- 1Department of Oncology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Medical Oncology, The Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China
Background: Interaction between cancer cells with microenvironment is essential for cancer progression, therapeutic resistance and prognosis. Chemokine CXCL1 shows variable roles in the development of cancers. DACH1 has been considered as a tumor suppressor and represses the expressions of several chemokines. The relationship between CXCL1 and DACH1 in non-small cell lung cancer (SCLC) deserves further investigation.
Methods: Immunohistochemistry staining was performed on tumor tissue microarrays from lung cancer patients to detect CXCL1 protein. The CXCL1 concentration in the serum of adenocarcinoma patients was measured by ELISA. The CXCL1 protein secreted by cancer cell lines was detected by SearchLight proteome array and human cytokine antibody array. The meta-analysis of CXCL1 expression form public databases was performed and correlation between CXCL1 and DACH1 was analyzed. Moreover, the association between clinicopathological features and prognosis with CXCL1 and DACH1 was analyzed by tissue array and KM-plotter from public database.
Results: The protein abundance of CXCL1 in lung cancer tissues was significantly higher than that in adjacent normal tissues. CXCL1 was closely related to TNM stage, tumor size, and lymph node metastasis and predicted worse overall survival in adenocarcinoma. The level of CXCL1 in the peripheral blood of adenocarcinoma patients also significantly elevated and positively related with clinical stage. The meta-analysis demonstrated that CXCL1 mRNA level was increased in lung cancer tissues and high level of CXCL1 indicated tumor progression in lung adenocarcinoma. In addition, public database analyses showed that CXCL1 negatively correlated with DACH1. Stable overexpressing DACH1 in cultured lung cancer cells remarkably decreased CXCL1 protein. Moreover, ectopic expression of DACH1 significantly inhibited the expression of CXCL1, Ki67, and cyclin D1 in tumor tissues compared with A549 cells with empty vector. Survival analysis showed that high CXCL1 and low DACH1 indicated poor overall survival and progression-free survival.
Conclusion: CXCL1 is closely associated with tumor progression and poor survival. DACH1 significantly inhibits the expression of CXCL1 and indicates good prognosis. Therefore, combined detection of CXCL1 and DACH1 could more precisely predict prognosis of lung adenocarcinoma.
Background
Lung cancer is the most common cause of cancer death in male and the second leading cause in female all over the word (1). Lung cancer is divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC includes adenocarcinoma (ADC), squamous cell carcinoma (SQC) and large cell lung cancer, etc. At present, surgery, radiotherapy and chemotherapy, and target therapy are the main therapeutic methods for NSCLC (2), which prolonged patients' survival. However, recurrence and resistance remain an urgent problem (3–5). Epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) -rearrangements are typically associated with tumorigenesis and prognosis of patients (6, 7). However, the mechanism of the oncogenesis and progression of NSCLC still requires more exploration. In a search for lung tumor suppressor by genomic landscape, DACH1 was described as a potential tumor suppressor in non-small cell lung cancer (NSCLC) (8).
Human Dachshund homolog 1 (DACH1) is a key member of retinal determination gene network (RDGN), which regulated cell proliferation, apoptosis, tumor growth and progression (9, 10). In comparison with normal tissues, DACH1 protein and mRNA expression level obviously decreased in several tumor tissues, including lung cancer (8, 11, 12). The reduction of DACH1 was mainly caused by genomic deletion and promoter region hypermethylation (13, 14). Functional studies identified DACH1 as a tumor suppressor in several cancer types, such as breast cancer, colorectal cancer, and renal cell carcinoma, and so on (15–17). Mechanism exploration showed that DACH1 participated in the negative regulation of cell cycle, epithelial-mesenchymal transition (EMT) and reduction of the subpopulation of cancer stem cell (CSC) (18–20). DACH1 also controlled secretion of multiple chemokine, including CXCL5, CXCL8, and IL-6 (21–23).
The chemokine (C-X-C motif) ligand 1 (CXCL1) belongs to CXC chemokine family. CXCL1 was regulated by multiple signal pathways and tumor microenvironment. For example, tumor necrosis factor (TNF) and vascular endothelial growth factor (VEGF) stimulated CXCL1 expression via JNK, p38 MAPK, and PI-3K/Akt signaling pathways in human lung carcinoma epithelial cells (24, 25). CXC receptor 2 (CXCR2) is a common receptor for CXC chemokines (26). The interaction of CXCL1 and CXCR2 promoted the development of malignant carcinoma, including proliferation, migration, angiogenesis, and therapy-resistance (27–30). Tumor-derived CXCL1 promoted the growth of lung cancer by recruiting neutrophils from peripheral blood into tumor tissues (31). The interaction between tumor cells with neutrophils increased the expression of metastasis-related genes (CXCR4, CXCR7, MMP12, MMP13, IL-6, TGF-β) (32). Consistent with the above result, the level of circulating CXCL1 in patients with metastasis was higher than patients in stage IA-IIB NSCLC (33). Besides, a study demonstrated that the expression of CXCL1 in tumor cells was elevated after paclitaxel and docetaxel treatment (34). Blocking CXCR2 enhanced chemotherapeutic response, suppressed tumor growth, angiogenesis, and metastasis (34, 35). In addition, low-dose radiation induced expression of CXC chemokines (CXCL1, CXCL2, CXCL6) in normal human fibroblasts (36).
