- 1Department of Pathology, University of Texas Southwestern Medical Center and Children’s Medical Center, Dallas, TX, USA
- 2Department of Pediatrics, University of Texas Southwestern Medical Center and Children’s Medical Center, Dallas, TX, USA
- 3Department of Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
- 4AmeriPath/Quest Diagnostics, Dallas, TX, USA
Approximately 20% of unselected cases and 30% cytogenetically diploid cases of acute myeloid leukemia (AML) and 80% of grade II–III gliomas and secondary glioblastomas carry mutations in the isocitrate dehydrogenase (IDH) 1 and 2 genes. IDH1/2 mutations prevent oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG) and modulate the function of IDH (neomorphic activity) thereby facilitating reduction of α-KG to D-2-hydroxyglutarate (D-2HG), a putative oncometabolite. D-2HG is thought to act as a competitive inhibitor of α-KG-dependent dioxygenases that include prolyl hydroxylases and chromatin-modifying enzymes. The end result is a global increase of cellular DNA hypermethylation and alterations of the cellular epigenetic state, which has been proposed to play a role in the development of a variety of tumors. In this review, we provide an update on potential molecular mechanisms linking IDH1/2 mutations and the resulting oncometabolite, D-2HG, with malignant transformation. In addition, in patients with AML and glioma we focus on the associations between IDH1/2 mutations and clinical, morphologic, cytogenetic, and molecular characteristics.
Introduction
There is increasing evidence that alterations in cellular metabolism are involved in the pathogenesis of many cancers. Notably, mutations in three different enzymes in the tricarboxylic acid cycle are associated with tumorigenesis. These enzymes are succinate dehydrogenease, fumarate hydratase, and isocitrate dehydrogenases (IDH1 and 2) (Soga, 2013). As many of the intermediates in the tricarboxylic acid cycle are important for synthesis of nucleotides, lipids, and amino acids, it is not surprising that alterations in this metabolic pathway may facilitate the development of cancers.
Somatic heterozygous mutations in IDH1 and IDH2 have been recognized recently in a number of cancers. The first reported mutation in an IDH-family gene was identified in a metastatic colon cancer in 2006 (Sjöblom et al., 2006). In 2008, sequencing of glioblastoma multiforme (GBM) tumor samples identified IDH1 mutations at R132 (IDH1R132) in 12% of tumors (Parsons et al., 2008). In 2009, targeted mutational analysis of a broader group of central nervous system tumors detected IDH1R132 mutations in 70% of grade II and III gliomas (Yan et al., 2009). Also in 2009, whole-genome sequencing of a patient with acute myeloid leukemia (AML) identified an IDH1R132 mutation (Mardis et al., 2009). Subsequent analyzes on a large number of AML patients treated on multiple clinical protocols confirmed the presence of IDH1R132 mutations and also identified IDH2R140 and IDH2R172 mutations in ∼20% of adult-onset AML with normal cytogenetics (Mardis et al., 2009; Abbas et al., 2010; Boissel et al., 2010; Green et al., 2010; Gross et al., 2010; Ho et al., 2010; Marcucci et al., 2010; Paschka et al., 2010; Schnittger et al., 2010; Thol et al., 2010a). These mutations also have been identified, albeit at much lower frequency, in myelodysplastic syndromes and myeloproliferative neoplasms (Abbas et al., 2010; Green and Beer, 2010; Paschka et al., 2010; Tefferi et al., 2010; Thol et al., 2010b). Recently, a number of other tumors have been identified to harbor IDH mutations including angioimmunoblastic T-cell lymphoma (Cairns et al., 2012), chondrosarcoma (Amary et al., 2011), and intrahepatic cholangiocarcinoma (Borger et al., 2012; Wang et al., 2012).
Isocitrate dehydrogenase 1 and 2 mutations reduce the affinity of their respective enzymes for isocitrate and increase their affinity for α-ketoglutarate (α-KG) and reduced nicotinamide adenine dinuceotide phosphate (NADPH). This reduced affinity impedes oxidative decarboxylation of isocitrate to α-KG and confers a novel enzymatic activity that facilitates reduction of α-KG to D-2-hydroxyglutarate (D-2HG) (Dang et al., 2009; Pietrak et al., 2011). Excess accumulation of D-2HG has been demonstrated in a subset of cases of glioma and AML with IDH1 or IDH2 mutations (Dang et al., 2009; Frezza et al., 2010; Gross et al., 2010; Ward et al., 2010; Andersson et al., 2011; Rakheja et al., 2011a; Choi et al., 2012). Others have suggested that overproduction of D-2HG promotes oncogenesis (Koivunen et al., 2012; Losman et al., 2013) and therefore IDH1 and IDH2 mutations are likely the integrally involved in the pathogenesis of malignant transformation (i.e., driver mutations) rather than epiphenomena.
In this review, we summarize the frequency and role of IDH1 and IDH2 mutations in gliomas and myeloid neoplasms, the latter with an emphasis on AML, and the association of these mutations with clinical, morphologic, cytogenetic, and molecular characteristics. We also provide an update on potential molecular mechanisms linking mutant IDH1 and IDH2 and their oncometabolite, D-2HG, with malignant transformation. The data suggest that IDH1/2 mutations constitute an early mutational event which affects the cellular epigenetic state in a subset of gliomas and AML, an important consideration for the development of therapeutic agents.
Biochemistry of Isocitrate Dehydrogenases
In mammalian cells, three classes of IDH isoenzymes exist: IDH1, IDH2, and IDH3 (Plaut et al., 1983). The human IDH1 enzyme is localized in the cytoplasm and peroxisomes and is encoded by IDH1 at chromosome band 2q33.3 (Narahara et al., 1985; Geisbrecht and Gould, 1999). The IDH2 enzyme is localized in mitochondria and is encoded by IDH2 at chromosome band 15q26.1 (Oh et al., 1996). The IDH1 and IDH2 enzymes are homodimeric, nicotinamide adenine dinucleotide phosphate-dependent, and catalyze the oxidative decarboxylation of isocitrate to α-KG, using NADP as a cofactor to yield NADPH (Haselbeck and McAlister-Henn, 1993). Mitochondrial NADPH participates in energy metabolism, as a part of the tricarboxylic acid cycle, and cytosolic NADPH, which is also produced in the pentose phosphate pathway, functions in anabolic processes and redox control. It is therefore reasonable to expect that changes in one or more of these processes occur in tumors that carry an IDH1 or IDH2 mutation. The NAD-dependent IDH3 is a heterotetramer composed of two alpha subunits (gene on chromosome 15), one beta subunit (gene on chromosome 20), and one gamma subunit (gene on chromosome X). IDH3 has not been shown to be mutated in cancer and therefore is not further addressed in this review.
