- 1State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangdong Institute of Microbiology, Guangzhou, China
- 2College of Food Science, South China Agricultural University, Guangzhou, China
Listeria monocytogenes is an important Gram-positive foodborne pathogen. However, limited information is available on the comprehensive investigation and potential risk of L. monocytogenes in fresh aquatic products, which are popular to consumers in China. This study aimed to determine the occurrence, virulence profiles, and population diversity of L. monocytogenes isolated from aquatic products in China. In total, 846 aquatic product samples were collected between July 2011 and April 2016 from 43 cities in China. Approximately 7.92% (67/846) aquatic product samples were positive for L. monocytogenes, 86.57% positive samples ranged from 0.3 to 10 MPN/g, whereas 5.97% showed over 110 MPN/g by the Most Probable Number method, which included two samples of products intended to be eaten raw. Serogroups I.1 (serotype 1/2a), I.2 (serotype 1/2b), and III (serotype 4c) were the predominant serogroups isolated, whereas serogroup II.1 (serotype 4b) was detected at much lower frequencies. Examination of antibacterial resistance showed that nine antibacterial resistance profiles were exhibited in 72 isolates, a high level susceptibility of 16 tested antibiotics against L. monocytogenes were observed, indicating these common antibacterial agents are still effective for treating L. monocytogenes infection. Multilocus sequence typing revealed that ST299, ST87, and ST8 are predominant in aquatic products, indicating that the rare ST299 (serotype 4c) may have a special ecological niche in aquatic products and associated environments. Except llsX and ptsA, the 72 isolates harbor nine virulence genes (prfA, actA, hly, plcA, plcB, iap, mpl, inlA, and inlB), premature stop codons (PMSCs) in inlA were found in four isolates, three of which belonged to ST9. A novel PMSC was found in 2929-1LM with a nonsense mutation at position 1605 (TGG→TGA). All ST87 isolates harbored the ptsA gene, whereas 8 isolates (11.11%) carried the llsX gene, and mainly belonged to ST1, ST3, ST308, ST323, ST330, and ST619. Taken together, these results first reported potential virulent L. monocytogenes isolates (ST8 and ST87) were predominant in aquatic products which may have implications for public health in China. It is thus necessary to perform continuous surveillance for L. monocytogenes in aquatic products in China.
Introduction
Listeria monocytogenes is an important foodborne pathogen worldwide, that can cause life-threatening listeriosis disease in vulnerable groups including pregnant women, fetuses, elderly people, and immunocompromised individuals, with a considerable mortality rate (20–30%) (Lomonaco et al., 2009). L. monocytogenes is capable of proliferating in different stressful environmental conditions, including high salinity, low temperature, and a wide range of pH values. Foodborne listeriosis poses a global economic and health burden due to the wide spread of L. monocytogenes in food and food processing environments.
In China, 147 clinical cases in total and 82 outbreak-related cases were reported in 28 provinces from January 1964 to December 2010 (Feng et al., 2013). In recent years, listeriosis diseases have been increasingly reported in China annually, especially in developed cities (Wang et al., 2015, 2018). This trend may be linked to the trend of direct consumption of fresh foods and ready-to-eat foods in China, especially in developed cities. However, there is no national monitoring system for listeriosis cases in China as it is not yet considered a notifiable disease. According to the report of the European Food Safety Authority (EFSA), the small number of listeriosis outbreaks may be linked to the lower consumption of fish and fish products compared to other foods in Europe (European Food Safety Authority [EFSA] and European Centre for Disease Control [ECDC], 2015). Similarly, as reported by Song et al. (2015), ready-to-eat (RTE) raw fish has the highest per serving risk of listeriosis among the five kinds of ready-to-eat foods based on risk assessment. This indicates that RTE fish and fish products may serve as a source of L. monocytogenes infection. Previous studies have focused on the prevalence of L. monocytogenes in aquatic products, but these studies only focused their investigation to limited cities/provinces, such as Jiangsu Province, Henan Province, Beijing City, and Zhanjiang City (Chen et al., 2013; Ma, 2015; Wu et al., 2017; Chui et al., 2018). However, little information is available on the comprehensive investigation and potential virulence of L. monocytogenes from aquatic products in China. According to data compiled by the National Bureau of Statistics of China, the average consumption of aquatic products per capita has increased annually and in 2016, it was 11.4 kg1 . In this context, it is necessary to systematically investigate the contamination level of L. monocytogenes in aquatic products in China.
Listeria monocytogenes has been differentiated into 13 serotypes based on its somatic (O) and flagellar (H) antigens, and are further grouped into five serovars, designated as serogroups I.1 (1/2a-3a), I.2 (1/2c-3c), II.1 (4b-4d-4e), II.2 (1/2b-3b-7), and III (4a-4c) (Doumith et al., 2004). A series of virulence factors participating in cellular infection cycles have been documented in L. monocytogenes. Listeria pathogenicity island-1 (LIPI-1, including prfA, hly, mpl, iap, plcB, plcC, and actA) and LIPI-2 (inlA and inlB) are considered as the two classic major pathogenicity islands in L. monocytogenes. L. monocytogenes is known to exhibit varied pathogenicity in intra-species isolates even though each isolate harbors both LIPI-1 and LIPI-2. In recent years, the llsX gene (encoding Listeriolysin, LLS, a hemolytic, and cytotoxic factor) belonging to LIPI-3, has been greatly associated with a subset of lineage I in human listeriosis (Cotter et al., 2008). Maury et al. (2016) identified that a cluster of six genes annotated as the cellobiose-family phosphotransferase system (PTS) and designated as LIPI-4, was highly associated with L. monocytogenes neuroinvasiveness and human maternal-neonatal (MN) infection. At present, LIPI-3 were found only in lineage I clonal complexes, 23 STs were found to carried LIPI-3 (ST1, ST1001, ST3, ST4, ST6, ST191, ST213, ST217, ST224, ST288, ST363, ST79, ST77, ST382, ST389, ST489, ST999, ST554, ST581, ST1000, ST380, ST778, and ST619), LIPI-4 presented in 11 STs belongs to lineage I and a single lineage III (ST4, ST87, ST213, ST217, ST363, ST382, ST388, ST663, ST1002, ST1166, and ST619) (Chen et al., 2018; Kim et al., 2018; Wang et al., 2018). The presence of these virulence genes may be an important determinant to their pathogenicity.
The objective of this study was to (i) comprehensively investigate the occurrence and contamination level of L. monocytogenes in fresh aquatic products; (ii) evaluate the potential virulence and antibacterial profiles of L. monocytogenes isolates; and (iii) explore the genetic diversity of L. monocytogenes isolates recovered from the Chinese retail aquatic system.
Materials and Methods
Samples
From July 2012 to April 2016, a total of 846 retail aquatic products were collected from rural markets (n = 245), open-air markets (n = 261), and large supermarkets (n = 340) from 43 cities of China. The marine aquatic products (n = 506) included squid (n = 150 samples), shrimp (n = 145), yellow croak (n = 100), weever (n = 24), shell fish (n = 23), saury (n = 20), octopus (n = 10), cuttlefish (n = 6), silvery pomfret (n = 3), salmon (n = 6), capelin (n = 3), and other species (n = 16). Freshwater aquatic products (n = 341) comprised the crucian carp (n = 117), grass carp (n = 82), Tilapia mossambica (n = 49), Megalobrama amblycephala (n = 29), Cyprinus carpio (n = 21), bighead carp (n = 7), Pelteobagrus fulvidraco (n = 5), silver carp (n = 5), Channa argus (n = 4), catfish (n = 2), and other species (n = 20). Except of packaged salmon products, all of marine aquatic products except shrimp were stored loosely in ice, while freshwater aquatic products were alive keeping in water. Among these samples, salmon, grass carp, and Tilapia mossambica were intended to be eaten raw. All samples were placed in insulated shipping coolers containing frozen gel packs, which were placed on the sides, middle, and the top of the samples. All samples were kept below 4°C during transportation and testing was initiated within 4 h after receiving the samples.
