tetraspanin-enriched microdomains (TEMs). PLoS One (2008) 3(10):e3313. doi:10.1371/journal.pone.0003313
36. Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, et al. Tetraspanin CD151 Mediates Papillomavirus Type 16 Endocytosis. J Virol (2013) 87(6):3435–46. doi:10.1128/JVI.02906-12
37. Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem (2003) 278(43):41624–30. doi:10.1074/jbc.M305289200
38. Farquhar MJ, Harris HJ, McKeating JA. Hepatitis C virus entry and the tetraspanin CD81. Biochem Soc Trans (2011) 39(2):532–6. doi:10.1042/BST0390532
39. Zhu Y-Z, Luo Y, Cao M-M, Liu Y, Liu X-Q, Wang W, et al. Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology (2012) 429(2):112–23. doi:10.1016/j.virol.2012.03.002
40. Fénéant L, Levy S, Cocquerel L. CD81 and hepatitis C virus (HCV) infection. Viruses (2014) 6(2):535–72. doi:10.3390/v6020535
41. Grigorov B, Reungoat E, Gentil Dit Maurin A, Varbanov M, Blaising J, Michelet M, et al. Hepatitis C virus infection propagates through interactions between Syndecan-1 and CD81 and impacts the hepatocyte glycocalyx. Cell Microbiol (2017) 19(5):e12711. doi:10.1111/cmi.12711
42. Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science (1998) 282(5390):938–41. doi:10.1126/science.282.5390.938
43. Earnest JT, Hantak MP, Park J-E, Gallagher T. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J Virol (2015) 89(11):6093–104. doi:10.1128/JVI.00543-15
44. Earnest JT, Hantak MP, Li K, McCray PB, Perlman S, Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog (2017) 13(7):e1006546. doi:10.1371/journal.ppat.1006546
45. He J, Sun E, Bujny MV, Kim D, Davidson MW, Zhuang X. Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog (2013) 9(10):e1003701. doi:10.1371/journal.ppat.1003701
46. Fu E, Pan L, Xie Y, Mu D, Liu W, Jin F, et al. Tetraspanin CD63 is a regulator of HIV-1 replication. Int J Clin Exp Pathol (2015) 8(2):1184–98.
47. Gordón-Alonso M, Yañez-Mó M, Barreiro O, Alvarez S, Muñoz-Fernández MA, Valenzuela-Fernández A, et al. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol (2006) 177(8):5129–37. doi:10.4049/jimmunol.177.8.5129
48. Hochdorfer D, Florin L, Sinzger C, Lieber D. Tetraspanin CD151 promotes initial events in human cytomegalovirus infection. J Virol (2016) 90(14):6430–42. doi:10.1128/JVI.00145-16
49. Stiles KM, Kielian M. Role of TSPAN9 in alphavirus entry and early endosomes. J Virol (2016) 90(9):4289–97. doi:10.1128/JVI.00018-16
50. Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, et al. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol (2013) 87(11):6062–72. doi:10.1128/JVI.00330-13
51. Day PM, Schelhaas M. Concepts of papillomavirus entry into host cells. Curr Opin Virol (2014) 4:24–31. doi:10.1016/j.coviro.2013.11.002
52. Florin L, Sapp M, Spoden GA. Host-cell factors involved in papillomavirus entry. Med Microbiol Immunol (2012) 201(4):437–48. doi:10.1007/s00430-012-0270-1
53. Bienkowska-Haba M, Sapp M. The Cytoskeleton in Papillomavirus Infection. Viruses (2011) 3(3):260–71. doi:10.3390/v3030260
54. Spoden G, Kühling L, Cordes N, Frenzel B, Sapp M, Boller K, et al. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol (2013) 87(13):7765–73. doi:10.1128/JVI.00370-13
55. Gräßel L, Fast LA, Scheffer KD, Boukhallouk F, Spoden GA, Tenzer S, et al. The CD63-Syntenin-1 complex controls post-endocytic trafficking of oncogenic human papillomaviruses. Sci Rep (2016) 6:32337. doi:10.1038/srep32337
56. Homsi Y, Schloetel J-G, Scheffer KD, Schmidt TH, Destainville N, Florin L, et al. The extracellular δ-domain is essential for the formation of CD81 tetraspanin webs. Biophys J (2014) 107(1):100–13. doi:10.1016/j.bpj.2014.05.028
57. Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol (1997) 71(3):2449–56.