Neomorphic Activity of IDH1/IDH2 Mutant Enzymes
Most cancer-associated enzyme mutations result in either catalytic inactivation or constitutive activation. A common feature of IDH1 and IDH2 mutations observed in AML and gliomas, however, is the apparent acquisition of enzymatic activity not shared by wild-type enzyme, known as neomorphic activity. The product of IDH1/IDH2 mutations, D-2HG, can be detected in tumor samples.
To date, all reported IDH1 or IDH2 mutations are heterozygous, with cancer cells retaining one wild-type copy of the respective IDH1 or IDH2 allele. All IDH mutations identified involve a single amino acid substitution at an arginine (R) residue, R132 in IDH1, or R140 or R172 in IDH2; no inactivating (frame-shift or protein-truncation) mutations have been found. These residues create hydrophilic interactions that allow the binding of isocitrate (Xu et al., 2004). The residues that are substituted for arginine are wide ranging, and include histidine, serine, cysteine, glycine, or leucine, suggesting that it is the replacement of arginine, and not the specific amino acid substituted, that supports tumorigenesis.
In vitro enzymatic analysis has confirmed that mutant IDH1 and IDH2 have altered substrate specificity and directionality (Dang et al., 2009; Zhao et al., 2009; Gross et al., 2010; Ward et al., 2010; Andersson et al., 2011; Pietrak et al., 2011). IDH1 R132 and IDH2 R172 are analogous residues that interact with the β-carboxyl of isocitrate. Normally, IDH functions as a homodimer; by contrast mutant IDH molecules in tumor cells form heterodimers with wild-type molecules. Whereas WT IDH converts isocitrate to α-KG, mutants of IDH can no longer catalyze this reaction and instead reduce α-KG to the D-stereoisomer of 2-hydroxyglutarate (D-2HG) (Figure 1). Structural comparisons of mutant and wild type IDH1 have revealed that mutations at R132 of IDH1 result in impaired enzyme affinity for substrate and dominantly inhibit wild-type IDH1 activity through the formation of catalytically inactive heterodimers. The R132 mutation also changes the orientation of the catalytic site so that the enzyme binds NADPH and α-KG, explaining the formation of the new product, rather than simply catalyzing the reaction in reverse. In support of the results of in vitro enzymatic analyzes, D-2HG levels are 100-fold higher in cases of glioma and AML that carry IDH1 or IDH2 mutations as compared with tumors with wild type IDH (Dang et al., 2009; Gross et al., 2010; Ward et al., 2010; Andersson et al., 2011). Therefore, detecting D-2HG in tumor samples can reliably predict patients with tumor-associated IDH mutations (Gross et al., 2010).
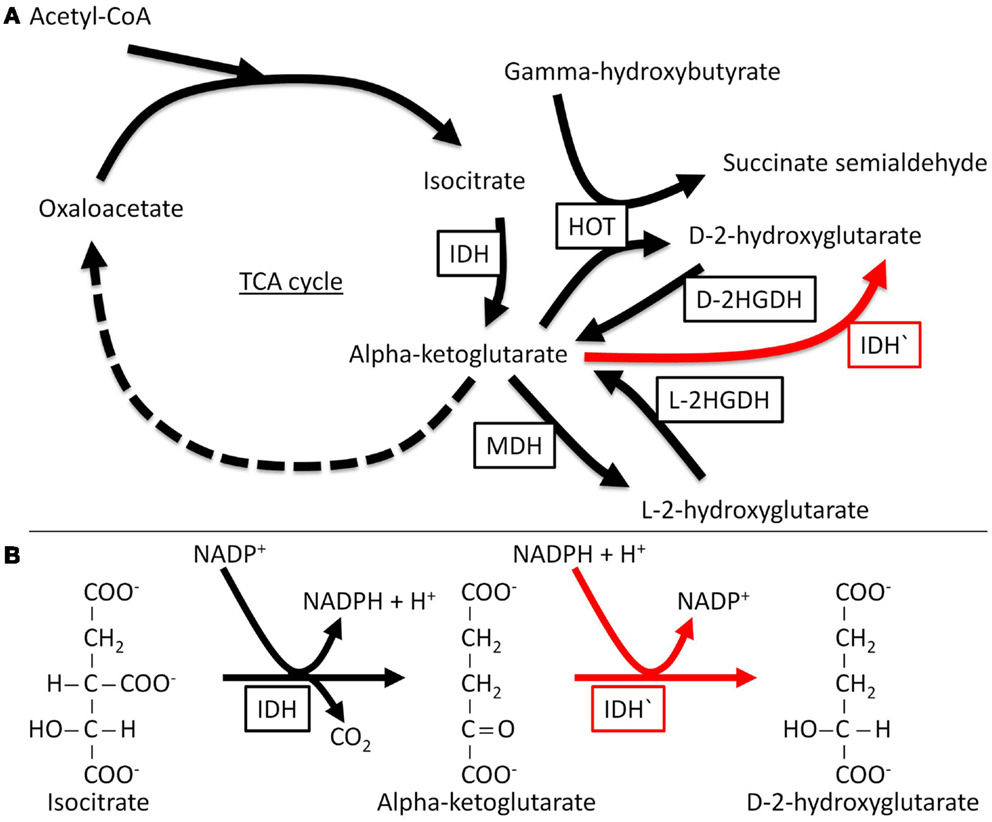
Figure 1. Normal and neomorphic reactions catalyzed by IDH. The normal enzyme catalyzes the nicotinamide adenine dinucleotide phosphate (NADP+)-dependent conversion of isocitrate to α-KG, while the mutant enzyme (neomorphic reaction in red) catalyzes the reduction of α-KG to D-2HG, a reaction that depends on NADPH (the reduced form of NADP+). D-2HGDH, D-2-hydroxyglutarate dehydrogenase; HOT, hydroxyacid oxoacid transhydrogenase; IDH, isocitrate dehydrogenase; IDH′, mutant isocitrate dehydrogenase; L-2HGDH, L-2-hydroxyglutarate dehydrogenase; MDH, malate dehydrogenase; TCA, tricarboxylic acid.
Mechanism of IDH Mutations in Tumorigenesis
The discovery of IDH mutations has led to renewed efforts to decipher the role of altered metabolic processes in cancer (Prensner and Chinnaiyan, 2011). The normal metabolic role of 2-hydroxyglutarate is not completely understood, but 2-hydroxyglutarate is not foreign to cells. It can be generated by specific α-KG reductase enzymes and oxidized back to α-KG by 2-hydroxyglutarate dehydrogenases (2HGDH). There are two enantiomers of 2-hydroxyglutarate with specific 2HGDH for each. Mutations in 2HGDH cause pathological accumulation of 2-hydroxyglutarate with different clinical features based on the enantiomer involved. Pathological accumulation of the L-2-hydroxyglutarate enantiomer (L-2HG) is known to occur in a rare inherited autosomal recessive disease characterized by encephalopathy and increased risk of brain tumors, including gliomas (Aghili et al., 2009). Accumulation of the D-2-hydroxyglutarate enantiomer (D-2HG) is associated with encephalopathy and cardiomyopathy, but not with tumors (Struys, 2006). Mutant IDH in cases of AML and glioma generates D-2HG and not the L-enantiomer.