The present study investigated the association between the expression profiles of CXCL1 and the clinicopathological features, as well as the prognostic value of CXCL1 in NSCLC. Besides, we also explored the relationship between DACH1 and CXCL1 in NSCLC.
Materials and Methods
Cell Culture and Transfection
Human lung cell lines (A549 and SKLU-1) were originally obtained from ATCC (Manassas, VA, USA) and we acquired from Dr. Guoan Chen in University of Michigan (37). These were cultured in RPMI-1640 medium (HyClone) with 10% fetal bovine serum (FBS) (Gibco), and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone) with 10% FBS. Cells were maintained at 37°C in a 5%CO2 humidified incubator. Plasmids encoding wild type DACH1 as previous description were subcloned to lentivirus expression vectors (38). DACH1 stable cell lines were characterized by inverted fluorescence microscopy and by immunofluorescence as previously described (11).
Immunohistochemistry Staining
Two commercially available tissue microarray (TMA) slides (BC041115c, US Biomax and HLug-Ade150Sur-02, Outdo Biotech) were purchased for immunohistochemistry (IHC). BC041115c, with core diameter of 5 mm containing 40 cases of squamous cell carcinomas, 48 adenocarcinomas, 3 adenosquamous carcinomas, 4 bronchioloalveolar carcinomas, 3 large cell carcinomas, 8 small cell carcinomas, 4 lung atypical carcinoids and 10 normal lung tissues was used to detect the expression of CXCL1. HLug-Ade150Sur with 75 matched pairs of human lung adenocarcinoma and adjacent lung tissues was used to evaluate the prognostic value of CXCL1 based on the detailed survival data. In addition, the protein level of DACH1, cyclin D1, Ki67, and CXCL1 in A549-vector and A549-DACH1 tumor tissues from xenograft mice models were examined by IHC. The specific primary antibodies include anti-CXCL1-antibody (ProteinTech), anti-DACH1-antibody (ProteinTech), anti-Ki67-antibody (Abcam), and anti-cyclin D1-antibody (MAIXIN-Bio). IHC staining was performed by Bios Biotech, Inc., with a 2-step protocol as previously described (39). Slide images were captured by Mv Image software.
Analysis and Quantification of Staining
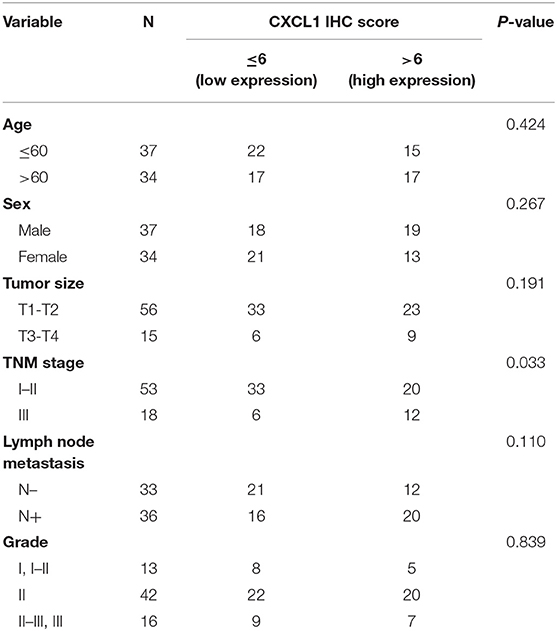
For quantification, at least three fields at 200× magnification of each spot were selected for IHC scoring. The immunohistochemical score were assessed independently by two experienced pathologists without knowledge of patients' characteristics. Scores were calculated on intensity and percentage of positive staining tumor cells in the whole tissue stains according to the Fromowitz Standard as described (40). Briefly, the staining intensity was scored as 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown) and 3 (strong staining, brown). The percentage of stained tumor cells were classified as 1 (0–25% staining), 2 (26–50% staining), 2 (51–75% staining), and 3 (76–100% staining). The multiplication for intensity and percentage was utilized to represent the protein levels of CXCL1. The final score was categorized as low (scores <6) or high (scores ≥6).
Enzyme-Linked Immunosorbent Assay (ELISA)
To evaluate the serum concentration of CXCL1, we collected the blood samples from about 100 patients who were diagnosed as ADC at various stages by April 2015 in Tongji hospital. The clinical samples collection and protocols were approved by the ethics committee of the Tongji Hospital of Huazhong University of Science and Technology. Serum samples from patients with obvious symptom of inflammation were excluded. The ELISA kit for CXCL1 was purchased from Cloud-Clone Corp. Diluted samples, including standard CXCL1 content, negative control and serums, are pipetted into 96 well plate pre-coated with anti-CXCL1 antibody. All procedures were performed following standard protocols with manual. After stopping the reaction, optical densities (OD) were measured at 450 nm using BioTek microplate reader. The intensity of this signal is directly proportional to the concentration of human CXCL1 present in the standard specimen. Standard curve demonstrated the linearity R2 = 0.9947.