Qualitative and Quantitative Analysis
Qualitative detection of L. monocytogenes was performed according to the National Food Safety Standard of China (4789.30-2010) with minor adaptations Ministry of Health of People’s Republic of China (2010). Briefly, 25 g of homogenized aquatic samples were added to 225 mL Listeria enrichment broth 1 (LB1) (Guangdong Huankai Co., Ltd., Guangzhou, China). The cultures in LB1 media were incubated at 30°C for 24 h, after which 0.1 mL LB1 enrichment culture was transferred to 10 mL Listeria enrichment broth 2 (LB2) at 30°C for 24 h. A loopful (about 10 μL) of the LB2 enrichment culture was streaked onto Listeria selective agar plates (Guangdong Huankai Co., Ltd.) and incubated at 37°C for 48 h. Three to five presumptive colonies were selected for the identification of L. monocytogenes using the Microgen ID Listeria identification system (Microgen, Camberley, United Kingdom) according to the manufacturer’s instructions.
The workflow for most probable number (MPN) was adapted according to a previous study by Gombas et al. (2003). Briefly, a 9-tube MPN method was used. The nine tubes were divided into three sets of three tubes. The first set of tubes was contained 10 mL of the sample homogenate, the second and third sets of tubes contained 10 mL of Fraser broth (Guangdong Huankai Co., Ltd., Guangzhou, China) inoculated with 1 and 0.1 mL of the homogenate, respectively. Three aliquots (10, 1, and 0.1 mL) of the sample homogenate were dispensed into three sets, representing 1.0, 0.1, and 0.01 g of the original sample, respectively. The tubes were incubated at 30 ± 2°C for 24 ± 2 h. Darkened Fraser tubes were subjected to streaking onto Listeria selective agar plates. If a Fraser broth tube did not darken, it was examined again after an additional 26 ± 2 h of incubation. The MPN value was determined based on the number of positive tube(s) in each of the three sets and the MPN table (U.S. Department of Agriculture, 1998).
Serogroup Analysis
Genomic DNA was extracted from L. monocytogenes using a Bacterial Genomic DNA Purification Kit (Magen Biotech Inc., Guangzhou, China) according to the manufacturer’s instructions. Serogroup analysis of 72 isolates was performed using multiplex PCR as described by Doumith et al. (2004) (Supplementary Table S1). Five distinct serogroups, I.1 (1/2a-3a), I.2 (1/2c-3c), II.1 (4b-4d-4e), II.2 (1/2b-3b-7), and III (4a-4c), were identified using multiplex PCR. The primers used are shown in Supplementary Table S2. PCR was performed with an initial denaturation step at 94°C for 3 min; 35 cycles of 94°C for 35 s, 53°C for 50 s, and 72°C for 60 s; and a final cycle of 72°C for 7 min in a thermocycler (Applied Biosystems, CA, United States). The amplicons (8 μl) were separated on 2% agarose gels in TAE buffer and then visualized by Goldview® staining (0.005%, v/v). All strains were serotyped by antigen serum agglutination according to the manufacturer’s instruction (Denka Seiken Co., Ltd., Tokyo, Japan).
Antimicrobial Susceptibility Test
All strains collected were analyzed by the KB method using breakpoints recommended by the National Committee for Clinical Laboratory Standards (Clinical and Laboratory Standards Institute, 2014) for Staphylococcus, except for ampicillin and penicillin G where specific Listeria breakpoints are defined (M45-A2 Vol. 30 No. 18) (Supplementary Table S3). In total, 16 antibiotic agents, including those used to treat human listeriosis, were tested by the KB method as follows: ampicillin (AMP; 10 μg), chloramphenicol (C; 30 μg), erythromycin (E; 15 μg), gentamicin (CN; 10 μg), kanamycin (K; 30 μg), rifampin (RD; 5 μg), doxycycline (DO, 30 μg), penicillin (P, 10 U), tetracycline (TE; 30 μg), vancomycin (VA; 30 μg), sulfamethoxazole with trimethoprim (SXT; 23.75/1.25 μg), sulbactam/ampicillin (SAM; 10/10 μg), meropenem (MEM; 10 μg), linezolid (LZD, 30 μg), and amoxycillin/clavulanic acid (AMC; 10 μg) (Oxoid, Basingstoke, United Kingdom). Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922 were used as quality control strains. Zones of inhibition were measured with precision calipers to the nearest 0.01 mm. Isolates exhibiting resistance to at least three classes of the antimicrobial agents tested, were considered multidrug-resistant strains.
Identification of Potential Hypervirulent Isolates
To evaluate the potential virulence of 72 L. monocytogenes isolates, the presence of virulence factors associated with infection cycle and invasiveness was detected by PCR. As shown in Supplementary Table S4, nine virulence factor genes belonging to LIPI-1 and LIPI-2 were detected by PCR. In addition, the presence of LIPI-3 and LIPI-4 genes (llsX and ptsaA, respectively) was also observed by PCR (Clayton et al., 2011; Maury et al., 2016). The premature stop codons (PMSCs) of inlA were determined by amplicon sequencing. The complete length of inlA (2403 bp) was sequenced in 72 isolates. External primers were used to amplify the complete inlA gene and internal primers were used for sequencing (Supplementary Table S3; Wu et al., 2016). The inlA sequences were assembled using DNAMAN software (version 8th). By comparing the obtained complete inlA sequence to that of the L. monocytogenes EGDe (Glaser et al., 2001), PMSC types were determined according to the site of PMSC-mutation in inlA gene as documented by Gelbicova et al. (2015).
Multilocus Sequence Typing
MLST analysis of L. monocytogenes was previously reported by Ragon et al. (2008) (Supplementary Table S5). Briefly, each 50 μL PCR contained 5.0 μL 10× PCR buffer (TAKARA, Dalian, China), 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 mM of each primer, 1.25 U Taq polymerase, and 1 μL genomic DNA. PCR was performed using the following program: 3 min initial denaturation at 94°C and 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 52°C (45°C for bglA) for 1 min and elongation at 72°C for 2 min, followed by a final elongation for 10 min at 72°C. The PCR products were purified and sequenced by Invitrogen (Thermo Fisher, Shanghai, China). An allele number was given according to each variant locus of each housekeeping gene; sequence types (STs) and clonal complexes (CCs) were assigned via the Listeria MLST database at the Pasteur Institute website2. A neighbor-joining tree of L. monocytogenes based on the MLST of seven housekeeping genes was constructed using MEGA 7.0 (Kumar et al., 2016) with 1000 bootstrap replications. Simpson’s indexes of discrimination (DI) for MLST were calculated to determine the ability of the MLST typing method according to a previous study reported by Hunter and Gaston (1988).
Data Analysis
The statistically significant difference of prevalence of L. monocytogenes between marine aquatic products and freshwater aquatic products was calculated using the Chi-square test. P-values of <0.05 were considered statistically significant.