As a result of IDH mutation, the oncometabolite of D-2HG is produced and α-KG is reduced. The combination of these two events may be important. At the high levels of D-2HG that have been observed in cases of AML and glioma (more than 10 mM) (Dang et al., 2009; Gross et al., 2010; Ward et al., 2010; Andersson et al., 2011), D-2HG may lead to DNA damage mediated by elevated levels of reactive oxygen species (Zhao et al., 2009; Ward et al., 2010), induction of redox stress owing to impairment of the respiratory chain, promotion of oncogenesis by promoting cytokine independence, and blocking differentiation in hematopoietic cells (Losman et al., 2013). Notably, D-2HG and α-KG are structurally similar except that the oxygen atom linked to C2 in α-KG is replaced by a hydroxyl group in D-2HG. This structural similarity suggests that D-2HG might exert its oncogenic effects through competitive inhibition with α-KG-dependant dioxygenases (Figueroa et al., 2010; Chowdhury et al., 2011; Xu et al., 2011). These enzymes include prolyl hydroxylases, and chromatin-modifying enzymes, such as histone demethylases and TET 5-methylcytosine hydroxylases.
TET2 is an α-KG-utilizing enzyme that hydroxylates 5-methylcytosine as a step in demethylation of DNA (Ito et al., 2010). Recently, all TET family members including TET2 were shown to catalyze the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (5-OH-MeC) (Ito et al., 2010). The fact that IDH1 and IDH2 mutations are mutually exclusive, and that IDH1/2 mutations are mutually exclusive with TET2 mutations, suggests that the biological effects of mutant IDH1/2 and TET are similar (Figueroa et al., 2010; Chou et al., 2011a; Patel et al., 2012). AMLs with TET2 or IDH mutation, and IDH mutated gliomas have more pronounced hypermethylation profiles than their non-mutated counterparts and share overlapping epigenetic signatures (Figueroa et al., 2010; Xu et al., 2011). Most importantly, expression of IDH1/2 mutants induces a global increase in global DNA hypermethylation and inhibits TET2-induced cytosine 5-hydroxymethylation due to reduction of α-KG. These data suggest that TET2 and IDH1/2 mutations characterize a distinctive group of AML cases in which the epigenetic state is altered. Many of the genes hypermethylated in the context of IDH1/2 mutated AML contain DNA-binding motifs for GATA1/GATA2 and EVI1, transcription factors known to play a role in leukemogenesis as well as normal myeloid differentiation (Figueroa et al., 2010).
Recent analysis of gliomas also has shown that IDH1 gene mutations represent a molecular basis for the epigenetic changes described above. In one study, the methylation profiles of immortalized primary human astrocytes in which either wild-type IDH1 or R132H IDH1 was introduced were analyzed using a genome-wide platform. Expression of wild-type IDH1 led to hypomethylation at several loci, compared with a marked increase in methylation seen in the R132H IDH1 mutants. The methylation profiles of the IDH1 mutants mirrored the CpG island methylator phenotype (CIMP), a unique phenotype seen in several tumors with extensive epigenetic changes (Turcan et al., 2012).
Histone lysine demethylases are another group of α-KG-dependent enzymes. In vitro, D-2HG can inhibit histone demethylases (Chowdhury et al., 2011; Xu et al., 2011). Experimental data suggest that inhibition of histone demethylases can induce DNA methylation (Lu et al., 2012). Methylated histones, such as H3K9me3 (trimethylation of lysine 9 of histone H3), may serve as a platform for recruiting a complex of heterochromatin protein 1 (HP1), histone methyltransferase, and DNA methytransferase. Direct cooperation between these enzymes could then provide a mechanism of coordinated histone and DNA methylation involved in epigenetic regulation of DNA (Estève et al., 2006).
Hypoxia-inducible factor 1α (HIF-1α) is a transcription factor that has functions linked to metabolism, angiogenesis, and tumorigenesis. HIF-1α protein levels are downregulated under normoxic conditions by prolyl hydroxylase-mediated hydroxylation and subsequent hydroxylation-targeted ubiquitination (Bruick and McKnight, 2001). As α-KG is required by prolyl hydroxylases, a reduction in α-KG levels in cancer cells with mutant IDH may lead to inhibition of prolyl hydroxylases and stabilization of HIF-1α (Zhao et al., 2009). An alternative mechanism proposed by Koivunen et al. (2012) suggests that D-2HG acts as a cofactor to promote the hydroxylase activity of the Eg1N prolyl-4-hydroxylase and subsequent downregulation of HIF1α, contributing to the pathogenesis of IDH mutant gliomas (Koivunen et al., 2012). These data seem incompatible with the finding that HIF1α protein levels are increased in IDH mutant tumors. However, these data may suggest that D-2HG quantitatively shifts the dose-response linking HIF activation to hypoxia, leading to a blunted HIF response for a given level of hypoxia. In support of this idea, HIF elevation in IDH mutant tumors is usually confined to areas of necrosis and presumed severe hypoxia (Williams et al., 2011).
Collectively, the data support the concept that IHD mutations promote oncogenesis through D-2HG-induced inhibition of α-KG-dependent enzymes, such as TET2 and histone demethylase. This idea further suggests a paradigm whereby oncogenic alterations in core cellular metabolic pathways could lead to neoplastic progression by dysregulating the epigenetic machinery in hematopoietic and glial progenitors. Mutations of IDH1 and IDH2 in combination with microenvironmental effects in certain tumor types are likely the driver mutations that are responsible for the malignant phenotype, rather than simply epiphenomena (Dang et al., 2009; Figueroa et al., 2010; Xu et al., 2011; Koivunen et al., 2012; Losman et al., 2013).
IDH Mutations in AML
Mardis and colleagues (Mardis et al., 2009) used next generation (massive parallel) DNA sequencing analysis of the genome of an AML patient with a normal karyotype and discovered an IDH1 mutation. Subsequently, this finding was validated in 187 AML patients in whom 8% had IDH1 mutations. To date, there have been ∼20 published studies on IDH1 and IDH2 mutations that have included ∼8,000 adult and ∼800 pediatric patients (Mardis et al., 2009; Abbas et al., 2010; Boissel et al., 2010; Chou et al., 2010, 2011b; Figueroa et al., 2010; Gross et al., 2010; Marcucci et al., 2010; Paschka et al., 2010; Schnittger et al., 2010; Thol et al., 2010a; Wagner et al., 2010; Ward et al., 2010; Andersson et al., 2011; Damm et al., 2011; Green et al., 2011; Pigazzi et al., 2011; Rakheja et al., 2011a; Patel et al., 2012). These studies have focused on the frequency and prognostic influence of IDH mutations in the context of other genetic mutations and prognostic markers.