Searchlight Proteome Array
CXCL1 in cell supernatant samples was measured using SearchLight proteome arrays (Pierce Biotechnology, Woburn, MA). Briefly, the samples including A549-vector, A549-DACH1 and SKLU-1-vector, SKLU-1-DACH1 were diluted to 1:5, 1:50 or 1:1,000 before 1 h incubation on the array plate that was pre-spotted with capture antibodies specific for CXCL1. Plate was decanted and washed three times before adding a cocktail of biotinylated detection antibody to each well. After incubating with detection antibody for 30 min, plate was washed three times and incubated for 30 min with streptavidin-horseradish peroxidase. Plate was again washed before adding SuperSignal Femto Chemiluminescent substrate. The plates were immediately imaged using the SearchLight imaging system, and data was analyzed using Array Vision software.
Human Cytokine Antibody Array
Human cytokine antibody array III kit (Ray Biotech Cat# AAH-CYT-3-4) was purchased to detect the CXCL1 level in the supernatant of lung cancer cell lines. The detected samples included cell-free supernatant of A549-vector, A549-DACH1, SKLU-1-vector, and SKLU-1-DACH1. The operation steps are carried out according to the protocol provided by the manufacturer, as previously described (11).
Meta-Analysis for CXCL1 on Published Gene Expression Omnibus Databases
We analyzed relevant Gene Expression Omnibus (GEO) databases for mRNA expression of CXCL1. We obtained these published datasets from ArrayExpress and Oncomine (41). And the included datasets must meet the following criteria: (a) the datasets were about human NSCLC cancer; (b) CXCL1 mRNA expression was measured in these databases; (c) these databases should include clinical characteristics of patients. As for datasets with the same population, the most recent or complete dataset was chosen. Finally, a total of 20 independent human NSCLC microarray databases with the mRNA expression of CXCL1 and clinical information were enrolled in this systematic analysis. The median expression of CXCL1 was used as cutoff value. overall survival (OS) and progression-free survival (PFS) were evaluated by Cox proportional hazard ratio (HR) and 95% confidence interval (95% CI). HR >1 indicated that high expression of CXCL1 predicted worse survival of patients. The meta-analysis was performed by the STATA software (Stata Corp LP, College Station, TX, USA). And statistical analysis was performed based on the guidelines of Meta-Analysis of Observational Studies.
Kaplan–Meier Plotter
Kaplan–Meier survival curves with HR and log-rank P-value were calculated and plotted on the online Kaplan–Meier plotter (http://kmplot.com/analysis). The background database, required from GEO, offers gene expression data and survival information. The Affymetrix ID for CXCL1 was 204470_at, which includes 2,435 cases of lung ADC and SQC. The follow up time was threshold at 150 months. The patients were divided by median expression of CXCL1. The Kaplan–Meier survival curves were downloaded from the website. And the blend curves were obtained from IBM SPSS Statistics 19.0 based on GSE31210. All curves were resized in Adobe Illustrator CS6.
Statistical Analysis
Statistical analysis between groups were calculated by Student's t-test and one-way ANOVA and the significance level was set at 0.05. Univariate cumulative survival analyses for PFS and OS were calculated using Kaplan–Meier method with the log-rank test. The correlation analysis was calculated according to Person χ2-test. Statistical analyses were conducted using SPSS 19.0 and GraphPad Prism 6.0. All data were presented as the mean ± SD.
Results
Upregulation of CXCL1 Protein in Various Kinds of Lung Cancer
We detected the expression of CXCL1 in normal and lung cancer tissues by IHC staining. The representative images of normal and cancer tissues were shown in Figures 1A–H. The results indicated that the expression of CXCL1 in cancer tissues was apparently higher than that of normal tissue. Among these kinds of lung cancers, the CXCL1 expression in adenocarcinoma was the highest (Figure 1I). However, the CXCL1 expressed in SCLC was significantly less than that of NSCLC. The average scores of normal, SCLC, adenocarcinoma and squamous cell carcinoma were 1.1 ± 0.23, 2.9 ± 0.81, 6.3 ± 0.27, and 5.7 ± 0.31, respectively.