Results
Occurrence and Contamination Level of L. monocytogenes
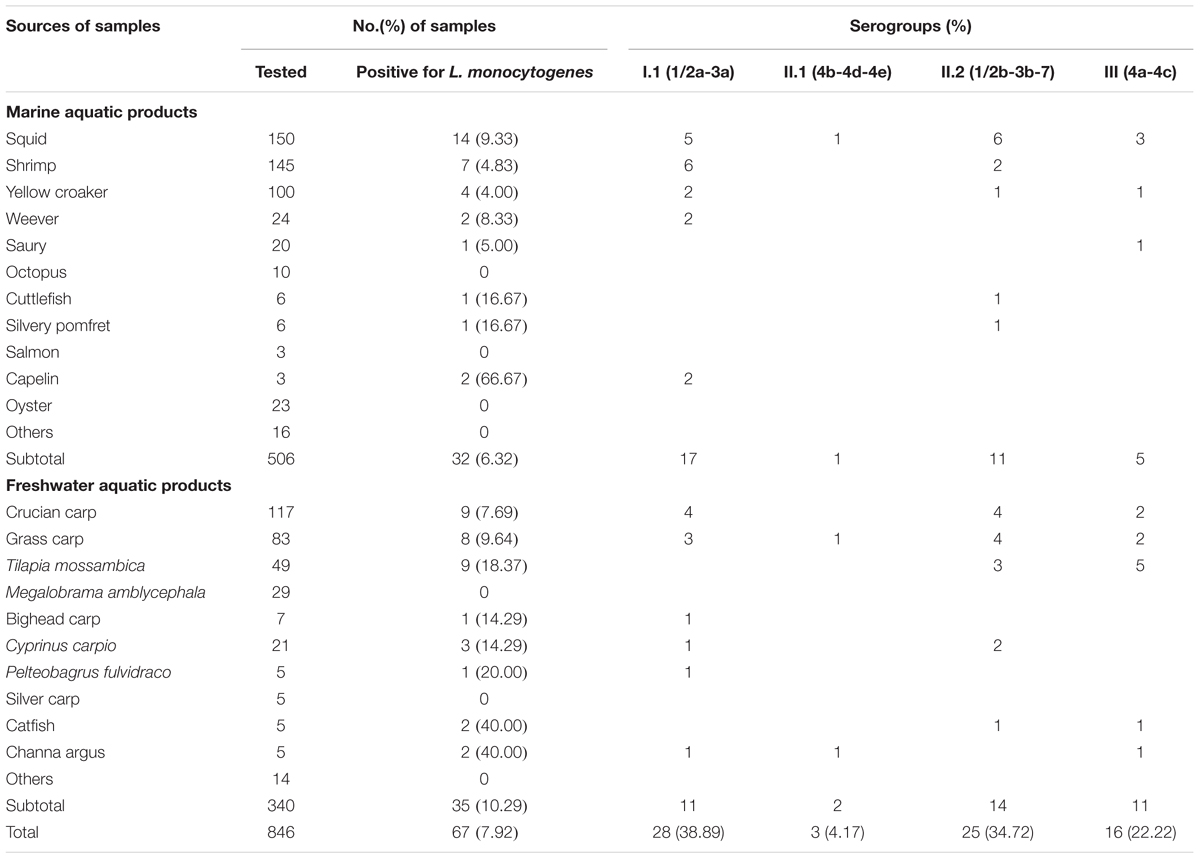
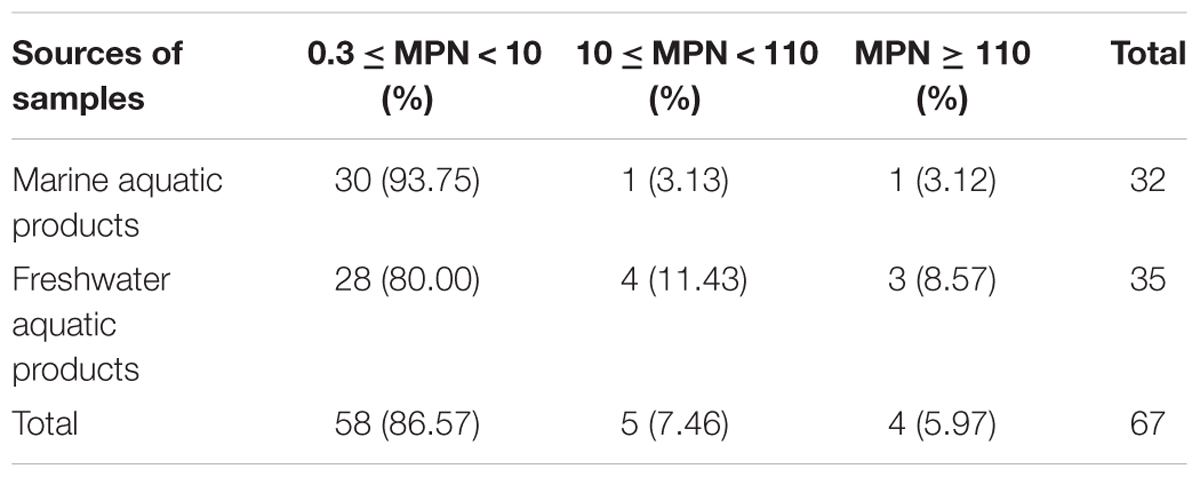
The overall prevalence of L. monocytogenes in freshwater and marine products tested during 2012–2016 is presented in Table 1. In total, 67 (7.92%) samples were positive for L. monocytogenes among 846 analyzed samples, 28 (8.24%) positive samples from large supermarket (n = 340), 25 (9.58%) samples from open-air markets (n = 261), 14 (5.71%) samples from rural markets (n = 245). Among 506 marine aquatic product and 340 freshwater aquatic product samples, the prevalence of L. monocytogenes in freshwater aquatic products (10.29%) was found to be significantly higher than that (6.32%) in marine aquatic products (p < 0.05). The contamination level by L. monocytogenes in aquatic products was determined by the MPN method. Approximately 86.57% of positive samples ranged from 0.3 to 10 MPN/g, five positive samples were ranged from 10 to 110 MPN/g, of which one sample from marine products and four samples from freshwater products, only four (5.97%) samples were over 110 MPN/g, including one sample from marine products and three samples from freshwater products. In addition, two of the samples with L. monocytogenes higher than 100 MPN/g, namely grass carp and Tilapia mossambica were intend to be eaten raw (Table 2 and Figure 1).
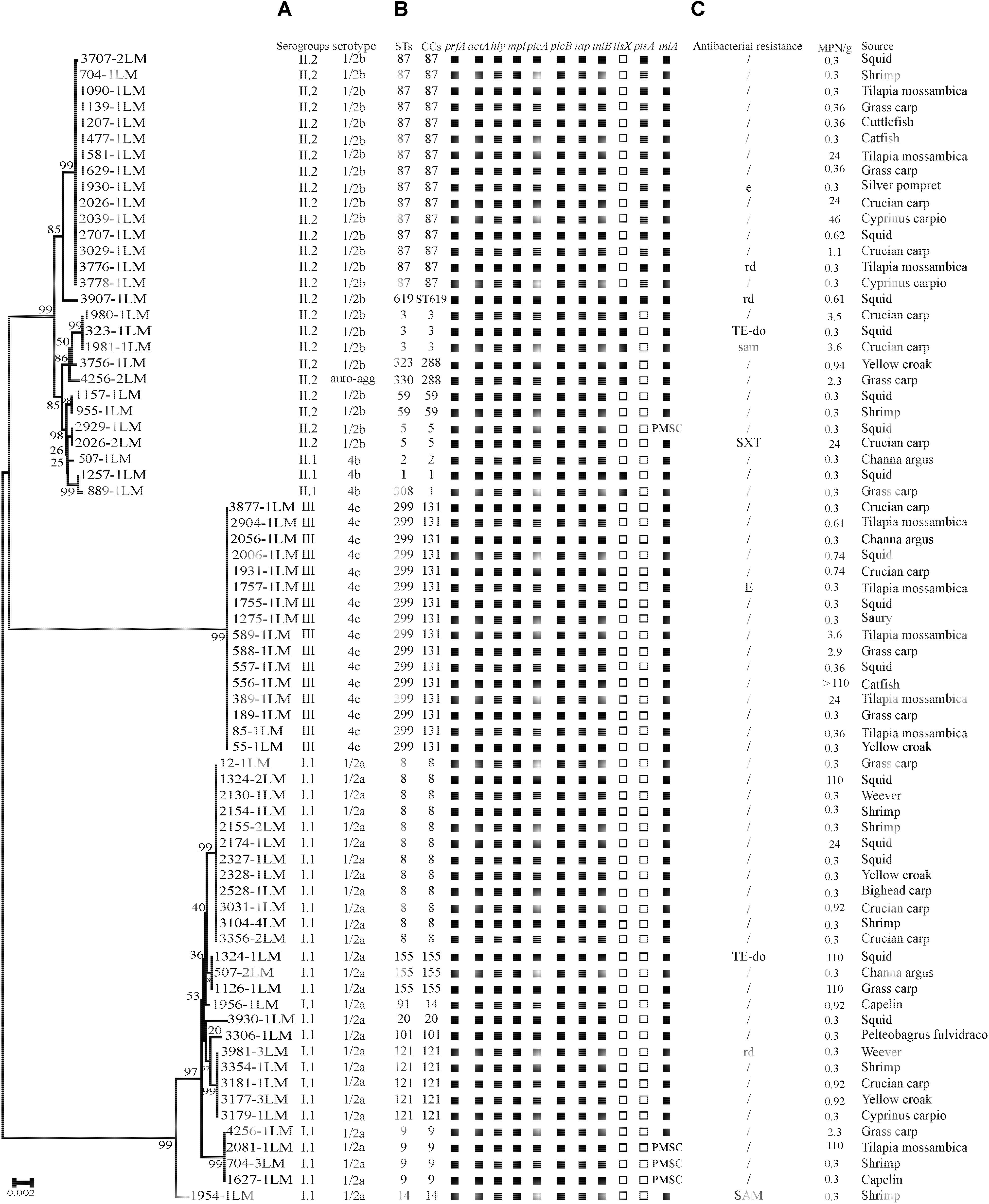
FIGURE 1. Genotypic and phenotypic characteristics of Listeria monocytogenes isolated from aquatic products in China. (A) Auto-agg: auto-aggregation. (B) PMSC, premature stop codons in inlA; solid squares indicate the presence of virulence genes or full-length inlA. Hollow squares indicate the absence of virulence genes. (C) E(e), erythromycin; DO(do), doxycycline; SXT(sxt), sulfamethoxazole with trimethoprim; OFX(ofx), ofloxacin; RD(rd), rifampin; SAM(sam), sulbactam/ampicillin; TE, tetracycline. A slash (/) indicates no resistance. Antibiotic abbreviations in uppercase indicate resistance, while those in lowercase indicate intermediate resistance. A neighbor-joining tree of L. monocytogenes based on the MLST of seven housekeeping genes was established in MEGA 7.0 with 1000 bootstrap replications. Bootstrap values are shown at the nodes.