Isocitrate dehydrogenase 1/2 mutations are almost mutually exclusive in AML, as only rare cases (<0.5%) harbor both IDH1 and IDH2 mutations (Abbas et al., 2010; Paschka et al., 2010; Green et al., 2011). This low frequency of concurrent mutations suggests that the biological effects of IDH1 and IDH2 are similar, and this idea is corroborated by the similar impact of these mutations on the distribution of cytosine methylation and the global DNA methylation profile in AML cells (Figueroa et al., 2010).
For IDH1R132 mutations in AML, as in other tumors, five major different amino acid substitutions for arginine (R) have been detected: cysteine (R132C), leucine (R132L), glycine (R132G), histidine (R132H), and serine (R132S) (Mardis et al., 2009; Abbas et al., 2010; Chou et al., 2010; Ho et al., 2010; Marcucci et al., 2010; Schnittger et al., 2010; Wagner et al., 2010; Patel et al., 2011a). R132C (∼30%) and R132H (∼50%) are the most common mutations in AML. The mutational profile is slightly different in gliomas, in which R132H (∼90%) is most common and R132C is uncommon (∼4%) (Yan et al., 2009; Chou et al., 2010). For IDH2R140 mutations, three different amino acid substitutions have been detected: glutamine (R140Q), leucine (R140L), and tryptophan (R140W). Of these mutations, R140Q is the most common by far, seen in ∼95% of AMLs with mutated IDH2 at R140. For IDH2R172 mutations, almost all involve replacement of arginine by lysine (R172K), except rare cases in which arginine is replaced by methionine (R172M) (Chou et al., 2011b).
In unselected adults with AML, in a total of 6,877 patients, IDH1 and IDH2 mutations have been identified in 7.3 and 9.7% of cases, respectively. The frequencies of IDH1 and IDH2 mutations are higher in cytogenetically normal (CN) versus abnormal AML patients (11 versus 3.5% for IDH1 mutations; and 16 versus 3.8% for IDH2 mutations), placing them among the most common molecular aberrations in CN-AML. Within the group of IDH2 mutations, IDH2R140 mutations are most common (∼80%) whereas IDH2R172 mutations occur in ∼2% of unselected AML and about 2.5% of CN-AML patients.
Acute myeloid leukemia patients with IDH1R132 or IDH2R140 mutations are more frequently older at diagnosis as compared with wild type AML patients. AML patients with IDH2R172 mutation are usually older with lower white blood cell (WBC) counts. The frequency of IDH1/2 mutations is substantially lower in pediatric (∼1–2%) than in adult patients with AML. IDH2R172 mutations have not been observed in children (Marcucci et al., 2010). In most studies to date, there has been little focus correlating IDH1/2 mutations with morphologic findings in AML. Most cases with IDH1/2 mutations have been classified as AML, not otherwise specified, with or without maturation. A rare morphologic subset of AML characterized by cuplike nuclei appears to commonly carry IDH1/2 mutations. In one study, 8 of 12 (67%) patients with AML with cuplike nuclei patients harbored either IDH1R132 or IDH2R140 mutations (Rakheja et al., 2011a). AML with cuplike nuclei also have a high frequency of NPM1 mutations and FLT3 internal tandem duplication (FLT3-ITD) (Chen et al., 2009).
Patients with AML carrying IDH1/2 mutations have a higher frequency of NPM1 mutations compared with AML wild type for IDH1/2. In general, IDH1/2 mutations are less frequent in AML patients with activating FLT3 mutations, CEBPA mutations, and are largely absent in patients with AML associated with recurrent chromosomal abnormalities, such as t(15;17), t(8;21), inv(16) (Chou et al., 2010, 2011b; Patel et al., 2011a). IDH2R172 mutation also appears to be virtually mutually exclusive with other genetic abnormalities (Marcucci et al., 2010; Paschka et al., 2010; Green et al., 2011; Patel et al., 2011a), thereby identifying a novel subset of patients among the ∼3% of CN-AML adult patients for whom no prognostic gene mutation has been reported to date.
The prognostic effect of IDH mutations in adult AML patients has been intensively studied, but remains a matter of discussion. In general, there have been no differences in response to therapy and survival between IDH1/2-mutated versus IDH1/2-wild type AML patients (Abbas et al., 2010; Chou et al., 2010; Thol et al., 2010a; Wagner et al., 2010; Marcucci et al., 2011). The impact of an IDH1 or IDH2 mutation, however, does seem to have prognostic importance if results are stratified according to cytogenetic data, FLT3-ITD and NPM1 mutation status, and type of IDH mutation (Boissel et al., 2010; Green et al., 2010, 2011; Marcucci et al., 2010; Paschka et al., 2010; Schnittger et al., 2010). IDH1 mutations may predict higher risk of relapse and shorter survival in the subset of CN-AML patients with mutated NPM1 and absence of FLT3-ITD (molecular low-risk group) (Marcucci et al., 2010; Paschka et al., 2010). In addition, there is some evidence that IDH2R172 mutations confer a poorer prognosis whereas IDH2R140 mutations have no impact on survival or may confer a favorable outcome (Boissel et al., 2010; Marcucci et al., 2010; Chou et al., 2011b; Green et al., 2011; Patel et al., 2012). Future prospective studies and clinical trials are needed to assess the prognostic impact of each type of IDH mutation in AML patients within the context of other molecular aberrations.
In pediatric AML patients, there is an association between NPM1 and IDH mutations, as has been described in adult AML patients. However, in contrast with adults, IDH mutations are not associated with a normal karyotype in childhood AML. Instead, IDH mutations have been observed in pediatric AML patients with good-risk cytogenetics, t(8;21), and t(15;17). In addition, neither type of IDH mutation occurs in children<3 years, affirming the distinctiveness of infant AML.
IDH Mutations in Myelodysplastic Syndromes and Myeloproliferative Neoplasms
Isocitrate dehydrogenase mutations occur at low frequency in patients with myelodysplastic syndromes (3.6–5%) and patients with Philadelphia chromosome/BCR-ABL1 negative myeloproliferative neoplasms in chronic phase (2–4%). Mutated cases have an increased frequency of progression to AML, ranging from 7.5 to 31% in different studies (Abbas et al., 2010; Green and Beer, 2010; Pardanani et al., 2010; Tefferi et al., 2010, 2011; Thol et al., 2010b). In patients with myeloproliferative neoplasms characterized by JAK2 and IDH1/2 mutations, it is tempting to speculate that these gene mutations have an additive or cooperative effect to facilitate leukemogenesis. The JAK2 mutation may offer a proliferative advantage whereas IDH1/2 mutations may disrupt epigenetic remodeling. Mutations in IDH1 or IDH2 mutations are absent or very rare in patients with chronic myelogenous leukemia in chronic phase (Abbas et al., 2010).