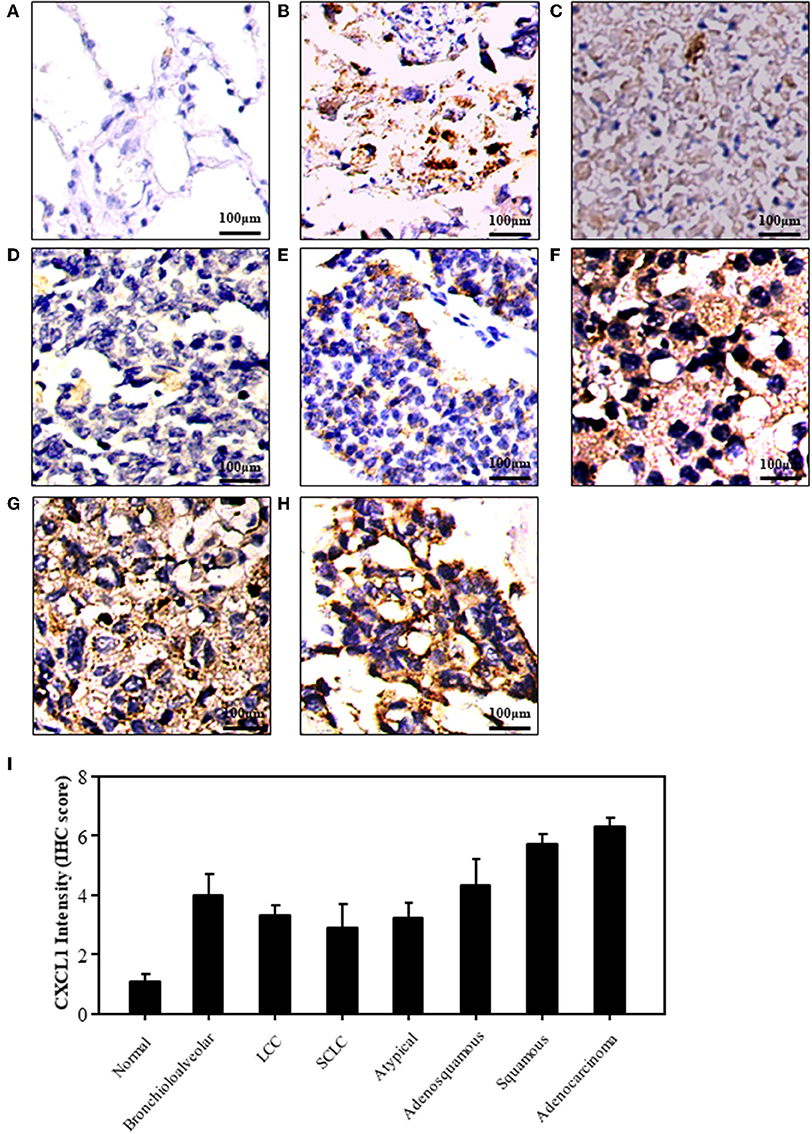
Figure 1. The expression of CXCL1 protein in lung cancers. (A) Normal tissue (B) Bronchioloalveolar cancer (C) lung large cell cancer (LCC) (D) small cell lung cancer (SCLC) (E) Atypical lung cancer (F) Adenosquamous cancer (G) Squamous cell carcinoma (H). Lung adenocarcinoma (I) CXCL1 IHC score.
Expression of CXCL1 Correlated With Progression and Prognosis of Lung ADC
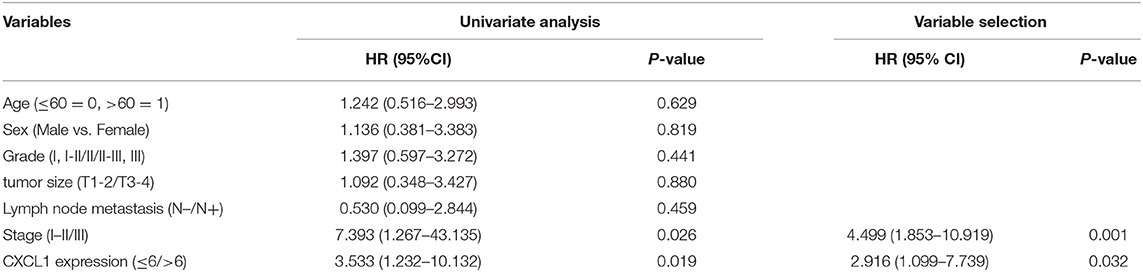
Considering the high protein abundance of CXCL1 in lung adenocarcinoma, we further analyzed the expression of CXCL1 in a TMA containing lung adenocarcinoma with matched adjacent lung tissue and clinicopathological parameters. Representative images of IHC staining for adjacent tissues and adenocarcinoma were shown in Figure 2. In comparison with adjacent lung tissues, the CXCL1 protein in lung adenocarcinoma was evidently elevated (P < 0.0001) (Figure 2A). The protein abundance of CXCL1 was positively correlated with tumor TNM stage (P = 0.0001), tumor size (P = 0.0032) and lymph node metastasis (P = 0.0234) (Figures 2B–D). However, there was no statistical difference with grade. According to the average score of staining, CXCL1 expression was divided into CXCL1 high (n = 32) and low (n = 39) subgroups. Medium OS time of the CXCL1 high and low subgroups were 30 ± 2.98 and 48 ± 2.14 months, respectively, which indicated that they had significant difference of survival (Kaplan–Meier log-rank test, P = 0.001, Figure 3). We also explored the relationship between CXCL1 expression and clinicopathological features of 71 ADC patients. As shown in Table 1, CXCL1 expression was only correlated with TNM stage. Besides, we investigated the correlation between cumulative OS and clinicopathological parameters by univariate Cox regression analysis including age, sex, grade, tumor size, lymph node metastasis, TNM stage and CXCL1 expression (Table 2). The results demonstrated that TNM stage (HR = 7.393; 95% CI 1.267–43.135; p = 0.026) and CXCL1 expression (HR = 3.533; 95% CI 1.232–10.132; p = 0.019) were prognostic factors for OS. Multi-various Cox analysis revealed that TNM stage (HR = 4.499; 95% CI 1.853–10.919; p = 0.001) and CXCL1 expression (HR = 2.916; 95% CI 1.099–7.739; p = 0.032) were independent prognostic factors for OS.