Serogroups
As shown in Table 1, four serogroups were identified among 72 L. monocytogenes isolates. Serogroup I.1 (1/2a-3a) (n = 28, 38.89%) was the most prevalent, followed by serogroup II.2 (1/2b-3b-7) (n = 25, 34.72%), and three isolates (4.17%) were identified as II.1 (4b-4d-4e). Sixteen (22.22%) isolates belonged to serogroup III (4a-4c), including four isolates from marine products and 12 isolates from freshwater products. All 72 strains were serotyped by antigen serum agglutination. Except for auto-aggregation of 4256-1LM isolate, 24 isolates were typed as 1/2b, 3 isolates as 4b, 16 isolates as 4c, and 28 isolates as 1/2a.
Presence of Virulence Genes
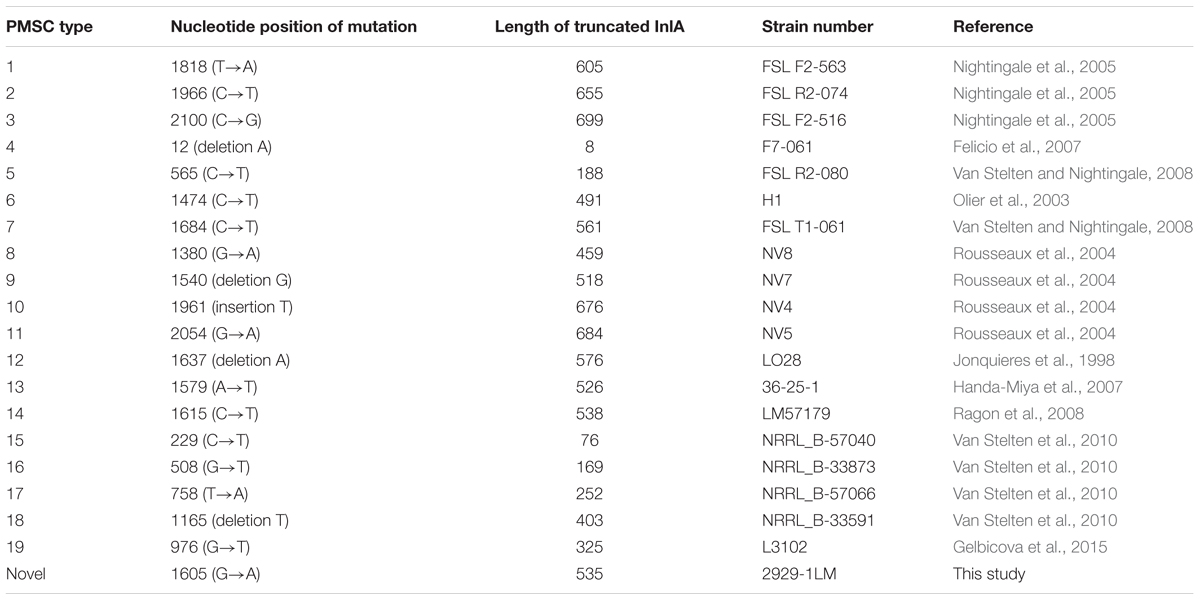
The presence of 11 known virulence genes was identified in the 72 isolates in this study by PCR. As shown in Figure 1, except llsX and ptsA, all 72 isolates were positive for nine virulence genes, including prfA, actA, hly, iap, mpl, plcA, plcB, inlA, and inlB. Eight isolates (11.11%) carried llsX and mainly belonged to ST1 (n = 1), ST3 (n = 3), ST308 (n = 1), ST323 (n = 1), ST330 (n = 1), and ST619 (n = 1). pstaA was harbored in 16 isolates, of which 15 isolates were from ST87 and one isolate belonged to ST619. pstaA was absent in the other isolates. Furthermore, the PMSCs in inlA were analyzed by sequencing. Compared to the full-length sequence of inlA in the reference L. monocytogenes EGDe (Glaser et al., 2001), a total of 68 isolates (94.44%) carried the complete codon sequence of inlA, including three isolates belonging to ST9 and one ST5 isolate. As shown in Figure 1, four isolates (1627-1LM, 2081-1LM, 704-3LM, and 2929-1LM) were found to carry PMSCs in inlA. In the isolate 1627-1LM, sequence analysis revealed a nonsense mutation at position 2054, which resulted in changing a glutamine codon to a stop codon (TGG→TAG). In 2081-1LM, a nonsense mutation at position 976 changed a glutamic acid (Glu) codon to a stop codon (GAA→TAA). Adenylic acid deletion at position 12 resulted in a PMSC in inlA of 704-3LM. A nonsense mutation was found at position 1605 (TGG→TGA) in the 2929-1LM isolate, and was identified as a novel type of point mutation in inlA based on the previously known PMSC mutation types of inlA (Gelbicova et al., 2015; Moura et al., 2016; Table 3).
Antibacterial Susceptibility Test
The results of antibacterial susceptibility of 72 isolates against 16 antibacterial agents are shown in Figure 2. Nine antibacterial resistance profiles were found in the 72 L. monocytogenes isolates. The number of antibiotic-resistant L. monocytogenes isolates was relatively low; most isolates were susceptible to the 16 tested antibacterial agents. None of the isolates exhibited resistance to nine antibacterial agents, which are ampicillin, penicillin, kanamycin, genmycin, amoxycillin/clavulanic acid, meropenem, vancomycin, chloramphenicol, and linezolid. None isolate was considered as multidrug resistant isolates, and were resistant to three classes of tested antibacterial agents (Figure 1).
