- 1Unité de Bactériologie Expérimentale, Institut Pasteur Dakar, Dakar, Sénégal
- 2Laboratoire de Microbiologie Fondamentale et Appliquée, Faculté de Médecine et Pharmacie et d’Odontologie, Université Cheikh Anta Diop, Dakar, Sénégal
- 3Laboratoire de Biologie Médicale, Institut Pasteur de Dakar, Dakar, Sénégal
- 4UMR IRD 224 – CNRS 5290 – Université de Montpellier – MIGEVEC, Centre IRD de Montpellier, Montpellier, France
- 5Laboratoire de Bactériologie, Centre Hospitalier Universitaire National de Fann, Dakar, Sénégal
- 6Département de Génie Chimique et Biologie Appliquée, Ecole Supérieure Polytechnique, Université Cheikh Anta Diop, Dakar, Sénégal
Vibrio cholerae O1 is the causative agent of cholera with classical and El Tor, two well-established biotypes. In last 20 years, hybrid strains of classical and El Tor and variant El Tor which carry classical ctxB have emerged worldwide. In 2004–2005, Senegal experienced major cholera epidemic with a number of cases totalling more than 31719 with approximately 458 fatal outcomes (CFR, 1.44%). In this retrospective study, fifty isolates out of a total of 403 V. cholerae biotype El Tor serovar Ogawa isolates from all areas in Senegal during the 2004–2005 cholera outbreak were randomly selected. Isolates were characterized using phenotypic and genotypic methods. The analysis of antibiotic resistance patterns revealed the predominance of the S-Su-TCY-Tsu phenotype (90% of isolates). The molecular characterization of antibiotic resistance revealed the presence of the SXT element, a self-transmissible chromosomally integrating element in all isolates. Most of V. cholerae isolates had an intact virulence cassette (86%) (ctx, zot, ace genes). All isolates tested gave amplification with primers for classical CT, and 10/50 (20%) of isolates carried classical and El Tor ctxB. The study reveals the presence of atypical V. cholerae O1 El Tor during cholera outbreak in Senegal in 2004–2005.
Introduction
Cholera is an epidemic diarrheal disease caused by toxigenic Vibrio cholerae, serogroup O1 or O139. There are two biotypes in the serogroup O1, classical and El Tor. The seventh pandemic of cholera were due to V. cholerae O1, biotype El Tor, began in Celebes (Islands) in 1961 and spread in West African countries in the early 1970s while the fifth and the sixth pandemics of cholera were caused by the classical biotype (Kaper et al., 1995). For over a decade, Africa has been the continent most affected by cholera in terms of the number of individuals infected and the frequency of outbreaks recorded1. In 2004–2005, cholera outbreaks occurred in eight countries in Africa with 125,082 cases and 2,230 deaths, CFR 1.78 (WHO, 2006). However, in some areas, the CFR exceeded 10%. In Senegal, the cholera outbreak has caused 31,719 cases with approximately 458 deaths (CFR, 1.44%) and the most affected regions were Diourbel and Dakar; the index case was a young Guinean living in a populous district in Dakar (Manga et al., 2008). Smaller scale epidemics have been reported from 2006 to 2008. However, no cases have been reported since July 2010, this is probably due to the monitoring implemented by the Senegalese Ministry for Health (Global Task Force on Cholera Control, 2006).
The pathogenicity of V. cholerae O1 and O139 isolates depends on a combination of factors including the coordinated expression of virulence factors, and the secretion of cholera toxin (CT). Molecular analysis of V. cholerae revealed the presence of two genetic elements in the genome of pathogenic strains: the lysogenic bacteriophage (CTXø), which hosts at least six genes including toxin genes: (ctxAB, ace, and zot encoding, respectively A and B subunits CT, accessory enterotoxin, and zonula occludens toxin), and the Vibrio cholerae pathogenicity island (VPI), which carries genes for the pilus colonization factor, toxin coregulated pilus (TCP) (Pearson et al., 1993; Waldor et al., 1997). Strains named “atypical El Tor” have traits of both classical and El Tor (Nair et al., 2002). Recently, several atypical El Tor strains have been reported, including Matlab variants (Safa et al., 2006), Mozambique variants (Ansaruzzaman et al., 2004), altered El Tor (Nair et al., 2006), and hybrid El Tor strains, harboring the classical CT allele ctxB1 (Goel et al., 2008; Safa et al., 2008). Safa et al. (2010) proposed the term “atypical El Tor” for all V. cholerae O1 El Tor that harbor classical traits.