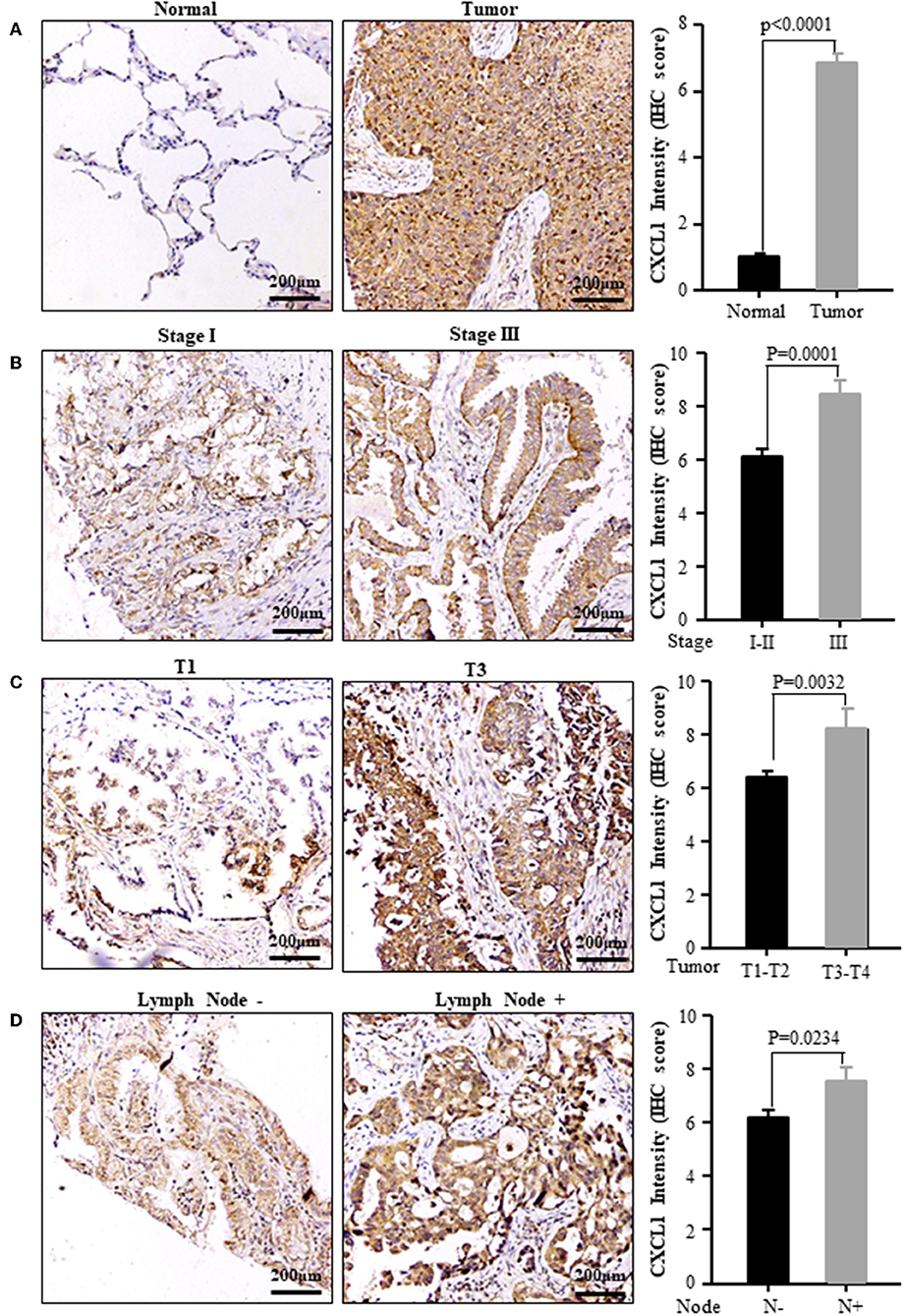
Figure 2. The expression of CXCL1 protein was positively correlated with the progression of ADC patients. (A) Normal vs. Tumor (B) TNM stage I vs. III (C) Tumor size T1 vs. T3 (D) Lymph node metastasis positive vs. negative. Left panel: representative images, right panel: CXCL1 IHC score.
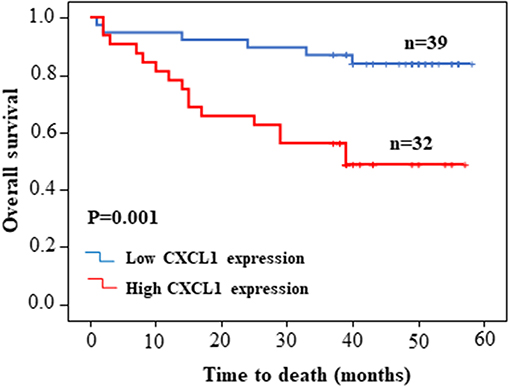
Figure 3. Kaplan–Meier survival curve of patients with low or high CXCL1 expression based on the survival data in tissue microarray slide.
CXCL1 Protein Increased in the Serum of Patients With Lung ADC
We also examined the serum CXCL1 concentration by commercial ELISA kit. The standard curve between CXCL1 amount and OD value was excellent linear with R2 = 0.9947 (Figure 4A). The tested serum samples were from 20 normal donors and 56 ADC patients with various tumor stages and histological grades. Sixteen of fifty-six patients were in early stage (I-II) and the rest were in III-IV stage. The results confirmed that CXCL1 expression in serum was considerably elevated in adenocarcinoma compared with normal samples (P = 0.0001, Figure 4B). High expression of CXCL1 tended to be associated with tumor progression, but it did not reach the statistical significance (Figure 4C).