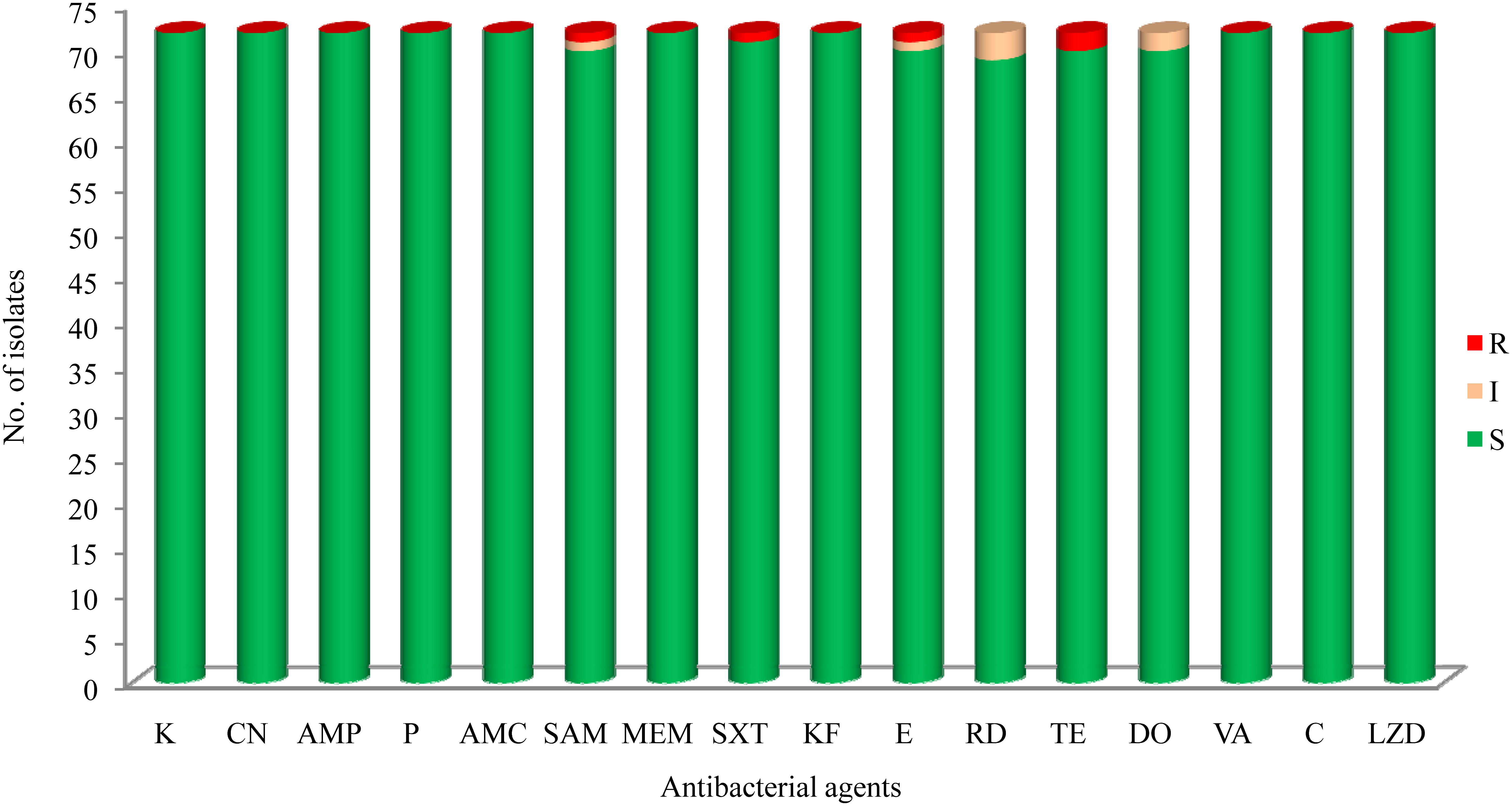
FIGURE 2. The results of common antibacterial agents against Listeria monocytogenes isolates∗. ∗C, chloramphenicol; K, kanamycin; CN, gentamicin; LEV, levofloxacin; OFX, ofloxacin; CIP, ciprofloxacin; AMP, ampicillin; P, penicillin; AMC, amoxycillin/clavulanic acid; SAM, sulbactam/ampicillin; MEM, meropenem; SXT, sulfamethoxazole with trimethoprim; KF, cephalothin; E, erythromycin; RD, rifampin; TE, tetracycline; DO, doxycycline; VA, vancomycin; LNZ, linezolid.
MLST Analysis
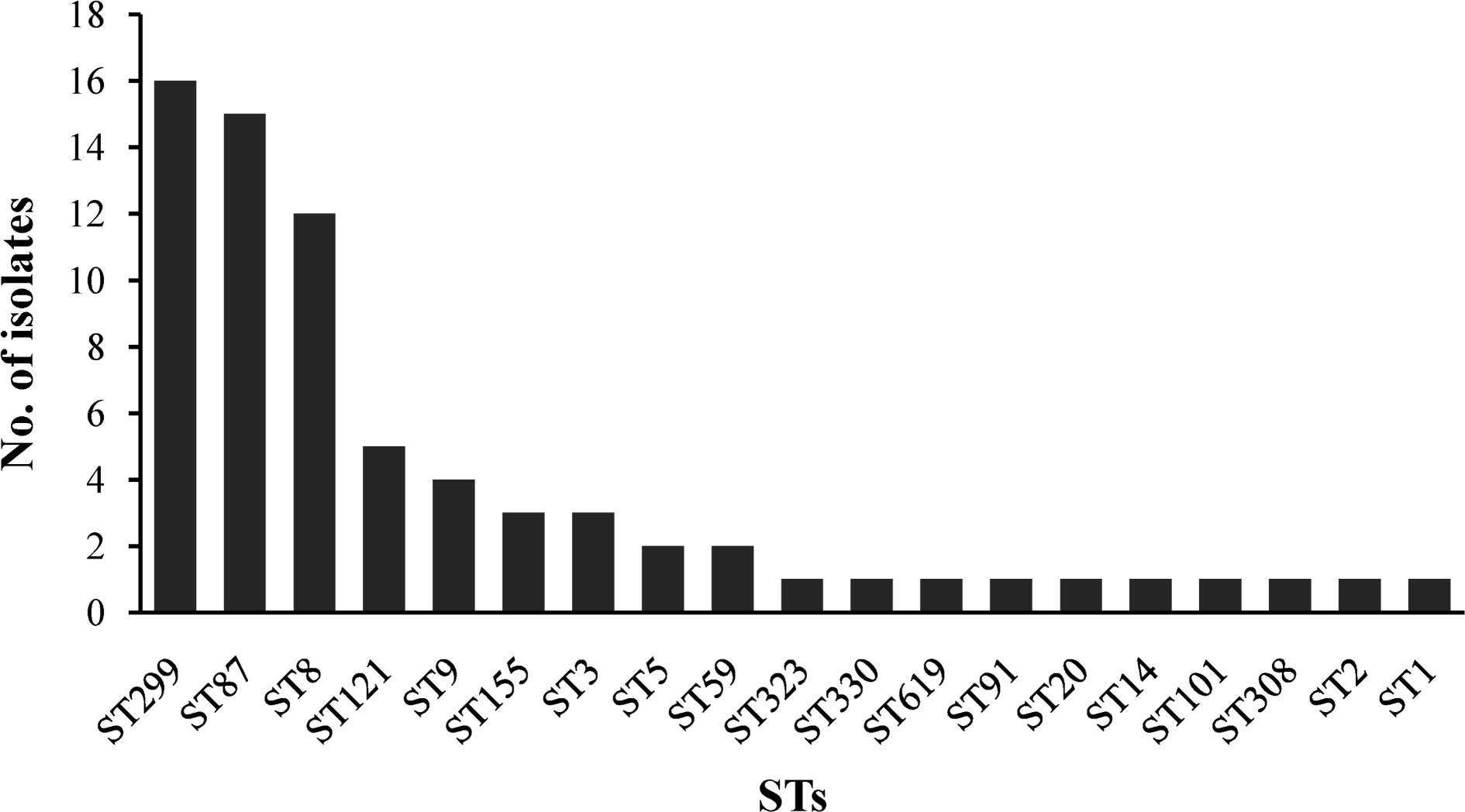
The 72 isolates were distributed into 19 different STs belonging to 16 complex clones (CCs), with a DI of 0.877. Ten STs that contained between 2 and 16 isolates and 9 singletons were included in the 19 STs. Of the major STs containing at least 12 isolates (ST299, ST87, and ST8), ST299 (n = 16, 22.22%) was the most prevalent, followed by ST87 (n = 15, 20.83%), and ST8 (n = 12, 16.67%). The top three STs (ST299, ST87, and ST8) comprised 59.72% of all isolates (Figure 3).
Discussion
Listeria monocytogenes, an important human foodborne pathogen, is of great public health concern. Because of its ability to resist various stress environments in food processing, the food chain is considered the main transmission route of L. monocytogenes to consumers. Fresh aquatic products are a popular food in China because of their taste and nutritional value. Some countries have formulated a safety criterion of <100 CFU/g for fishery products (including smoked fish) (Anonymous, 2005; International Commission on Microbiological Specifications for Foods [ICMSF], 2011), there are no qualitative and quantitative criteria or regulations for L. monocytogenes in fresh fish products in China. It is therefore necessary to explore the potential risk of L. monocytogenes infection upon consumption of fresh fish in China. However, little information on comprehensive investigation of L. monocytogenes contamination in fresh aquatic products in China was available. This study thus aimed to determine the occurrence, antibacterial susceptibility, and genotypic characteristics of L. monocytogenes isolates recovered from fresh aquatic products in the Chinese food system.