IDH Mutations in Gliomas
The first report of an IDH mutation in gliomas occurred in 2008 after over 20,000 protein coding genes were analyzed in 22 human GBM samples (Parsons et al., 2008). In this study, IDH1R132 point mutations were detected in 12% of GBM samples. The authors observed that IDH1 mutations occurred in younger adults, in most of the secondary GBM samples, and that mutation was associated with increased overall survival. The fact that IDH1 mutations occurred in secondary GBM prompted the same group to determine the frequency of IDH1 mutations in the low-grade gliomas. Targeted sequencing of IDH1 and IDH2 in 445 central nervous system tumors revealed IDH1/2 mutations in 90% of cases of diffuse astrocytoma (grade II), 84% of oligodendroglioma (grade II), 73% of anaplastic astrocytoma (grade III), 94% of anaplastic oligodendroglioma (WHO grade III), and 85% of secondary GBM (grade IV) (Yan et al., 2009). Several follow-up studies encompassing thousands of central nervous system neoplasms have found similar frequencies of IDH1 and IDH2 mutations (Hartmann et al., 2009; Nobusawa et al., 2009; van den Bent et al., 2010; Jha et al., 2011; Thon et al., 2012). Similar to what has been observed in AML patients, IDH1 and IDH2 gene mutations in gliomas are mutually exclusive, heterozygous, and restricted to the R132 site of IDH1 and the R172 site of IDH2. The discovery of IDH mutations in gliomas has allowed for further characterization of the sequence of events in glioma pathogenesis. The detection of IDH mutations in most low-grade gliomas and secondary GBM suggests that IDH1 mutation is an early event in pathogenesis. Cases of primary GBM that do not show IDH mutations are genetically distinct.
Potential Utility of Detecting IDH Mutations in Gliomas
Detection of an IDH mutation, either directly in surgical specimens or indirectly by measuring D-2HG levels in the brain, has practical implications. The presence of IDH mutation in a GBM in a patient without a prior history of a brain lesion would support a diagnosis of secondary GBM, which portends a better, albeit still dismal prognosis. Like primary GBM, grade I central nervous system tumors, such as pilocytic astrocytoma, and non-neoplastic brain tissue lack IDH mutations. Distinguishing rare infiltrating neoplastic cells from reactive gliosis in surgical biopsy specimens can be difficult, and the presence of IDH mutation would support a neoplastic process. An additional implication includes distinguishing diffuse astrocytoma (grade II) from pilocytic astrocytoma (grade I). Although associated with a better prognosis, the presence of IDH mutations in gliomas does not predict a response to therapy (Capper et al., 2010; Preusser et al., 2011).
Potential Utility of IDH Mutations as a Follow-Up Marker
Isocitrate dehydrogenase 1 and 2 mutations appear to be stable. In one study of 225 AML patients with wild type IDH at diagnosis, not a single patient acquired an IDH mutation during clinical follow-up (Chou et al., 2010). In addition, in patients with IDH mutated AML at diagnosis, the same mutation persisted in over 95% of IDH1-mutated and IDH2-mutated AML cases at relapse (Chou et al., 2010, 2011b; Schnittger et al., 2010; Thol et al., 2010a). From a biological perspective, this stability suggests that IDH mutations may be a primary event that is involved early in leukemogenesis and/or maintenance of the leukemic phenotype. Recent data (Dinardo et al., 2013) suggest that elevated levels of D-2HG in serum, higher than 700 ng/ml could segregate patients with and without IDH mutations. Furthermore, IDH mutant patients with D-2HG levels>200 ng/ml at complete remission experienced shorter overall survival compared to those with D-2HG<200 ng/ml. These data confirm that serum measurement of an oncometabolite can provide useful information for diagnosis, treatment response, and prognosis.
Methods of Detecting IDH Mutations and the Oncometabolite D-2HG
Isocitrate dehydrogenase mutations can be detected by a number of methods including polymerase chain reaction (PCR)-based assays (Felsberg et al., 2010; Meyer et al., 2010; Patel et al., 2011b; Rakheja et al., 2011a), expression of IDH1 mutant protein in tumor tissue as detected by mutation specific antibody using immunohistochemistry (Andrulis et al., 2010; Capper et al., 2010), and indirectly by detecting the putative oncometabolite, D-2HG, by mass-spectrometry in vitro or by magnetic resonance spectroscopy in vivo (Gross et al., 2010; Rakheja et al., 2011b; Choi et al., 2012; Sahm et al., 2012).
Polymerase chain reaction-based assays include amplification of genomic DNA at exon 4 of IDH1 (IDH1R132) and IDH2 (IDH2R140 or IDH2R172) followed by Sanger sequencing, restriction endonuclease digestion and high-resolution melting curve analysis, and pyrosequencing. Unfortunately, these methods have a relatively low sensitivity, for example, ∼20% for heterozygous mutations by Sanger sequencing. More sensitive methods, such as allele-specific PCR, also can be performed to detect of IDH1/IDH2 mutations. More recently, next generation sequencing methods have been applied to the detection of IDH1/2 mutations. With coverage of 500 times, the sensitivity of this approach is ∼5%. These more sensitive assays can be used to detect mutations in treated AML patients in whom bone marrow samples have low blast counts as well as for the detection of early relapse or minimal residual disease.
Currently, only IDH1 mutant protein, IDH1R132 can be detected by immunohistochemistry using the H09 clone (Dianova, Hamburg, Germany) (Andrulis et al., 2010; Capper et al., 2010; Sahm et al., 2012). This antibody has been examined in gliomas, but has not yet been evaluated comprehensively in the myeloid neoplasms. Future development of a monoclonal antibody targeted at the IDH2 mutant proteins, IDH2R140 or IDH2R172, would be helpful and convenient for detecting these mutations.
In the laboratory at Children’s Medical Center of Dallas, exon 4 and flanking intronic regions of IDH1 and IDH2 are sequenced (Rakheja et al., 2011a,b,c). PCR amplification is followed by Sanger sequencing. At MD Anderson Cancer Center, a Sanger based assay has been recently transitioned to a next generation sequencing assay used to detect IDH1/2 mutations in AML patients at time of initial diagnosis. The metabolites, D-2HG and L-2-hydroxyglutaric acid (L-2HG), are measured using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (Rakheja et al., 2011a,b,c).
Detection of D-2HG in tumor tissue by mass-spectrometry in vitro or by magnetic resonance spectroscopy in vivo offers an advantage in that potentially all mutations are detectable, as all mutations generate this oncometabolite (Gross et al., 2010; Rakheja et al., 2011b,c; Choi et al., 2012; Sahm et al., 2012). This non-invasive detection of D-2HG may prove to be valuable in diagnosis and providing prognostic biomarker. It should be noted that archival formalin-fixed paraffin-embedded tumor specimens may not be optimal for detecting D-2HG because this metabolite can be lost during the routine embedding process (Sahm et al., 2012).