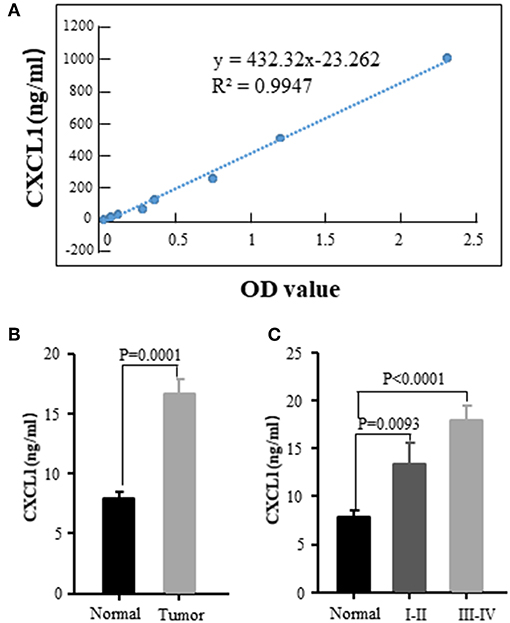
Figure 4. Elevated CXCL1 protein in serum of patients with ADC. (A) Standard curve of CXCL1 with OD value (B) Serum CXCL1 in healthy donors and lung ADC patients (C) Serum CXCL1 protein was positively correlated with TNM stage.
High Level of mRNA CXCL1 Predicted Progression and Worse Clinical Outcomes for Patients With NSCLC
In order to verify the prognostic value of CXCL1 in lung cancer, we analyzed 20 published GEO databases (summarized in Table 3) mainly containing NSCLC patients and corresponding clinicopathological parameters. The patients were divided into CXCL1 high and low based on the median CXCL1 mRNA value. Our results of meta-analysis indicated that higher expression of mRNA CXCL1 was strongly correlation with worse OS whether it is in NSCLC (HR: 1.22, 95%CI: 1.07–1.40, P = 0.962, and I2 = 0.0%) (Figure 5A) or ADC (HR: 1.27, 95%CI: 1.07–1.49, P = 0.643, and I2 = 0.0%) (Figure 5B) or SQC (HR: 1.36, 95%CI: 1.01–1.84, P = 0.982, and I2 = 0.0%) (Figure 5C). However, only in ADC, the high mRNA expression of CXCL1 contributed to shorter PFS (HR: 1.62, 95%CI: 1.03–2.55, P = 0.211, and I2 = 33.6%) (Figure 5D). Furthermore, we found that mRNA CXCL1 was increased in TNM stage of III-IV in NSCLC (OR: 1.51, 95%CI:1.19–1.93, P = 0.637, and I2 = 0.0%) (Figure 5E) and ADC (OR: 1.69, 95%CI:1.25–2.28, P = 0.386, and I2 = 6.0%) (Figure 5F), but not in SQC. Similarly, mRNA expression of CXCL1 was elevated in ADC patients with lymph node metastasis (OR: 1.37, 95%CI: 0.99–1.90, P = 0.462, and I2 = 0.0%) (Figure 5G). There was no statistical significance between CXCL1 mRNA level and histological grade. The mRNA expression of CXCL1 in SQC was higher than that in ADC (Figure 5H).
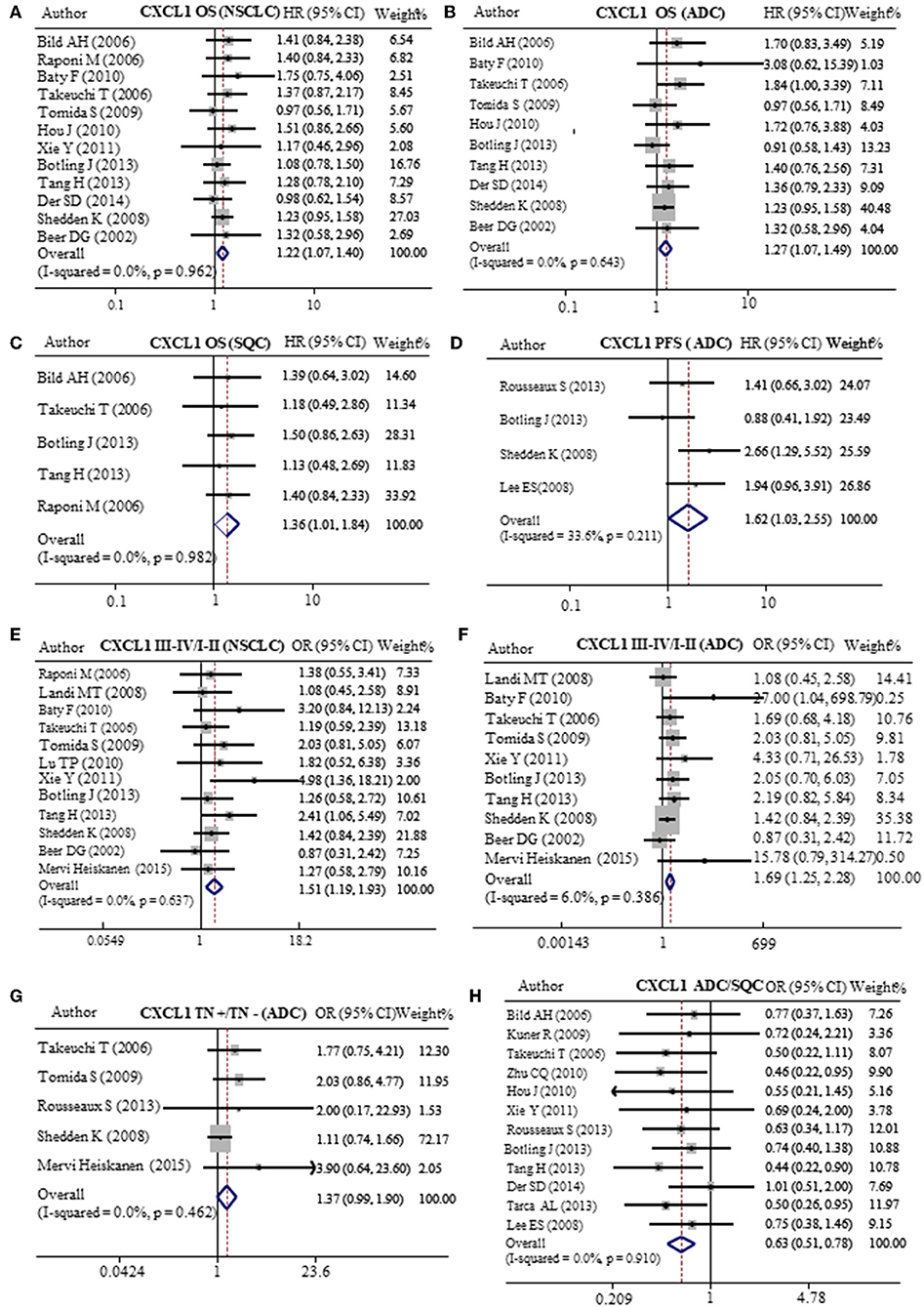