Vibrio cholerae O1 strains isolated in Africa are known to be resistant to many antibiotics (Ceccarelli et al., 2006; Quilici et al., 2010). Drug-resistance of bacteria is mainly linked to the mobilization and the dissemination of resistance genes through genetic determinants such as plasmids, integrons, and transposons. The SXT element is a self-transmissible mobile genetic element belonging to the family of integrating and conjugative elements (ICEs) that originally was discovered in a V. cholerae O139 isolate from India (MO10) which is resistant to streptomycin (Sm), trimethoprim (Tm), sulfamethoxazole (Su), and chloramphenicol (Wozniak et al., 2009; Ceccarelli et al., 2011b). The SXT element is always integrated into the 5′ end of the chromosomal gene prfC and able to replicate with the host chromosome (Waldor et al., 1997). Capture and spread of antibiotic resistance determinants by integrons is an effective route of antimicrobial resistance dissemination among Gram-negative bacteria (Mazel, 2006). Several of integrons have been described based on integrase gene. The class 1 integron is widely spread among V. cholerae isolates with various types of resistance gene cassettes (Dalsgaard et al., 2001; Ceccarelli et al., 2006). Class 4 integron named superintegrons is a component SXT element found in several bacteria in particular in V. cholerae. Superintegrons are the ancestors of multiresistant integrons (Mazel, 2006). Integrons and ICEs have been found in V. cholerae isolated in Mozambique, Iran, and India and they have largely contributed to the spread of antibiotic resistance (Amita et al., 2003; Adabi et al., 2009; Pugliese et al., 2009).
In Senegal, there is little data available on the genetic determinants of virulence and antibiotic-resistance among epidemic V. cholerae isolates. Despite the description of multiresistant isolates during the lastest cholera outbreaks in Senegal in 1994 (Aidara et al., 1998) and in 2004–2006 (Manga et al., 2008), the molecular mechanisms of antibiotic resistance have never been studied.
The objective of this work was to characterize the genetic determinants of virulence and antibiotic-resistance in V. cholerae O1 isolated during the latest cholera outbreak in Senegal.