Implications for Future Treatment
Isocitrate dehydrogenase 1/2 mutations are thought to be one of the driver mutations in AML and early events in the pathogenesis of gliomas. The common feature of IDH1 and IDH2 mutations is the ability of the respective enzymes to exhibit neomorphic activity, not characteristic of the wild type enzymes, that facilitate production of D-2HG. Therefore, targeted therapies that inhibit the neomorphic function of mutant IDH enzymes or DNA hypomethylating agents might reverse the associated epigenetic patterning and may promote myeloid or glial differentiation and improve outcome in patients with IDH1/2-mutated tumors. Recently, IDH inhibitors have been shown to produce cytostatic effects and cellular differentiation in leukemia and glioma cells (Rohle et al., 2013; Wang et al., 2013). It is currently unknown, however, whether these inhibitors can induce a permanent state of differentiation. The survival of viable tumor cells still containing a potentially transforming constellation of mutations makes it important to determine whether the therapeutic effects will persist over long time frames.
Conclusions and Future Perspectives
IDH1R132, IDH2R140, and IDH2R172 mutations represent a novel class of point mutations in patients with AML and glioma. In AML patients, it is possible that IDH1R132, IDH2R140, and IDH2R172 mutations represent molecular or clinically distinctive subgroups, with IDH1R132 and IDH2R140 more frequently accompanied by normal cytogenetics and NPM1 mutation, whereas IDH2R172 is frequently the only mutation detected and portends a poor prognosis. In glioma patients, IDH mutations are present in grade II and III gliomas as well as secondary GBM. Whether these different mutation types represent distinctive subgroups of glioma patients is less understood. As both IDH1 and IDH2 mutations result in the generation of the putative oncometabolite, D-2HG, this oncometabolite can be measured directly in tumor samples and serum. Screening for the presence of D-2HG could be used as an assay to detect IDH mutations, monitor therapeutic response, and potentially uncover novel IDH mutations. Importantly, expression of mutant IDH1/2 induces a global increase in DNA hypermethylation and inhibits TET2-induced cytosine 5-hydroxymethylation, suggesting that TET2 and IDH1/2 mutations constitute a distinct mutational class in these tumors in which the epigenetic state is altered. Furthermore, this biologic effect is an important consideration for developing a therapeutic agent that can target dysregulated IDH enzymes in addition to induction of DNA hypomethylation. Inhibitors to mutant IDH have been shown recently to produce cytostatic effects and cellular differentiation in leukemia and glioma cells and seem promising. Future studies are important to determine whether these inhibitors can induce a permanent state of differentiation, and their therapeutic effects and toxicity in clinical trials. Discovering IDH mutations is an example of a pathologic finding that links disruption of metabolism to oncogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbas, S., Lugthart, S., Kavelaars, F. G., Schelen, A., Koenders, J. E., Zeilemaker, A., et al. (2010). Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood 116, 2122–2126. doi:10.1182/blood-2009-11-250878
Aghili, M., Zahedi, F., and Rafiee, E. (2009). Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J. Neurooncol. 91, 233–236. doi:10.1007/s11060-008-9706-2
Amary, M. F., Bacsi, K., Maggiani, F., Damato, S., Halai, D., Berisha, F., et al. (2011). IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 224, 334–343. doi:10.1002/path.2913
Andersson, A. K., Miller, D. W., Lynch, J. A., Lemoff, A. S., Cai, Z., Pounds, S. B., et al. (2011). IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia 25, 1570–1577. doi:10.1038/leu.2011.133
Andrulis, M., Capper, D., Luft, T., Hartmann, C., Zentgraf, H., and von Deimling, A. (2010). Detection of isocitrate dehydrogenase 1 mutation R132H in myelodysplastic syndrome by mutation-specific antibody and direct sequencing. Leuk. Res. 34, 1091–1093. doi:10.1016/j.leukres.2010.02.014
Boissel, N., Nibourel, O., Renneville, A., Gardin, C., Reman, O., Contentin, N., et al. (2010). Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the acute leukemia French association group. J. Clin. Oncol. 28, 3717–3723. doi:10.1200/JCO.2010.28.2285
Borger, D. R., Tanabe, K. K., Fan, K. C., Lopez, H. U., Fantin, V. R. S. K., Hezel, A. F., et al. (2012). Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17, 72–79. doi:10.1634/theoncologist.2011-0386
Bruick, R. K., and McKnight, S. L. A. (2001). Conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340. doi:10.1126/science.1066373
Cairns, R. A., Iqbal, J., Lemonnier, F., Kucuk, C., de Leval, L., Jais, J. P., et al. (2012). IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 119, 1901–1903. doi:10.1182/blood-2011-11-391748
Capper, D., Sahm, F., Hartmann, C., Meyermann, R., von Deimling, A., and Schittenhelm, J. (2010). Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy-induced changes. Am. J. Surg. Pathol. 34, 1199–1204. doi:10.1097/PAS.0b013e3181e7740d
Chen, W., Konoplev, S., Medeiros, L. J., Koeppen, H., Leventaki, V., Vadhan-Raj, S., et al. (2009). Cuplike nuclei (prominent nuclear invaginations) in acute myeloid leukemia are highly associated with FLT3 internal tandem duplication and NPM1 mutation. Cancer 115, 5481–5489. doi:10.1002/cncr.24610
Choi, C., Ganji, S. K., DeBerardinis, R. J., Hatanpaa, K. J., Rakheja, D., Kovacs, Z., et al. (2012). 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 18, 624–629. doi:10.1038/nm.2682
Chou, W. C., Chou, S. C., Liu, C. Y., Chen, C. Y., Hou, H. A., and Kuo, Y. Y. (2011a). TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 118, 3803–3810. doi:10.1182/blood-2011-02-339747
Chou, W. C., Lei, W. C., Ko, B. S., Hou, H. A., Chen, C. Y., Tang, J. L., et al. (2011b). The prognostic impact and stability of Isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia 25, 246–253. doi:10.1038/leu.2010.267
Chou, W. C., Hou, H. A., Chen, C. Y., Tang, J. L., Yao, M., Tsay, W., et al. (2010). Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood 115, 2749–2754. doi:10.1182/blood-2009-11-253070
Chowdhury, R., Yeoh, K. K., Tian, Y. M., Hillringhaus, L., Bagg, E. A., Rose, N. R., et al. (2011). The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469. doi:10.1038/embor.2011.43