58. McMillan N, Payne E, Frazer I, Evander M. Expression of the [alpha] 6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology (1999) 261(2):271–9. doi:10.1006/viro.1999.9825
59. Yoon C, Kim K, Park S, Cheong S. [alpha] 6 Integrin is the main receptor of human papillomavirus type 16 VLP. Biochem Biophys Res Commun (2001) 283(3):668–73. doi:10.1006/bbrc.2001.4838
60. Surviladze Z, Dziduszko A, Ozbun MA. Essential roles for soluble virion-associated heparan sulfonated proteoglycans and growth factors in human papillomavirus infections. PLoS Pathog (2012) 8(2):e1002519. doi:10.1371/journal.ppat.1002519
61. Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, et al. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One (2012) 7(8):e43519. doi:10.1371/journal.pone.0043519
62. Dziduszko A, Ozbun MA. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol (2013) 87(13):7502–15. doi:10.1128/JVI.00519-13
63. Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans (2011) 39(6):1667–73. doi:10.1042/BST20110745
64. Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) (2005) 20(4):218–24. doi:10.1152/physiol.00015.2005
65. Hemler ME. Specific tetraspanin functions. J Cell Biol (2001) 155(7):1103–7. doi:10.1083/jcb.200108061
66. Kabanova A, Marcandalli J, Zhou T, Bianchi S, Baxa U, Tsybovsky Y, et al. Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat Microbiol (2016) 1(8):16082. doi:10.1038/nmicrobiol.2016.82
67. Hetzenecker S, Helenius A, Krzyzaniak MA. HCMV induces macropinocytosis for host cell entry in fibroblasts. Traffic (2016) 17(4):351–68. doi:10.1111/tra.12355
68. Haspot F, Lavault A, Sinzger C, Sampaio KL, Stierhof Y-D, Pilet P, et al. Human cytomegalovirus entry into dendritic cells occurs via a macropinocytosis-like pathway in a pH-independent and cholesterol-dependent manner. PLoS One (2012) 7(4):e34795. doi:10.1371/journal.pone.0034795
69. Li Q, Fischer E, Cohen JI. Cell surface THY-1 contributes to human cytomegalovirus entry via a macropinocytosis-like process. J Virol (2016) 90(21):9766–81. doi:10.1128/JVI.01092-16
70. Bonander N, Jamshad M, Hu K, Farquhar MJ, Stamataki Z, Balfe P, et al. Structural characterization of CD81-Claudin-1 hepatitis C virus receptor complexes. Biochem Soc Trans (2011) 39(2):537–40. doi:10.1042/BST0390537
71. Zona L, Tawar RG, Zeisel MB, Xiao F, Schuster C, Lupberger J, et al. CD81-receptor associations – impact for hepatitis C virus entry and antiviral therapies. Viruses (2014) 6(2):875–92. doi:10.3390/v6020875
72. Schäfer G, Blumenthal MJ, Katz AA. Interaction of human tumor viruses with host cell surface receptors and cell entry. Viruses (2015) 7(5):2592–617. doi:10.3390/v7052592
73. Gerold G, Meissner F, Bruening J, Welsch K, Perin PM, Baumert TF, et al. Quantitative proteomics identifies serum response factor binding protein 1 as a host factor for hepatitis C virus entry. Cell Rep (2015) 12(5):864–78. doi:10.1016/j.celrep.2015.06.063
74. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med (2011) 17(5):589–95. doi:10.1038/nm.2341
75. Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol (2008) 82(10):5007–20. doi:10.1128/JVI.02286-07
76. Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, et al. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol (2012) 86(20):10935–49. doi:10.1128/JVI.00750-12
77. Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe (2013) 13(3):302–13. doi:10.1016/j.chom.2013.02.006
78. Zheng K, Kitazato K, Wang Y. Viruses exploit the function of epidermal growth factor receptor. Rev Med Virol (2014) 24(4):274–86. doi:10.1002/rmv.1796
79. Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol (2009) 7(6):439–50. doi:10.1038/nrmicro2147
80. König R, Stertz S, Zhou Y, Inoue A, Hoffmann H-H, Bhattacharyya S, et al. Human host factors required for influenza virus replication. Nature (2010) 463(7282):813–7. doi:10.1038/nature08699
81. Karlas A, Machuy N, Shin Y, Pleissner K-P, Artarini A, Heuer D, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature (2010) 463(7282):818–22. doi:10.1038/nature08760
82. Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol (2015) 89(4):1954–64. doi:10.1128/JVI.02615-14
83. Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic (2007) 8(2):89–96. doi:10.1111/j.1600-0854.2006.00515.x
84. Raaben M, Jae LT, Herbert AS, Kuehne AI, Stubbs SH, Chou Y-Y, et al. NRP2 and CD63 are host factors for lujo virus cell entry. Cell Host Microbe (2017) 22(5):688–96.e5. doi:10.1016/j.chom.2017.10.002
85. Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res (2009) 315(9):1584–92. doi:10.1016/j.yexcr.2008.09.020
86. Schulze U, Brast S, Grabner A, Albiker C, Snieder B, Holle S, et al. Tetraspanin CD63 controls basolateral sorting of organic cation transporter 2 in renal proximal tubules. FASEB J (2016) 31(4):1421–33. doi:10.1096/fj.201600901R
87. Yoshida T, Kawano Y, Sato K, Ando Y, Aoki J, Miura Y, et al. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic (2008) 9(4):540–58. doi:10.1111/j.1600-0854.2008.00700.x
88. Park JH, Park S, Yang J-S, Kwon OS, Kim S, Jang SK. Discovery of cellular proteins required for the early steps of HCV infection using integrative genomics. PLoS One (2013) 8(4):e60333. doi:10.1371/journal.pone.0060333
89. Li G, Endsley MA, Somasunderam A, Gbota SL, Mbaka MI, Murray JL, et al. The dual role of tetraspanin CD63 in HIV-1 replication. Virol J (2014) 11(1):23. doi:10.1186/1743-422X-11-23
90. Lindern von J, Rojo D, Grovit-Ferbas K, Yeramian C, Deng C, Herbein G, et al. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J Virol (2003) 77(6):3624–33. doi:10.1128/JVI.77.6.3624-3633.2003
91. Latysheva N, Muratov G, Rajesh S, Padgett M, Hotchin NA, Overduin M, et al. Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol Cell Biol (2006) 26(20):7707–18. doi:10.1128/MCB.00849-06
92. Broniarczyk J, Bergant M, Goździcka-Józefiak A, Banks L. Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology (2014) 460-461:83–90. doi:10.1016/j.virol.2014.05.005
93. Broniarczyk J, Pim D, Massimi P, Bergant M, Goździcka-Józefiak A, Crump C, et al. The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Sci Rep (2017) 28(7):45159. doi:10.1038/srep45159
94. Hurley JH, Boura E, Carlson L-A, Różycki B. Membrane budding. Cell (2010) 143(6):875–87. doi:10.1016/j.cell.2010.11.030
95. Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nature (2012) 14(7):677–85. doi:10.1038/ncb2502
96. Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun (2014) 18:5. doi:10.1038/ncomms4477
97. Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol (2014) 24(1):19–25. doi:10.1016/j.tcb.2013.10.009
98. Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol (2003) 162(3):443–55. doi:10.1083/jcb.200304008
99. Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol (2007) 81(15):7873–84. doi:10.1128/JVI.01845-06
100. Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, et al. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology (2009) 6(1):28. doi:10.1186/1742-4690-6-28
101. Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell (2010) 102(6):335–50. doi:10.1042/BC20090165
102. Mazurov D, Heidecker G, Derse D. HTLV-1 Gag protein associates with CD82 tetraspanin microdomains at the plasma membrane. Virology (2006) 346(1):194–204. doi:10.1016/j.virol.2005.10.033
103. Mazurov D, Heidecker G, Derse D. The inner loop of tetraspanins CD82 and CD81 mediates interactions with human T cell lymphotrophic virus type 1 Gag protein. J Biol Chem (2007) 282(6):3896–903. doi:10.1074/jbc.M607322200
104. Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol (2007) 9(4):939–51. doi:10.1111/j.1462-5822.2006.00842.x
105. Makino A, Yamayoshi S, Shinya K, Noda T, Kawaoka Y. Identification of amino acids in Marburg virus VP40 that are important for virus-like particle budding. J Infect Dis (2011) 204(Suppl 3):S871–7. doi:10.1093/infdis/jir309
106. Wang L, Liu L, Che Y, Wang L, Jiang L, Dong C, et al. Egress of HSV-1 capsid requires the interaction of VP26 and a cellular tetraspanin membrane protein. Virol J (2010) 7(1):156. doi:10.1186/1743-422X-7-156
107. Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, et al. CD63 regulates Epstein-Barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-κB signaling. J Virol (2017) 91(5):e02251-16. doi:10.1128/JVI.02251-16