Figure 5. Meta-analysis of mRNA CXCL1 expression in lung cancers. Relative risk of CXCL1 mRNA expression to OS in NSCLC (A), ADC (B), and SQC (C). (D) Relative risk of CXCL1 mRNA expression to PFS in ADC. The forest plot of relative mRNA expression of CXCL1 between III–IV and I–II patients in NSCLC (E) and ADC (F). (G) The forest plot of relative mRNA expression of CXCL1 between lymph node metastasis positive and negative patients in ADC (H) OR value of CXCL5 in ADC vs. SQC.
DACH1 Reduced the Expression of CXCL1 in vitro
Our previous study has demonstrated that DACH1 was a tumor suppressor factor and negatively correlated with CXCL5 (22). And CXCL5 contributed to the tumorigenesis and poor prognosis in NSCLC (62). Both CXCL1 and CXCL5 belong to CXC chemokine family. Therefore, we analyzed lung cancer cell line dataset (GSE32474) and tumor tissue dataset (GSE30219) and found that CXCL1 inversely correlated with DACH1 (Figure 6A, R = 0.684, P = 0.042; R = 0.222, P = 0.006). To test whether DACH1 directly regulated CXCL1, two lung cancer cell lines A549 and SKLU-1 were transferred with control or DACH1 plasmid, respectively. We collected the supernatant of A549-vector, A549-DACH1, SKLU-1-vector and SKLU-1-DACH1 cell lines and detected the protein level of CXCL1 by SearchLight proteome array. The results demonstrated that DACH1 significantly reduced the expression of CXCL1 in A549-DACH1 and SKLU-1-DACH1 cells (Figure 6B). In addition, the human cytokine array III also proved that CXCL1 was indeed suppressed by DACH1 both in A549 and SKLU-1 cells. (Figure 6C).
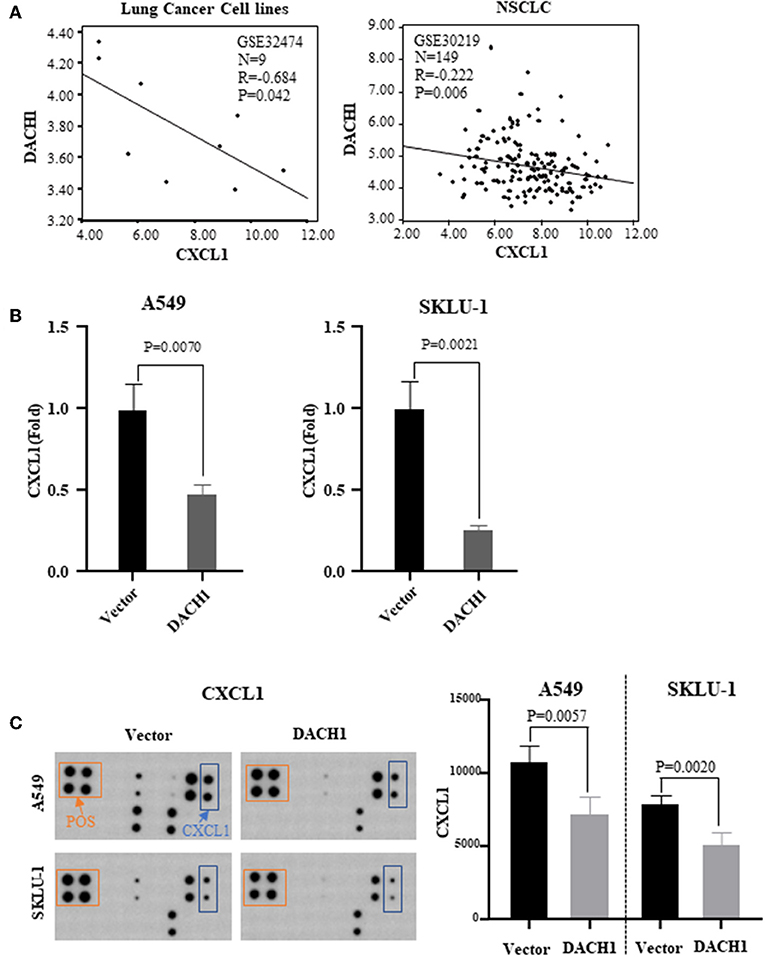
Figure 6. Correlation between CXCL1 and DACH1. (A) CXCL1 negatively correlated with DACH1 in mRNA level based on published databases. SearchLight proteome array (B) and human cytokine array III (C) demonstrated that CXCL1 protein was suppressed by DACH1 both in A549 and SKLU-1 cell lines.