Materials and Methods
Bacterial Isolates
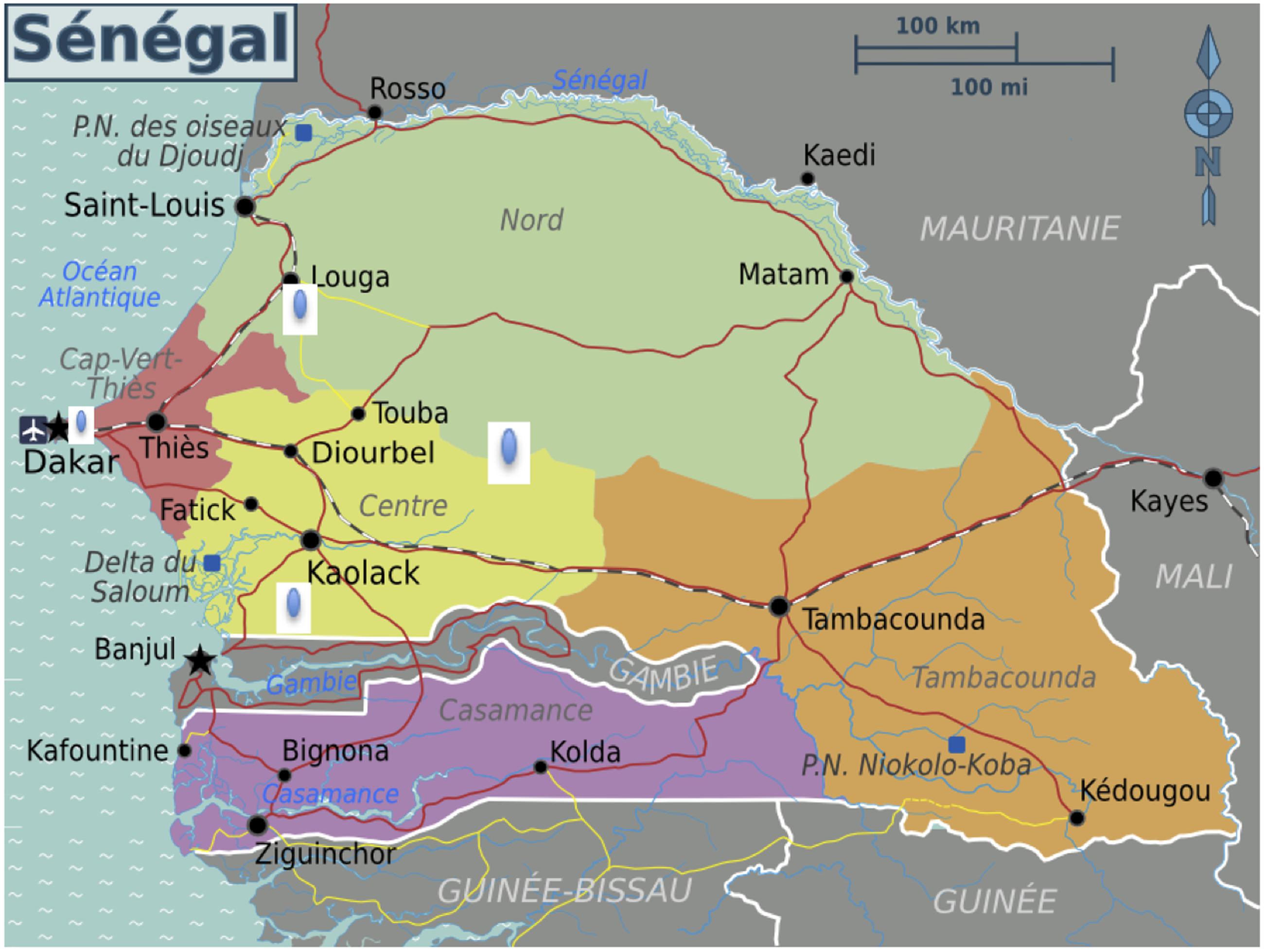
A total of 403 V. cholerae serogroup O1 strains were isolated in different areas in Senegal between November 2004 to May 2005 from patients with acute diarrhea. A sampling of fifty isolates were randomly selected to represent the most affected population: Dakar (39); Diourbel (09); Kaolack (01); Louga (01) (Figure 1).
All isolates were identified with the API 20E (Biomérieux, Marcy l’Etoile, France) and serotyped using anti-Ogawa, anti-Inaba antisera.
Susceptibility to Antibiotics
Antimicrobial susceptibility testing was performed using the Kirby Bauer disk diffusion method on Müller Hinton agar. The following antibiotics were tested: ampicillin (AM, 10 μg), amoxicillin-clavulanic acid (AMC, 20 μg/10 μg), cefotaxime (CTX, 30 μg), streptomycin (S, 10 μg), tetracycline (TCY, 30 UI), chloramphenicol (CHL, 30 μg), nalidixic acid (NA, 30 μg), pefloxacin (5 μg), trimethoprim (T, 5 μg), sulfamethoxazole (Su, 200 μg), trimethoprim-sulfamethoxazole (TSu, 1.25 μg/23.25 μg). The diameter of inhibition zones was interpreted following the CLSI recommendations for enterobacteria2.
DNA Extraction
Total DNA was obtained by using QIAamp DNA Mini Kit (Qiagen SA, Courtaboeuf, France).
PCR Amplifications
Detection of Antibiotic Resistance and Virulence Molecular Markers
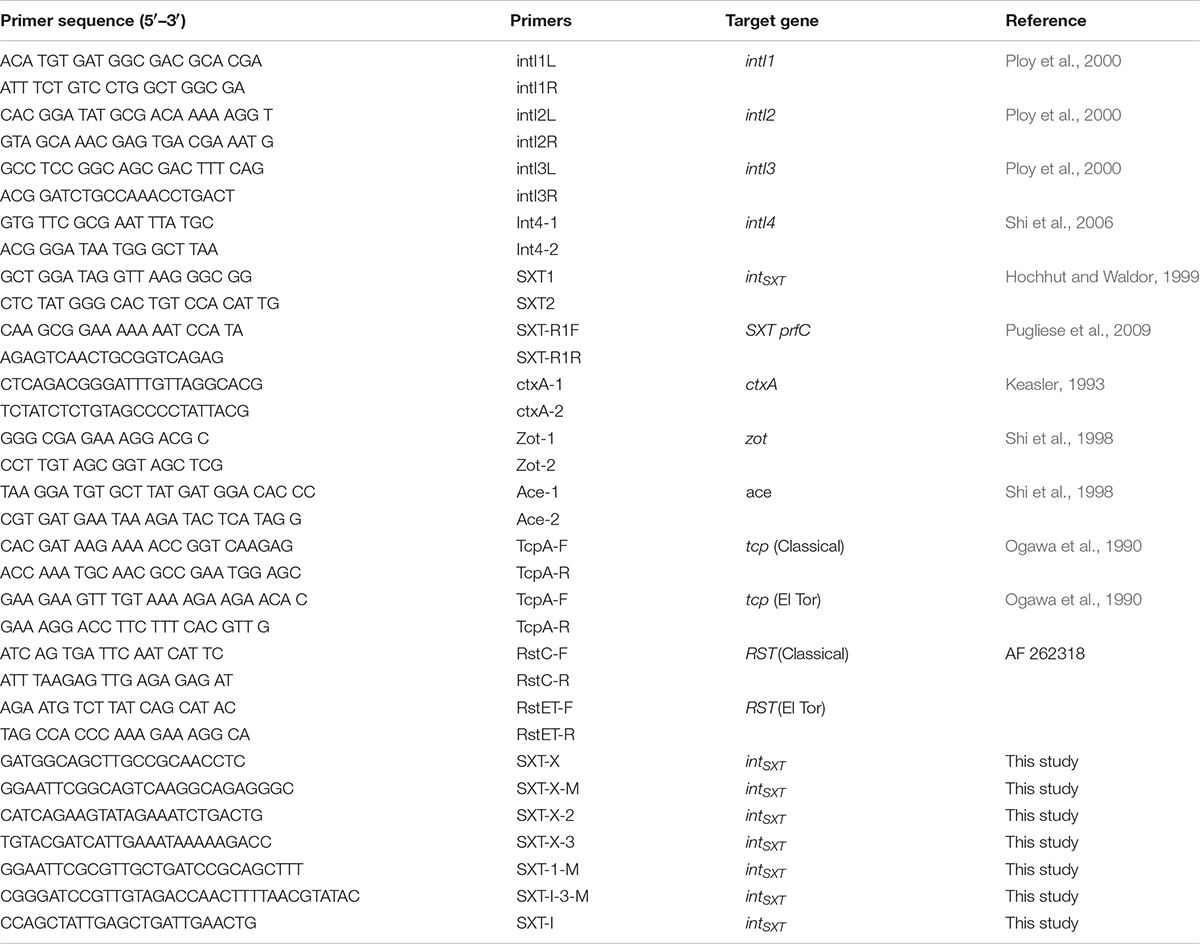
PCR analysis was performed for detection of genetic determinants of antibiotic resistance and virulence. Amplification was carried out and virulence with primers described elsewhere (Ploy et al., 2000; Hochhut et al., 2001; Shi et al., 2006), and GenBank accession number AF 099172. For the detection of virulence genes, primers used as those described by Ogawa et al. (1990), Keasler (1993), and Shi et al. (1998) (Table 1) and GenBank accession numbers (AF262318, GQ485654). Chromosomal integration was detected by amplification of the right SXT element chromosome junction (attP-prfC gene sequence) (Table 1).
All amplified DNA fragments were resolved by conventional electrophoresis in 1% agarose gel, stained with ethidium bromide and visualized under UV light.