In this study, 67 samples (7.92%) out of 846 fresh aquatic products were positive for L. monocytogenes. This is in agreement with the results of a study conducted in Beijing City (Ma, 2015), whereas several previous studies reported a lower prevalence of L. monocytogenes in different cities in China, ranging from 1.57 to 4.88% (Chen et al., 2013; Wu et al., 2017; Chui et al., 2018). These discrepancies may be due to different sample composition of aquatic products and/or the difference farming, storage and processing of aquatic products. In addition, the prevalence of L. monocytogenes in freshwater aquatic products was found to be significantly higher than that in marine aquatic products. Quantitative analysis also showed that only four positive samples were over 110 MPN/g, of which one was a marine fish sample and three were freshwater aquatic products, two samples (Tilapia mossambica, and grass carp) were intended to be eaten by raw, indicating potential risk was present in aquatic products. In addition, these results indicate that contamination with this pathogen may be higher in freshwater fish, which may be associated with the different storage temperature between marine aquatic products and freshwater aquatic products. This result is consistent with the results of Wieczorek and Osek (2017).
In recent years, the emerging prevalence of antibiotic resistance in L. monocytogenes has been reported in Asia as well as globally (Pagadala et al., 2012; Chen et al., 2015; Obaidat et al., 2015; Sharma et al., 2017; Akrami-Mohajeri et al., 2018; Wilson et al., 2018). The findings of this study suggested that the prevalence of antibiotic resistance among L. monocytogenes isolated from fish products in China is relatively low (Figure 2). Notably, unlike the high susceptibility of ampicillin and penicillin against L. monocytogenes in this study, a high level of resistance was also reported in ready-to-eat foods in Gondar Town, Ethiopia (66.7%) (Garedew et al., 2015), dairy-based food products in Lebanon (90.0%) (Harakeh et al., 2009), milk and dairy products in Yazd, central Iran (Akrami-Mohajeri et al., 2018), and retail foods in Turkey (72.2%) (Cetinkaya et al., 2014). Penicillin is the first-line antibiotic therapy alone or with gentamicin for treating L. monocytogenes infection, but sulfamethoxazole and trimethoprim is an optional choice for penicillin-allergic patients, and erythromycin is used in pregnant women. In this study, we found a high level of susceptibility to common antibiotics including gentamicin, sulfamethoxazole and trimethoprim, and erythromycin, against L. monocytogenes isolates, which is consistent with previous studies (Shen et al., 2013; Chen et al., 2014, 2015; Wieczorek and Osek, 2017). Although our data are limited, antibiotic-resistant L. monocytogenes isolates are relatively low in this study (Figure 1), further studies are required to surveil the antibiotic susceptibility of foodborne L. monocytogenes.
The pathogenicity of L. monocytogenes is known to differ at an intra-species level. Serotype 4b, 1/2a, and 1/2b account for 95% of the strains isolated from human listeriosis (Cartwright et al., 2013; Althaus et al., 2014). In this study, the majority of L. monocytogenes belonged to serogroup I.1 (serotype 1/2a, 38.89%), II.2 (serotype 1/2b, 34.72%), and III (serotype 4c, 22.22%), which is consistent with the previous results obtained in fish from several countries (Fallah et al., 2013; Jamali et al., 2015; Terentjeva et al., 2015). Wang et al. (2015) reported serotype 1/2b (belonged to ST87 and ST3) with a frequency of 64.3% of L. monocytogenes was predominant in human listeriosis cases in China, similar results was also reported in Zigong, Sichuan province, conducted by Wang et al. (2018), indicating potential hypervirulent isolates were present in aquatic products in China. The three most common STs were ST299, ST87, and ST8 in this study, Wang et al. (2012) reported ST9 (29.1%), ST8 (11.7%), and ST87 (10.7%) of L. monocytogenes predominated in animal foods in China, similar results of ST9 (72%) was found in raw pork and associated environments in China (Li et al., 2018), ST2 was predominated in some Brazilian dairy industries and retail products. Serogroup III (4a-4c) was mainly isolated from ruminants (Orsi et al., 2011), and 22.22% of isolates in this study were identified as serogroup III (ST299), which is in agreement with previous observations in Beijing, China (Ma, 2015). These results indicating that the rare ST299 (serotype 4c) may have a special ecological niche in aquatic products and associated environments, such as aquacultural water, containers, and chilling rooms, in China. Therefore, a comprehensive study is needed to explore whether ST299 (serotype 4c) were predominant in aquatic products and associated environments. To date, several well-known virulence genes of L. monocytogenes have been documented in previous studies, and have been used for assessing potential risk, by detecting the presence of these genes. In the present study, the 72 isolates harbored the nine classical virulence genes belonging to LIPI-1 and LIPI-2, including prfA, hly, actA, plcA, plcB, mpl, iap, inlA, and inlB. However, four isolates showed PMSCs in inlA, which significantly attenuates the ability to invade host cells (Nightingale et al., 2008; Van Stelten et al., 2010). According to the PMSC type of inlA reported in previous studies (Gelbicova et al., 2015; Moura et al., 2016), a novel type of PMSC in inlA was found at position 1605 (TGG→TGA) in isolate 2929-1LM. Interestingly, three fourths of isolates with PMSCs belonged to ST9 in this study. In contrast, none of the ST121 isolates was identified as PMSC, which was most identified as PMSCs in food and associated environments in the other countries (Hein et al., 2011; Ciolacu et al., 2015; Palma et al., 2017). In recent years, LIPI-3 genes were identified as important virulence factors that acts to target the host gut microbiota, responsible for the majority of listeriosis outbreaks (Cotter et al., 2008; Quereda et al., 2017), and human central nervous system (CNS) or MN listeriosis was mainly attributed to LIPI-4 (Maury et al., 2016). Eight isolates (11.11%) carried the llsX gene and mainly belonged to ST1, ST3, ST308, ST323, ST330, and ST619, whereas 15 isolates belonging to ST87 showed the presence of LIPI-4 in this study. LIPI-3 were mainly present in lineage I, included 21 STs belongs to 18 CCs (CC1, CC3, CC4, CC6, CC191, CC213, CC217, CC224, CC288, CC363, CC379, CC382 CC389, CC489, CC554, CC581, CC1000, and CC380) (Chen et al., 2018; Hilliard et al., 2018; Kim et al., 2018; Wang et al., 2018). While LIPI-4 were only reported in CC4 isolates by Maury et al. (2016), recent studies reported that LIPI-4 presented in several other lineage I CCs and in a single lineage III CC in China, the United States, and Ireland, namely ST4, ST87, ST213, ST217, ST363, ST382, ST388, ST663, ST1002, ST1166, and ST619 (Chen et al., 2018; Hilliard et al., 2018; Kim et al., 2018; Wang et al., 2018). Importantly, isolate 3907LM belonging to ST619 carried both LIPI-3 and LIPI-4. Isolates belonging to ST8/CC8 are also potential hypervirulent to humans, and are known to cause listeriosis in China as well as other countries (Althaus et al., 2014; Jensen et al., 2016; Oxaran et al., 2017; Wang et al., 2018). Unlike western countries, in which the majority of clinical isolates mainly belong to CC1, CC2, CC6, CC4, and CC101, which have been demonstrated to be strongly associated with human listeriosis (Maury et al., 2016; Hilliard et al., 2018; Kim et al., 2018), CC87 is predominant in human listeriosis in China (Huang et al., 2015; Wang et al., 2018). These results demonstrate that potential virulent strains isolated from aquatic products pose a potential health risk to consumers in China.