Damm, F., Thol, F., Hollink, I., Zimmermann, M., Reinhardt, K., and den Heuvel-Eibrink, M. M. (2011). Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia 25, 1704–1710. doi:10.1038/leu.2011.142
Dang, L., White, D. W., Gross, S., Bennett, B. D., Bittinger, M. A., Driggers, E. M., et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744. doi:10.1038/nature08617
Dinardo, C. D., Propert, K. J., Loren, A. W. P. E., Sun, Z., Levine, R. L., Straley, K. S., et al. (2013). Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood 121, 4917–4924. doi:10.1182/blood-2013-03-493197
Estève, P. O., Chin, H. G., Smallwood, A., Feehery, G. R., Gangisetty, O., Karpf, A. R., et al. (2006). Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20, 3089–3103. doi:10.1101/gad.1463706
Felsberg, J., Wolter, M., Seul, H., Friedensdorf, B., Göppert, M., Sabel, M. C., et al. (2010). Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol. 119, 501–507. doi:10.1007/s00401-010-0647-4
Figueroa, M. E., Abdel-Wahab, O., Lu, C., Ward, P. S., Patel, J., Shih, A., et al. (2010). Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567. doi:10.1016/j.ccr.2010.11.015
Frezza, C., Tennant, D. A., and Gottlieb, E. (2010). IDH1 mutations in gliomas: when an enzyme loses its grip. Cancer Cell 17, 7–9. doi:10.1016/j.ccr.2009.12.031
Geisbrecht, B. V., and Gould, S. J. (1999). The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J. Biol. Chem. 274, 30527–30533. doi:10.1074/jbc.274.43.30527
Green, A., and Beer, P. (2010). Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N. Engl. J. Med. 362, 369–370. doi:10.1056/NEJMc0910063
Green, C. L., Evans, C. M., Hills, R. K., Burnett, A. K., Linch, D. C., and Gale, R. E. (2010). The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood 116, 2779–2782. doi:10.1182/blood-2010-02-270926
Green, C. L., Evans, C. M., Zhao, L., Hills, R. K., Burnett, A. K., Linch, D. C., et al. (2011). The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 118, 409–412. doi:10.1182/blood-2010-12-322479
Gross, S., Cairns, R. A., Minden, M. D., Driggers, E. M., Bittinger, M. A., Jang, H. G., et al. (2010). Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344. doi:10.1084/jem.20092506
Hartmann, C., Meyer, J., Balss, J., Capper, D., Mueller, W., Christians, A., et al. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 118, 469–474. doi:10.1007/s00401-009-0561-9
Haselbeck, R. J., and McAlister-Henn, L. (1993). Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 268, 12116–12122.
Ho, P. A., Alonzo, T. A., Kopecky, K. J., Miller, K. L., Kuhn, J., Zeng, R., et al. (2010). Molecular alterations of the IDH1 gene in AML: a children’s oncology group and southwest oncology group study. Leukemia 24, 909–913. doi:10.1038/leu.2010.56
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion. ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi:10.1038/nature09303
Jha, P., Suri, V., Sharma, V., Singh, G., Sharma, M. C., Pathak, P., et al. (2011). IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp. Mol. Pathol. 91, 385–393. doi:10.1016/j.yexmp.2011.04.017
Koivunen, P., Lee, S., Duncan, C. G., Lopez, G., Lu, G. R. S., Losman, J. A., et al. (2012). Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488. doi:10.1038/nature10898
Losman, J. A., Looper, R. E., Koivunen, P., Lee, S. S. R., McMahon, C., Cowley, G. S., et al. (2013). (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625. doi:10.1126/science.1231677
Lu, C., Ward, P. S., Kapoor, G. S., Rohle, D., Turcan, S., Abdel-Wahab, O., et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478. doi:10.1038/nature10860
Marcucci, G., Maharry, K., Wu, Y. Z., Radmacher, M. D., Mrózek, K., Margeson, D., et al. (2010). IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J. Clin. Oncol. 28, 2348–2355. doi:10.1200/JCO.2009.27.3730
Marcucci, G., Mrozek, K., Radmacher, M. D., Garzon, R., and Bloomfield, C. D. (2011). The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood 117, 1121–1129. doi:10.1182/blood-2010-09-191312
Mardis, E. R., Ding, L., Dooling, D. J., Larson, D. E., McLellan, M. D., Chen, K., et al. (2009). Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066. doi:10.1056/NEJMoa0903840
Meyer, J., Pusch, S., Balss, J., Capper, D., Mueller, W., Christians, A., et al. (2010). PCR- and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol. 20, 298–300. doi:10.1111/j.1750-3639.2009.00327.x
Narahara, K., Kimura, S., Kikkawa, K., Takahashi, Y., Wakita, Y., Kasai, R., et al. (1985). Probable assignment of soluble isocitrate dehydrogenase (IDH1) to 2q33.3. Hum. Genet. 71, 37–40. doi:10.1007/BF00295665
Nobusawa, S., Watanabe, T., Kleihues, P., and Ohgaki, H. (2009). IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin. Cancer Res. 15, 6002–6007. doi:10.1158/1078-0432.CCR-09-0715
Oh, I. U., Inazawa, J., Kim, Y. O., Song, B. J., and Huh, T. L. (1996). Assignment of the human mitochondrial NADP(+)-specific isocitrate dehydrogenase (IDH2) gene to 15q26.1 by in situ hybridization. Genomics 38, 104–106. doi:10.1006/geno.1996.0602
Pardanani, A., Lasho, T. L., Finke, C. M., Mai, M., McClure, R. F., and Tefferi, A. (2010). IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia 24, 1146–1151. doi:10.1038/leu.2010.77
Parsons, D. W., Jones, S., Zhang, X., Lin, J. C., Leary, R. J., Angenendt, P., et al. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812. doi:10.1126/science.1164382
Paschka, P., Schlenk, R. F., Gaidzik, V. I., Habdank, M., Krönke, J., Bullinger, L., et al. (2010). IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 28, 3636–3643. doi:10.1200/JCO.2010.28.3762
Patel, J. P., Gönen, M., Figueroa, M. E., Fernandez, H., Sun, Z., Racevskis, J., et al. (2012). Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089. doi:10.1056/NEJMoa1112304
Patel, K. P., Ravandi, F., Ma, D., Paladugu, A., Barkoh, B. A., Medeiros, L. J., et al. (2011a). Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am. J. Clin. Pathol. 135, 35–45. doi:10.1309/AJCPD7NR2RMNQDVF
Patel, K. P., Barkoh, B. A., Chen, Z., Ma, D., Reddy, N., and Medeiros, L. J. (2011b). Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J. Mol. Diagn. 13, 678–686. doi:10.1016/j.jmoldx.2011.06.004
Pietrak, B., Zhao, H., Qi, H., Quinn, C., Gao, E., and Boyer, J. G. (2011). A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry 21, 4804–4812. doi:10.1021/bi200499m