ctxB Typing by MAMA PCR
Mismatch Amplification Mutation Assay (MAMA) based PCR was performed to detect the presence of ctxB classical and or El Tor biotype V. cholerae O1 isolates, using specific primers described elsewhere (Morita et al., 2008).
Cloning and Sequencing
The integrase SXT fragment (intSXT) was purified with the QIAquick kit (Qiagen SA, Courtaboeuf, France), and cloned with the pGEMT vector (Promega, Madison, WI, USA), transformed into XL1-Blue competent cells (Stratagene, Garden Grove, CA, USA).
The insert of the recombinant plasmid was sequenced with dye terminator on ABI Prism automatic sequencer as described by the manufacturers. The sequences were analyzed by nucleotide BLAST search at the National Center for Biotechnology Information (NCBI) website3. Primers were used for cloning and sequencing are listed on Table 1.
Conjugation Experiments
Conjugation experiments were used to transfer resistance determinants from V. cholerae O1 isolates into nalidixic acid-resistant E. coli C1 strain. Mating experiments were carried out by mixing volumes of Luria Bertani (LB) broth in a ratio 2:1 overnight culture of donor and recipient strains. The cultures were transferred to LB agar plates containing trimethoprim (32 μg/ml), sulfamethoxazole (160 μg/ml), and nalidixic acid (50 μg/ml). To confirm the transfer of antibiotic resistance genes, transconjugants were tested for sensitivity to antibiotic and by PCR.
This study was carried out in accordance with the recommendations of Senegalese Ethical Committee, with informed consent from all subjects. The isolates used in this study were taken for the purposes of research, and the protocol was approved by the Senegalese Ethical Committee (N°0046/MSAS/DRPS/CNERS)4.
Results
Antimicrobial Susceptibility
In this retrospective study, we show that isolates were resistant to at least four antibiotics including streptomycin (S), trimethoprim (T), sulfamethoxazole (Su), trimethoprim-sulfamethoxazole (Tsu). The analysis of antibiotic resistance patterns reveals three phenotypes: S-Su-T-TSu (90%), S-Su-T-TSu-C (8%), S-Su-T-TSu-AM (2%). All isolates were resistant to trimethoprim-sulfamethoxazole but susceptible to tetracyclines.