Conclusion
In conclusion, L. monocytogenes is an important foodborne pathogen worldwide. Of fresh aquatic products samples, which are popular among consumers in China, approximately 7.92% were positive for L. monocytogenes. Of these, 86.57% positive samples ranged from 0.3 to 10 MPN/g and 5.97% showed over 110 MPN/g by the MPN method, two samples (Tilapia mossambica, and grass carp) were intended to be eaten by raw. Serogroup I.1 (serotype 1/2a), I.2 (serotype 1/2b), and III (serotype 4c) were predominant in aquatic products and most of the common antibacterial agents are still effective for treating L. monocytogenes infection. The three most common STs were ST299, ST87, and ST8, indicating that the rare ST299 (serotype 4a, 4c) may have a special ecological niche in aquatic products and associated environments in China. PMSCs in inlA were found in four isolates, of which three belonged to ST9. A novel PMSC at position 1605 (TGG→TGA) in inlA was found in 2929-1LM. Several STs (ST87, ST8, ST1, ST3, ST308, ST323, ST330, and ST619) found in aquatic products have been previously reported in listeriosis outbreaks and sporadic cases. Although the occurrence of L. monocytogenes is low, potential virulent L. monocytogenes isolates may pose a potential risk to consumers. This study emphasizes the need for continuous surveillance for this pathogen in aquatic products in China.
Author Contributions
QW, JZ, and MC conceived and designed the experiments. MC, JC, and YC performed the experiments. LX, HZ, SW, and QY conducted bioinformatics analyses. MC, QW, JZ, and TL drafted the manuscript. QW, JB, and JW reviewed the manuscript. All authors read and approved the final manuscript.
Funding
We would like to acknowledge the financial support from the National Natural Science Foundation of China (31471664 and 31701718), the China Postdoctoral Science Foundation (2016M602447), Natural Science Foundation of Guangdong Province, China (2017A030313173), Pearl River S&T Nova Program of Guangzhou (201710010018), GDAS’ Special Project of Science and Technology Development (2017GDASCX-0201 and 2017GDASCX-0810), and the Science and Technology Planning Project of Guangdong Province (2016A010105012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the team of curators of the Institute Pasteur MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr/.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02215/full#supplementary-material
Footnotes
- ^http://www.stats.gov.cn/tjsj/ndsj/2017/indexch.htm
- ^http://bigsdb.pasteur.fr/listeria/listeria.html
References
Akrami-Mohajeri, F., Derakhshan, Z., Ferrante, M., Hamidiyan, N., Soleymani, M., Conti, G. O., et al. (2018). The prevalence and antimicrobial resistance of Listeria spp in raw milk and traditional dairy products delivered in Yazd, central Iran (2016). Food Chem. Toxicol. 114, 141–144. doi: 10.1016/j.fct.2018.02.006
Althaus, D., Lehner, A., Brisse, S., Maury, M., Tasara, T., and Stephan, R. (2014). Characterization of Listeria monocytogenes strains isolated during 2011-2013 from human infections in Switzerland. Foodborne Pathog. Dis. 11, 753–758. doi: 10.1089/fpd.2014.1747
Anonymous (2005). Commission Regulation (EC) No. 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs, O. J. EU L338. ιιBrussels: European Commission, 1–26.
Cartwright, E. J., Jackson, K. A., Johnson, S. D., Graves, L. M., Silk, B. J., and Mahon, B. E. (2013). Listeriosis outbreaks and associated food vehicles, United States, 1998-2008. Emerg. Infect. Dis. 19, 1–9. doi: 10.3201/eid1901.120393
Cetinkaya, F., Mus, T. E., Yibar, A., Guclu, N., Tavsanli, H., and Cibik, R. (2014). Prevalence, serotype identification by multiplex polymerase chain reaction and antimicrobial resistance patterns of Listeria monocytogenes isolated from retail foods. J. Food Saf. 34, 42–49. doi: 10.1111/jfs.12093
Chen, J., Jiang, L., Lv, Y., Liang, Q., Ding, X., He, Z., et al. (2013). Molecular epidemiology and pathogenic potential of Listeria monocytogenes in aquatic products. J. Chin. Inst. Food Sci. Technol. 3, 182–189.
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H., et al. (2018). Prevalence, potential virulence, and genetic diversity of Listeria monocytogenes isolates from edible mushrooms in Chinese markets. Front. Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, M., Wu, Q., Zhang, J., Wu, S., and Guo, W. (2015). Prevalence, enumeration, and pheno- and genotypic characteristics of Listeria monocytogenes isolated from raw foods in South China. Front. Microbiol. 6:1026. doi: 10.3389/fmicb.2015.01026
Chen, M., Wu, Q., Zhang, J., Yan, Z., and Wang, J. (2014). Prevalence and characterization of Listeria monocytogenes isolated from retail-level ready-to-eat foods in South China. Food Control 38, 1–7. doi: 10.1016/j.foodcont.2013.09.061
Chui, H., Wu, L., Qiu, Z., and Zhang, X. (2018). Survey of foodborne pathogens contamination in aquatic products in Henan, 2016. Chin. J. Health Lab. Technol. 28, 338–342.
Ciolacu, L., Nicolau, A. I., Wagner, M., and Rychli, K. (2015). Listeria monocytogenes isolated from food samples from a Romanian black market show distinct virulence profiles. Int. J. Food Microbiol. 209, 44–51. doi: 10.1016/j.ijfoodmicro.2014.08.035
Clayton, E. M., Hill, C., Cotter, P. D., and Ross, R. P. (2011). Real-time PCR assay to differentiate Listeriolysin S-positive and -negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 77, 163–171. doi: 10.1128/AEM.01673-10
Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing: 24th Informational Supplement(M100- S24). Wayne, PA: Clinical and Laboratory Standards Institute.
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4:e1000144. doi: 10.1371/journal.ppat.1000144
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/jcm.42.8.3819-3822.2004
European Food Safety Authority [EFSA] and European Centre for Disease Control [ECDC] (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329.
Fallah, A. A., Saei-Dehkordi, S. S., and Mahzounieh, M. (2013). Occurrence and antibiotic resistance profiles of Listeria monocytogenes isolated from seafood products and market and processing environments in Iran. Food Control 34, 630–636. doi: 10.1016/j.foodcont.2013.06.015
Felicio, M. T., Hogg, T., Gibbs, P., Teixeira, P., and Wiedmann, M. (2007). Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity, including virulence-attenuated isolates. Appl. Environ. Microbiol. 73, 3887–3895. doi: 10.1128/AEM.02912-06
Feng, Y. F., Wu, S. Y., Varma, J. K., Klena, J. D., Angulo, F. J., and Ran, L. (2013). Systematic review of human listeriosis in China, 1964-2010. Trop. Med. Int. Health 18, 1248–1256. doi: 10.1111/tmi.12173
Garedew, L., Taddese, A., Biru, T., Nigatu, S., Kebede, E., Ejo, M., et al. (2015). Prevalence and antimicrobial susceptibility profile of Listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 15:100. doi: 10.1186/s12866-015-0434-4
Gelbicova, T., Kolackova, I., Pantucek, R., and Karpiskova, R. (2015). A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 38, 293–296.
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., et al. (2001). Comparative genomics of Listeria species. Science 294, 849–852. doi: 10.1126/science.1063447
Gombas, D. E., Chen, Y., Clavero, R. S., and Scott, V. N. (2003). Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66, 559–569.
Handa-Miya, S., Kimura, B., Takahashi, H., Sato, M., Ishikawa, T., Igarashi, K., et al. (2007). Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 117, 312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003
Harakeh, S., Saleh, I., Zouhairi, O., Baydoun, E., Barbour, E., and Alwan, N. (2009). Antimicrobial resistance of Listeria monocytogenes isolated from dairy-based food products. Sci. Total Environ. 407, 4022–4027. doi: 10.1016/j.scitotenv.2009.04.010
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stessl, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi: 10.1128/AEM.02159-10
Hilliard, A., Leong, D., O’Callaghan, A., Culligan, EP., Morgan, C. A., DeLappe, N., et al. (2018). Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9:E171. doi: 10.3390/genes9030171
Huang, Y. T., Ko, W. C., Chan, Y. J., Lu, J. J., Tsai, H. Y., Liao, C. H., et al. (2015). Disease burden of invasive listeriosis and molecular characterization of clinical isolates in Taiwan, 2000-2013. PLoS One 10:e0141241. doi: 10.1371/journal.pone.0141241
Hunter, P. R., and Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 26, 2465–2466
International Commission on Microbiological Specifications for Foods [ICMSF] (2011). Microorganisms in Foods 8. Use of Data for Assessing Process Control and Product Acceptance. Springer. International Commission on Microbiological Specifications for Foods. New York, NY: Springer.