Pigazzi, M., Ferrari, G., Masetti, R., Falini, B., Martinolli, F., Basso, G., et al. (2011). Low prevalence of IDH1 gene mutation in childhood AML in Italy. Leukemia 25, 173–174. doi:10.1038/leu.2010.229
Plaut, G. W., Cook, M., and Aogaichi, T. (1983). The subcellular location of isozymes of NADP-isocitrate dehydrogenase in tissues from pig, ox and rat. Biochim. Biophys. Acta 760, 300–308. doi:10.1016/0304-4165(83)90177-0
Prensner, J. R., and Chinnaiyan, A. M. (2011). Metabolism unhinged: IDH mutations in cancer. Nat. Med. 17, 291–293. doi:10.1038/nm0311-291
Preusser, M., Wöhrer, A., Stary, S., Höftberger, R., Streubel, B., and Hainfellner, J. A. (2011). Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J. Neuropathol. Exp. Neurol. 70, 715–723. doi:10.1097/NEN.0b013e31822713f0
Rakheja, D., Konoplev, S., Su, M., Wheeler, D., Muzny, D. M., Ruvolo, V. R., et al. (2011a). High incidence of IDH mutations in acute myeloid leukaemia with cuplike nuclei. Br. J. Haematol. 155, 125–128. doi:10.1111/j.1365-2141.2011.08646.x
Rakheja, D., Boriack, R. L., Mitui, M., Khokhar, S., Holt, S. A., and Kapur, P. (2011b). Papillary thyroid carcinoma shows elevated levels of 2-hydroxyglutarate. Tumour Biol. 32, 325–333. doi:10.1007/s13277-010-0125-6
Rakheja, D., Mitui, M., Boriack, R. L., and DeBerardinis, R. J. (2011c). Isocitrate dehydrogenase 1/2 mutational analyses and 2-hydroxyglutarate measurements in Wilms tumors. Pediatr. Blood Cancer 56, 379–383. doi:10.1002/pbc.22697
Rohle, D., Popovici-Muller, J., Palaskas, N. T. S., Grommes, C., Campos, C., Tsoi, J., et al. (2013) An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630. doi:10.1126/science.1236062
Sahm, F., Capper, D., Pusch, S., Balss, J., Koch, A., Langhans, C. D., et al. (2012). Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol. 22, 26–31. doi:10.1111/j.1750-3639.2011.00506.x
Schnittger, S., Haferlach, C., Ulke, M., Alpermann, T., Kern, W., and Haferlach, T. (2010). IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood 116, 5486–5496. doi:10.1182/blood-2010-02-267955
Sjöblom, T., Jones, S., Wood, L. D., Parsons, D. W., Lin, J., Barber, T. D., et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274. doi:10.1126/science.1133427
Soga, T. (2013). Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 104, 275–281. doi:10.1111/cas.12085
Struys, E. A. (2006). D-2-hydroxyglutaric aciduria: unravelling the biochemical pathway and the genetic defect. J. Inherit. Metab. Dis. 29, 21–29. doi:10.1007/s10545-006-0317-9
Tefferi, A., Jimma, T., Sulai, N. H., Lasho, T. L., Finke, C. M., and Knudson, R. A. (2011). IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia 26, 475–480. doi:10.1038/leu.2011.253
Tefferi, A., Lasho, T. L., Abdel-Wahab, O., Guglielmelli, P., Patel, J., Caramazza, D., et al. (2010). IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 24, 1302–1309. doi:10.1038/leu.2010.113
Thol, F., Damm, F., Wagner, K., Göhring, G., Schlegelberger, B., Hoelzer, D., et al. (2010a). Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood 116, 614–616. doi:10.1182/blood-2010-03-272146
Thol, F., Weissinger, E. M., Krauter, J., Wagner, K., Damm, F., Wichmann, M., et al. (2010b). IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 95, 1668–1674. doi:10.3324/haematol.2010.025494
Thon, N., Eigenbrod, S., Kreth, S., Lutz, J., Tonn, J. C., Kretzschmar, H., et al. (2012). IDH1 mutations in grade II astrocytomas are associated with unfavorable progression-free survival and prolonged postrecurrence survival. Cancer 118, 452–460. doi:10.1002/cncr.26298
Turcan, S., Rohle, D., Goenka, A., Walsh, L. A., Fang, F., Yilmaz, E., et al. (2012). IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483. doi:10.1038/nature10866
van den Bent, M. J., Dubbink, H. J., Marie, Y., Brandes, A. A., Taphoorn, M. J., Wesseling, P., et al. (2010). IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European organization for research and treatment of cancer brain tumor group. Clin. Cancer Res. 16, 1597–1604. doi:10.1158/1078-0432.CCR-09-2902
Wagner, K., Damm, F., Göhring, G., Görlich, K., Heuser, M., Schäfer, I., et al. (2010). Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J. Clin. Oncol. 28, 2356–2364. doi:10.1200/JCO.2009.27.6899
Wang, F., Travins, J., DeLaBarre, B. P.-L. V., Schalm, S., Hansen, E., Straley, K., et al. (2013). Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626. doi:10.1126/science.1234769
Wang, P., Dong, Q., Zhang, C., Kuan, P. F., Liu, Y., Jeck, W. R. A. J., et al. (2012). Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32, 3091–3100. doi:10.1038/onc.2012.315
Ward, P. S., Patel, J., Wise, D. R., Abdel-Wahab, O., Bennett, B. D., Coller, H. A., et al. (2010). The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234. doi:10.1016/j.ccr.2010.01.020
Williams, S. C., Karajannis, M. A. C. L., Golfinos, J. G., von Deimling, A., and Zagzag, D. (2011). R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1α upregulation in adult glioma. Acta Neuropathol. 121, 279–281. doi:10.1007/s00401-010-0790-y
Xu, W., Yang, H., Liu, Y., Yang, Y., Wang, P., Kim, S. H., et al. (2011). Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30. doi:10.1016/j.ccr.2010.12.014
Xu, X., Zhao, J., Xu, Z., Peng, B., Huang, Q., Arnold, E., et al. (2004). Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 279, 33946–33957. doi:10.1074/jbc.M404298200
Yan, H., Parsons, D. W., Jin, G., McLendon, R., Rasheed, B. A., Yuan, W., et al. (2009). IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773. doi:10.1056/NEJMoa0808710
Keywords: IDH mutation, NPM1 mutation, acute myeloid leukemia, glioma
Citation: Rakheja D, Medeiros LJ, Bevan S and Chen W (2013) The emerging role of d-2-hydroxyglutarate as an oncometabolite in hematolymphoid and central nervous system neoplasms. Front. Oncol. 3:169. doi: 10.3389/fonc.2013.00169
Received: 15 January 2013; Accepted: 13 June 2013;
Published online: 02 July 2013.
Edited by:
Lorenzo Galluzzi, Institut National de la Santé et de la Recherche Medicale, FranceReviewed by:
Keith R. Laderoute, SRI International, USATimothy A. Chan, Memorial Sloan Kettering Cancer Center, USA
Copyright: © 2013 Rakheja, Medeiros, Bevan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Weina Chen, AmeriPath/Quest Diagnostics, 4350 Alpha Road, Dallas, TX 75244, USA e-mail: wchen@ameripath.com