Detection of Genetic Determinants of Antimicrobial Resistance: Detection of Integrons, Resistance Genes, and SXT Element
All isolates were negative for class 1, 2, and 3 integrons. A 900 bp PCR product of the intI4 gene was obtained for all isolates.
The amplification of SXT integrase revealed an amplicon size of 3 kb in all isolates, different from the expected size, i.e., 592 bp. The SXT integrase from senegalese isolates was identical to a fragment of V. cholerae KN14, isolated in Kenya GenBank accession number (AB535680). The integration of the SXT element in the chromosome was highlighted by the amplification of the right SXT element-chromosome junction (attP-prfC) which producted a 785 bp PCR product.
To identify genes hosted by the ICE circulating in Senegal, PCR analysis revealed the presence of the following antibiotic resistances genes (sulI, floR, strA, and dfrA1), except dfr18 gene.
Detection of Virulence Markers
The ctxB, zot, ace genes in the CTX element were present, respectively, in 98, 92, and 88% of isolates. All isolates gave positive results for tcpA (classical and El Tor) (Figure 2) and for bacteriophage rstR repressor gene of the El Tor and classical types. The detection of virulence genes revealed the presence of the genome of filamentous bacteriophage CTXø.
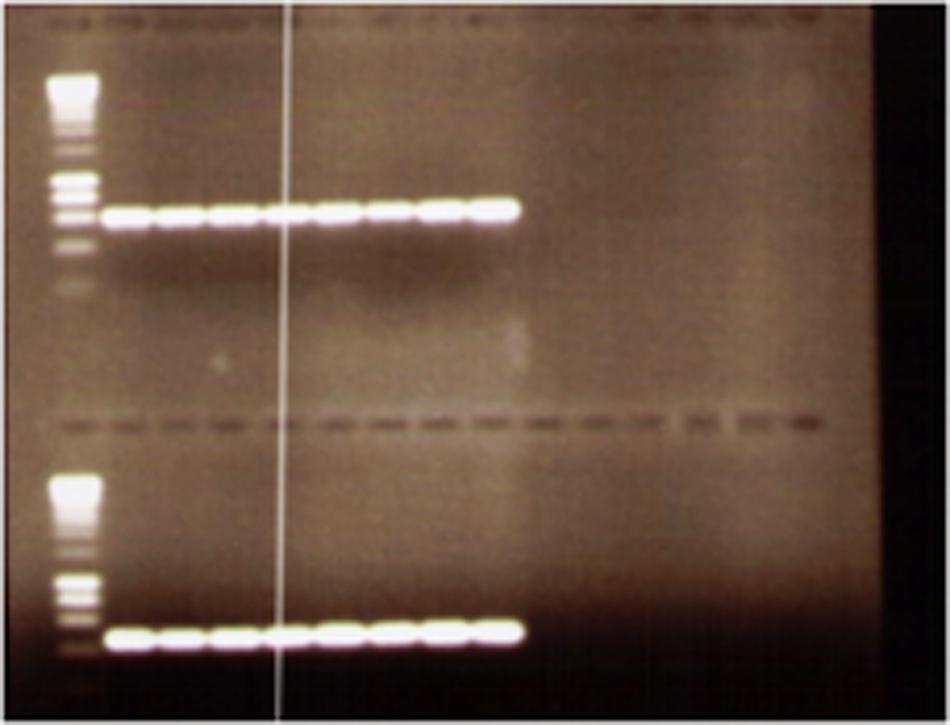
FIGURE 2. Agarose gel electrophoresis of examples of PCR products of Vibrio cholerae O1 isolates using tcpA (classical and El Tor).
ctxB Typing
All isolates tested gave amplification with primers for classical CT allele ctxB1, and 10/50 (20%) of isolates gave amplicons with primers specific for classical and El Tor CT allele ctxB1.
Conjugation Experiments
Mating experiments revealed transfer of the resistance determinants to chloramphenicol, streptomycin, sulfamethoxazole, and trimethoprim by conjugation. To identify a possible ICE-mediated resistance, transconjugants were tested for SXT-related elements. Antimicrobial susceptibility testing of V. cholerae transconjugants showed that resistance profiles were expressed by each of the transconjugants. The PCRs for detection of the intSXT element-integrase gene gave an amplicon of the same size of 3 Kbp.
Discussion
Senegalese isolates are still susceptible to quinolones while resistance to nalidixic acid or reduced sensitivity to a fluoroquinolone was described in Africa and India (Quilici et al., 2010; Ismail et al., 2013; Kutar et al., 2013).