Jamali, H., Paydar, M., Ismail, S., Looi, C. Y., Wong, W. F., Radmehr, B., et al. (2015). Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 15:144. doi: 10.1186/s12866-015-0476-7
Jensen, A. K., Björkman, J. T., Ethelberg, S., Kiil, K., Kemp, M., and Nielsen, E. M. (2016). Molecular typing and epidemiology of human listeriosis cases, Denmark, 2002-2012. Emerg. Infect. Dis. 22, 625–633. doi: 10.3201/eid2204.150998
Jonquieres, R., Bierne, H., Mengaud, J., and Cossart, P. (1998). The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66, 3420–3422.
Kim, S. W., Haendiges, J., Keller, E. N., Myers, R., Kim, A., Lombard, J. E., et al. (2018). Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS One 13:e0197053. doi: 10.1371/journal.pone.0197053
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H., Wang, P., Lan, R., Luo, L., Cao, X., Wang, Y., et al. (2018). Risk factors and level of Listeria monocytogenes contamination of raw pork in retail markets in China. Front. Microbiol. 9:1090. doi: 10.3389/fmicb.2018.01090
Lomonaco, S., Decastelli, L., Nucera, D., Gallina, S., Manila Bianchi, D., and Civera, T. (2009). Listeria monocytogenes in Gorgonzola: subtypes, diversity and persistence over time. Int. J. Food Microbiol. 128, 516–520. doi: 10.1016/j.ijfoodmicro.2008.10.009
Ma, A. (2015). Isolation and Characterization of Listeria Monocytogenes Isolated from Raw Meat and Aquatic Product Samples. Beijing: Chiense center for disease control and prevention.
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Ministry of Health of People’s Republic of China (2010). National Food Safety Standard Food Microbiological Examination: Listeria monocytogenes (GB4789.30-2010). Beijing: Ministry of Health of People’s Republic of China.
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Nightingale, K. K., Ivy, R. A., Ho, A. J., Fortes, E. D., Njaa, B. L., Peters, R. M., et al. (2008). inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74, 6570–6583. doi: 10.1128/aem.00997-08
Nightingale, K. K., Windham, K., Martin, K. E., Yeung, M., and Wiedmann, M. (2005). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005
Obaidat, M. M., Bani Salman, A. E., Lafi, S. Q., and Al-Abboodi, A. R. (2015). Characterization of Listeria monocytogenes from three countries and antibiotic resistance differences among countries and Listeria monocytogenes serogroups. Lett. Appl. Microbiol. 60, 609–614. doi: 10.1111/lam.12420
Olier, M., Pierre, F., Rousseaux, S., Lemaitre, J. P., Rousset, A., Piveteau, P., et al. (2003). Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71, 1217–1224. doi: 10.1128/Iai.71.3.1217-1224.2003
Orsi, R. H., den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Oxaran, V., Lee, S. H. I., Chaul, L. T., Corassin, C. H., Barancelli, G. V., Alves, V. F., et al. (2017). Listeria monocytogenes incidence changes and diversity in some Brazilian dairy industries and retail products. Food Microbiol. 68, 16–23. doi: 10.1016/j.fm.2017.06.012
Pagadala, S., Parveen, S., Rippen, T., Luchansky, J. B., Call, J. E., Tamplin, M. L., et al. (2012). Prevalence, characterization and sources of Listeria monocytogenes in blue crab (Callinectes sapidus) meat and blue crab processing plants. Food Microbiol. 31, 263–270. doi: 10.1016/j.fm.2012.03.015
Palma, F., Pasquali, F., Lucchi, A., Cesare, A., and Manfreda, G. (2017). Whole genome sequencing for typing and characterisation of Listeria monocytogenes isolated in a rabbit meat processing plant. Ital. J. Food Saf. 6:6879. doi: 10.4081/ijfs.2017.6879
Quereda, J. J., Nahori, M. A., Meza-Torres, J., Sachse, M., Titos-JimeÂnez, P., Gomez-Laguna, J., et al. (2017). Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. mBio 8:e00259-17. doi: 10.1128/mBio.00259-17
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Rousseaux, S., Olier, M., Lemaitre, J. P., Piveteau, P., and Guzzo, J. (2004). Use of PCR-Restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade caco-2 cells. Appl. Environ. Microbiol. 70, 2180–2185. doi: 10.1128/70.4.2180-2185.2004
Sharma, S., Sharma, V., Dahiya, D. K., Khan, A., Mathur, M., and Sharma, A. (2017). Prevalence, virulence potential, and antibiotic susceptibility profile of Listeria monocytogenes isolated from bovine raw milk samples obtained from Rajasthan, India. Foodborne Pathog. Dis. 14, 132–140. doi: 10.1089/fpd.2016.2118
Shen, J., Rump, L., Zhang, Y., Chen, Y., Wang, X., and Meng, J. (2013). Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 35, 58–64. doi: 10.1016/j.fm.2013.02.014
Song, X., Pei, X., Xu, H., Yang, D., and Zhu, J. (2015). Risk ranking of Listeria monocytogenes contaminated ready-to-eat foods at retail for sensitive population in China. Chin. J. Food Hyg. 27, 447–450.
Terentjeva, M., Eizenberga, I., Valcina, O., Novoslavskij, A., Strazdina, V., and Berzins, A. (2015). Prevalence of foodborne pathogens in freshwater fish in Latvia. J. Food Prot. 78, 2093–2098. doi: 10.4315/0362-028x.Jfp-15-121
U.S. Department of Agriculture (1998). “Isolation and identification of Listeria monocytogenes from meat, poultry and egg products,” in Microbiology Guide Book Chap. 8, 3rd Edn, eds B. P. Dey and C. P. Lattuada (Washington, DC: Government Printing Office).
Van Stelten, A., and Nightingale, K. K. (2008). Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74, 7365–7375. doi: 10.1128/AEM.01138-08
Van Stelten, A., Simpson, J., Ward, T., and Nightingale, K. (2010). Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis Cases. Appl. Environ. Microbiol. 76, 2783–2790. doi: 10.1128/AEM.02651-09
Wang, H., Luo, L., Zhang, Z., Deng, J., Wang, Y., Miao, Y., et al. (2018). Prevalence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from listeriosis cases. Front. Med. 12, 104–112. doi: 10.1007/s11684-017-0593-9
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., Wang, X., et al. (2015). Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg. Microbes Infect. 4:e50. doi: 10.1038/emi.2015.50
Wang, Y., Zhao, A., Zhu, R., Lan, R., Jin, D., Cui, Z., et al. (2012). Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 12:119. doi: 10.1186/1471-2180-12-119
Wieczorek, K., and Osek, J. (2017). Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 64, 164–171. doi: 10.1016/j.fm.2016.12.022
Wilson, A., Gray, J., Chandry, P. S., and Fox, E. M. (2018). Phenotypic and Genotypic analysis of antimicrobial resistance among Listeria monocytogenes isolated from Australian food production chains. Genes 9:E80. doi: 10.3390/genes9020080
Keywords: Listeria monocytogenes, multi-locus sequence typing, antibiotic resistance, aquatic products, llsX, cellobiose family PTS, premature stop codon
Citation: Chen M, Cheng J, Wu Q, Zhang J, Chen Y, Xue L, Lei T, Zeng H, Wu S, Ye Q, Bai J and Wang J (2018) Occurrence, Antibiotic Resistance, and Population Diversity of Listeria monocytogenes Isolated From Fresh Aquatic Products in China. Front. Microbiol. 9:2215. doi: 10.3389/fmicb.2018.02215
Received: 29 May 2018; Accepted: 30 August 2018;
Published: 19 September 2018.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Marcello Trevisani, Università degli Studi di Bologna, ItalyBeatrix Stessl, Veterinärmedizinische Universität Wien, Austria
Copyright © 2018 Chen, Cheng, Wu, Zhang, Chen, Xue, Lei, Zeng, Wu, Ye, Bai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, wuqp203@163.com
 Moutong Chen
Moutong Chen Jianheng Cheng1
Jianheng Cheng1 Qingping Wu
Qingping Wu Jumei Zhang
Jumei Zhang Liang Xue
Liang Xue Haiyan Zeng
Haiyan Zeng Shi Wu
Shi Wu Juan Wang
Juan Wang