108. Hurwitz SN, Cheerathodi MR, Nkosi D, York SB, Meckes DG. Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of Epstein-Barr virus LMP1. J Virol (2018) 92(5):e01969-17. doi:10.1128/JVI.01969-17
109. Krementsov DN, Rassam P, Margeat E, Roy NH, Schneider-Schaulies J, Milhiet P-E, et al. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic (2010) 11(11):1401–14. doi:10.1111/j.1600-0854.2010.01111.x
110. Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J Virol (2011) 85(19):9749–66. doi:10.1128/JVI.00743-11
111. Mariani C, Desdouits M, Favard C, Benaroch P, Muriaux DM. Role of Gag and lipids during HIV-1 assembly in CD4(+) T cells and macrophages. Front Microbiol (2014) 5:312. doi:10.3389/fmicb.2014.00312
112. Nydegger S, Foti M, Derdowski A, Spearman P, Thali M. HIV-1 egress is gated through late endosomal membranes. Traffic (2003) 4(12):902–10. doi:10.1046/j.1600-0854.2003.00145.x
113. Mücksch F, Laketa V, Müller B, Schultz C, Kräusslich H-G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife (2017) 6:e25287. doi:10.7554/eLife.25287
114. Krementsov DN, Weng J, Lambelé M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology (2009) 6:64. doi:10.1186/1742-4690-6-64
115. Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, et al. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol (2008) 82(2):1021–33. doi:10.1128/JVI.01044-07
116. Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol (2009) 83(15):7467–74. doi:10.1128/JVI.00163-09
117. Schmid E, Zurbriggen A, Gassen U, Rima B, Meulen ter V, Schneider-Schaulies J. Antibodies to CD9, a tetraspan transmembrane protein, inhibit canine distemper virus-induced cell-cell fusion but not virus-cell fusion. J Virol (2000) 74(16):7554–61. doi:10.1128/JVI.74.16.7554-7561.2000
118. Singethan K, Topfstedt E, Schubert S, Duprex WP, Rima BK, Schneider-Schaulies J. CD9-dependent regulation of Canine distemper virus-induced cell-cell fusion segregates with the extracellular domain of the haemagglutinin. J Gen Virol (2006) 87(Pt 6):1635–42. doi:10.1099/vir.0.81629-0
119. Singethan K, Müller N, Schubert S, Lüttge D, Krementsov DN, Khurana SR, et al. CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic (2008) 9(6):924–35. doi:10.1111/j.1600-0854.2008.00737.x
120. Rocha-Perugini V, Suárez H, Alvarez S, López-Martín S, Lenzi GM, Vences-Catalán F, et al. CD81 association with SAMHD1 enhances HIV-1 reverse transcription by increasing dNTP levels. Nat Microbiol (2017) 2(11):1513–22. doi:10.1038/s41564-017-0019-0
121. Ménager MM, Littman DR. Actin dynamics regulates dendritic cell-mediated transfer of HIV-1 to T cells. Cell (2016) 164(4):695–709. doi:10.1016/j.cell.2015.12.036
122. Ono A, Freed E. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol (2004) 78(3):1552–63. doi:10.1128/JVI.78.3.1552-1563.2004
123. Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol (2006) 80(18):9039–52. doi:10.1128/JVI.01013-06
124. Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in “ESCRTing” HIV-1. Nat Rev Microbiol (2007) 5(12):912–6. doi:10.1038/nrmicro1790
125. Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med (2012) 2(7):a006924. doi:10.1101/cshperspect.a006924
126. McKnight KL, Xie L, González-López O, Rivera-Serrano EE, Chen X, Lemon SM. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci U S A (2017) 114(25):6587–92. doi:10.1073/pnas.1619519114
Keywords: tetraspanin, microdomain, virus, entry, endocytosis, trafficking, budding, receptor
Citation: Florin L and Lang T (2018) Tetraspanin Assemblies in Virus Infection. Front. Immunol. 9:1140. doi: 10.3389/fimmu.2018.01140
Received: 26 February 2018; Accepted: 07 May 2018;
Published: 25 May 2018
Edited by:
María Yáñez-Mó, Universidad Autonoma de Madrid, SpainReviewed by:
Markus Thali, University of Vermont, United StatesStephanie Charrin, INSERM UMR935 Modèles de cellules souches malignes et thérapeutiques, France (Emily Whitaker and Menenlaos Symeonides contributed to the review of Markus Thali)
Copyright: © 2018 Florin and Lang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luise Florin, lflorin@uni-mainz.de
 Luise Florin
Luise Florin Thorsten Lang
Thorsten Lang