Our results showed that class 1, 2, and 3 integrons were not involved in the spread of resistance among Senegalese V. cholerae O1 isolates, even though they have been detected in Mozambican V. cholerae O1 isolates and other Gram-negative enteric bacteria in Senegal (Hochhut and Waldor, 1999; Dalsgaard et al., 2001; Gassama et al., 2004; Gassama-Sow et al., 2006). According to Mazel (2006), the IntI4 gene associated with superintegron is characterized by a large number of gene cassettes, closely associated with genome evolution rather than the capture of antibiotic resistance.
All genes (sulI, floR, strA, and dfrA1) excepted dfrA18 gene were found in our isolates suggesting that the SXT isolated in Senegal is closely related to the SXTET. Kenyan isolates also lacked the dfrA18 gene (Kiiru et al., 2009). Since the emergence of SXT in V. cholerae O139, several studies on V. cholerae O1 have found this ICE as responsible for the dissemination of antibiotic resistance in Africa and Asia (Dalsgaard et al., 2001; Amita et al., 2003; Opintan et al., 2008; Adabi et al., 2009; Pugliese et al., 2009). ICEs of the SXT/R391 family are usually found in atypical O1 El Tor V. cholerae epidemic strains; they confer a narrow antibiotic resistance profile (Wozniak et al., 2009). Conjugation experiments revealed that although the isolates carry the SXT/R391-like elements which confers resistance to streptomycin, trimethoprim sulfamethoxazole and chloramphenicol, they lack multiple resistant integrons.
Further studies are needed to characterize and completely sequenced the SXT-related ICE in senegalese isolates.
Vibrio cholerae O1 strains isolated during the lastest outbreak (2004–2005) in Senegal were “atypical” as appointed by Safa et al. The presence of these atypical isolates may explain the disease severity After 2001, atypical V. cholerae O1 strains have emerged in India and spread worldwide, particularly in Africa. Indeed, atypical V. cholerae O1 strains were described in Mozambique (B33) (Ansaruzzaman et al., 2004), and Angola (Ceccarelli et al., 2011a). The appereance of atypical strains in Senegal is enigmatic, and suggest that probably these new strains followed the same West African path used by cholera to enter Africa in the early 1970s. The presence of intsxt identical to a fragment of V. cholerae KN14 could confirm this hypothesis. The global replacement of El Tor prototype by atypical strains indicates the evolution of V. cholerae O1. Our study revealed that atypical strains are also in the process of replacing El Tor strains; this phenonenon has been described in Eastern Africa (Ceccarelli et al., 2011a). This global replacement is believed to be due to unknown environmental factors and phages contribution (Faruque and Mekalanos, 2012).
Conclusion
Based to our results, atypical V. cholerae O1 El Tor strains were responsible for cholera outbreak in Senegal in 2004–2005.
According to our study integrons were not involved in the spread of resistance among senegalese isolates of V. cholerae O1 even though they have been detected in other Gram-negative enteric bacteria in Senegal. However, the detection of the SXT element in all isolates, irrespective of their resistance phenotypes, could have a clinical significance and should be monitored to avoid dissemination in other bacteria. The understanding of the basis of antimicrobial resistance patterns could inform guidelines for empirical treatment to reduce injudicious antimicrobial use. Further studies should be conducted to characterize the SXT element identified in Senegalese V. cholerae isolates. The genetic changes occurred in V. cholerae O1 El Tor strains need to be monitored to prevent severe cholera outbreaks in Africa.
Author Contributions
BS-B participated in the molecular genetic studies and drafted the manuscript. MD carried out the molecular genetic studies. AW participated in the molecular genetic studies. AS participated on the identification of isolates. GC helped in drafting the manuscript. AIS helped on the collection and the identification of isolates in all sites. CB participated in the design and coordination of the study. AG-S designed the study, and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grant from Institut Pasteur Paris and Pasteur Institute in Dakar. We thank the Research Unit « Unité postulante Plasticité du Génome Bactérien », Institut Pasteur de Paris, particularly Didier Mazel and Anne-Marie Guérout for the sequencing.
Footnotes
- ^ http://www.who.int/wer
- ^ http://www.clsi.org/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ www.healthresearchweb.org/fr/senegal/ethics_1057
References
Adabi, M., Bakhshi, B., Goudarzi, H., Zahraei, S. M., and Pourshafie, M. R. (2009). Distribution of class I integron and sulfamethoxazole trimethoprim constin in Vibrio cholerae isolated from patients in Iran. Microb. Drug Resist. 15, 179–184. doi: 10.1089/mdr.2009.0885
Aidara, A., Koblavi, S., Boye, C. S., Raphenon, G., Gassama, A., Grimont, F., et al. (1998). Phenotypic and genotypic characterization of Vibrio cholerae isolates from a recent cholera outbreak in Senegal: comparison with isolates from Guinea-Bissau. Am. J. Trop. Med. Hyg. 58, 163–167. doi: 10.4269/ajtmh.1998.58.163
Amita, M., Chowdhury, S. R., Thungapathra, M., Ramamurthy, T., Nair, G. B., and Ghosh, A. (2003). Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9, 500–502. doi: 10.3201/eid0904.020317
Ansaruzzaman, M., Bhuiyan, N. A., Nair, G. B., Sack, D. A., Lucas, M., Deen, J. L., et al. (2004). Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10, 2057–2059. doi: 10.3201/eid1011.040682
Ceccarelli, D., Bani, S., Cappuccinelli, P., and Colombo, M. M. (2006). Prevalence of aadA1 and dfrA15 class 1 integron cassettes and SXT circulation in Vibrio cholerae O1 isolates from Africa. J. Antimicrob. Chemother. 58, 1095–1097. doi: 10.1093/jac/dkl352
Ceccarelli, D., Spagnoletti, M., Bacciu, D., Cappuccinelli, P., and Colombo, M. M. (2011a). New V. cholerae atypical El Tor variant emerged during the 2006 epidemic outbreak in Angola. BMC Microbiol. 11:130. doi: 10.1186/1471-2180-11-130
Ceccarelli, D., Spagnoletti, M., Bacciu, D., Dani-Poleg,Y., Mendiratta, D. K., et al. (2011b). ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int. J. Med. Microbiol. 301, 318–324. doi: 10.1016/j.ijmm.2010.11.005
Dalsgaard, A., Forslund, A., Sandvang, D., Arntzen, L., and Keddy, K. (2001). Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48, 827–838. doi: 10.1093/jac/48.6.827
Faruque, S. M., and Mekalanos, J. J. (2012). Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3, 556–565. doi: 10.4161/viru.22351
Gassama, A., Aïdara-Kane, A., Chainier, D., Denis, F., and Ploy, M.-C. (2004). Integron-associated antibiotic resistance in enteroaggregative and enteroinvasive Escherichia coli. Microb. Drug Resist. 10, 27–30. doi: 10.1089/107662904323047763
Gassama-Sow, A., Diallo, M. H., Boye, C. S., Garin, B., Sire, J. M., Sow, A. I., et al. (2006). Class 2 integron-associated antibiotic resistance in Shigella sonnei isolates in Dakar, Senegal. Int. J. Antimicrob. Agents 27, 267–270. doi: 10.1016/j.ijantimicag.2005.10.016
Global Task Force on Cholera Control (2006). Cholera country profile?: senegal. World Heal. Organ. 352, 1–2.
Goel, A. K., Jain, M., Kumar, P., Bhadauria, S., Kmboj, D. V, and Singh, L. (2008). A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J. Infect. 57, 280–281. doi: 10.1016/j.jinf.2008.06.015
Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R., and Waldor, M. K. (2001). Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45, 2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001
Hochhut, B., and Waldor, M. K. (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32, 99–110.
Ismail, H., Smith, A. M., Tau, N. P., Sooka, A., Keddy, K. H. and Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (2013). Cholera outbreak in South Africa, 2008–2009: Laboratory analysis of Vibrio cholerae O1 strains. J. Infect. Dis. 208, S39–S45. doi: 10.1093/infdis/jit200
Keasler, S. (1993). Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341, 1661.
Kiiru, J. N., Saidi, M. S., Goddeeris, B. M., Wamae, N. C., Butaye, P., and Kariuki, S. M. (2009). Molecular characterisation of Vibrio cholerae O1 strains carrying an SXT/R391-like element from cholera outbreaks in Kenya: 1994-2007. BMC Microbiol. 9:275. doi: 10.1186/1471-2180-9-275
Kutar, B. M. R. N. S., Rajpara, N., Upadhyay, H., Ramamurthy, T., and Bhardwaj, A. K. (2013). Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS ONE 8:e56477. doi: 10.1371/journal.pone.0056477
Manga, N. M., Ndour, C. T., Diop, S. A., Dia, N. M., Ka-Sall, R., Diop, B. M., et al. (2008). Cholera in Senegal from 2004 to 2006: lessons learned from successive outbreaks. Med. Trop. 68, 589–592.
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
Morita, M., Ohnishi, M., Arakawa, E., Bhuiyan, N. A., Nusrin, S., Alam, M., et al. (2008). Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol. Immunol. 52, 314–317. doi: 10.1111/j.1348-0421.2008.00041.x
Nair, G. B., Faruque, S. M., Bhuiyan, N. A., Siddique, A. K., Sack, D. A, and Kamruzzaman, M. (2002). New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. 40, 3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002
Nair, G. B., Qadri, F., Holmgren, J., Svennerholm, A. M., Safa, A., Bhuiyan, N. A., et al. (2006). Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44, 4211–4213. doi: 10.1128/JCM.01304-06
Ogawa, A., Kato, J. I., Watanabe, H., Nair, B. G., and Takeda, T. (1990). Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler’s diarrhea. Infect. Immun. 58, 3325–3329.
Opintan, J. A., Newman, M. J., Nsiah-Poodoh, O. A., and Okeke, I. N. (2008). Vibrio cholerae O1 from Accra, Ghana carrying a class 2 integron and the SXT element. J. Antimicrob. Chemother. 62, 929–933. doi: 10.1093/jac/dkn334
Pearson, G. D. N., Woods, A., Chiang, S. L., and Mekalanos, J. J. (1993). CTX genetic element encodes a site-specific recombination system and an intestinal colonisation factor. Proc. Natl. Acad. Sci. U.S.A. 90, 3750–3754.
Ploy, M. C., Denis, F., Courvalin, P., and Lambert, T. (2000). Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44, 2684–2688.
Pugliese, N., Maimone, F., Scrascia, M., Materu, S. F., and Pazzani, C. (2009). SXT-related integrating conjugative element and IncC plasmids in Vibrio cholerae O1 strains in Eastern Africa. J. Antimicrob. Chemother. 63, 438–442. doi: 10.1093/jac/dkn542
Quilici, M.-L., Massenet, D., Gake, B., Bwalki, B., and Olson, D. M. (2010). Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg. Infect. Dis. 16, 1804–1805. doi: 10.3201/eid1611.100568
Safa, A., Bhuyian, N. A., Nusrin, S., Ansaruzzaman, M., Alam, M., Hamabata, T., et al. (2006). Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. J. Med. Microbiol. 55, 1563–1569. doi: 10.1099/jmm.0.46689-0
Safa, A., Nair, G. B., and Kong, R. Y. C. (2010). Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18, 46–54. doi: 10.1016/j.tim.2009.10.003
Safa, A., Sultana, J., Dac Cam, P., Mwansa, J. C., and Kong, R. Y. (2008). Vibrio cholerae O1 hybrid El tor strains, Asia, and Africa. Emerg. Infect. Dis. 14, 987–988. doi: 10.3201/eid1406.080129
Shi, L., Fujihara, K., Sato, T., Ito, H., Garg, P., Chakrabarty, R., et al. (2006). Distribution and characterization of integrons in various serogroups of Vibrio cholerae strains isolated from diarrhoeal patients between 1992 and 2000 in Kolkata, India. J. Med. Microbiol. 55, 575–583. doi: 10.1099/jmm.0.46339-0
Shi, L., Miyoshi, S., Hiura, M., Tomochika, K., Shimada, T., and Shinoda, S. (1998). Detection of genes encoding cholera toxin (CT), zonula occludens toxin (ZOT), accessory cholera enterotoxin (ACE) and heat-stable enterotoxin (ST) in Vibrio mimicus clinical strains. Microbiol. Immunol. 42, 823–828. doi: 10.1111/j.1348-0421.1998.tb02357.x
Waldor, M. K., Rubin, E. J., Pearson, G. D., Kimsey, H., and Mekalanos, J. J. (1997). Regulation, replication, and integration functions of the Vibrio cholerae CTXϕ are encoded by region RS2. Mol. Microbiol. 24, 917–926. doi: 10.1046/j.1365-2958.1997.3911758.x
Keywords: Vibrio cholerae, O1 virulence, antibioresistance, SXT element, Senegal
Citation: Sambe-Ba B, Diallo MH, Seck A, Wane AA, Constantin de Magny G, Boye CS-B, Sow AI and Gassama-Sow A (2017) Identification of Atypical El Tor V. cholerae O1 Ogawa Hosting SXT Element in Senegal, Africa. Front. Microbiol. 8:748. doi: 10.3389/fmicb.2017.00748
Received: 30 June 2016; Accepted: 11 April 2017;
Published: 15 May 2017.
Edited by:
Pendru Raghunath, Texila American University, GuyanaReviewed by:
Biswajit Maiti, Nitte University, IndiaAshima Kushwaha Bhardwaj, Indian Institute of Advanced Research, India
Ahmed Askora, Zagazig University, Egypt
Copyright © 2017 Sambe-Ba, Diallo, Seck, Wane, Constantin de Magny, Boye, Sow and Gassama-Sow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy Gassama-Sow, gassama@pasteur.sn
 Bissoume Sambe-Ba
Bissoume Sambe-Ba Mamadou H. Diallo1
Mamadou H. Diallo1 Guillaume Constantin de Magny
Guillaume Constantin de Magny Amy Gassama-Sow
Amy Gassama-Sow