DACH1 Reduced the Expression of CXCL1, Cyclin D1, and Ki67 in vivo
Previous study showed that ectopic expression of DACH1 inhibited tumor growth (11). IHC staining was performed to evaluate the protein level of DACH1, CXCL1, cyclin D1, and Ki67 in nude mice xenograft tumor tissues from cells expressing vector control and DACH1. Representative images of IHC staining and scoring results for DACH1, CXCL1, cyclin D1, and Ki67 were shown in Figure 7. Our results demonstrated that overexpression of DACH1 (P < 0.001) obviously decreased the expression of CXCL1 (P < 0.001), cyclin D1 (P < 0.001), and Ki67 (P < 0.001), suggesting that DACH1 not only reduced the expression of CXCL1, but also inhibited proliferation biomarkers cyclin D1 and Ki67 in vivo at protein level.
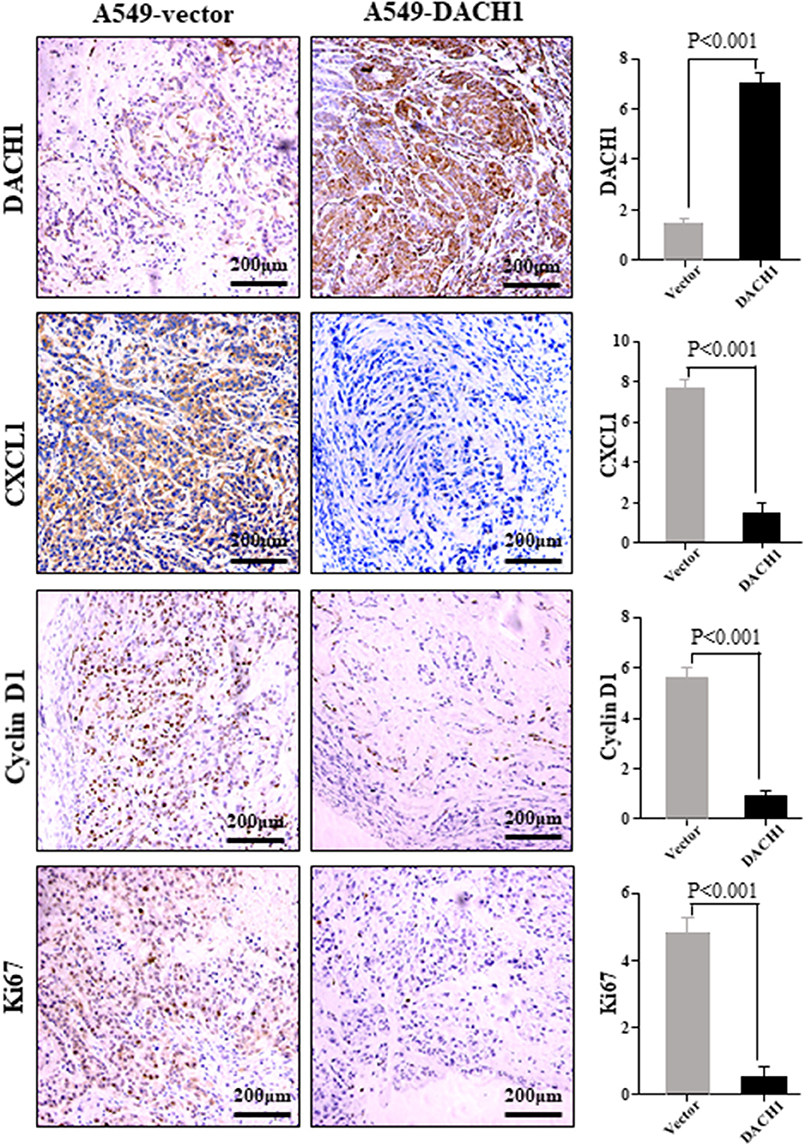
Figure 7. Representative images of DACH1, CXCL1, cyclin D1, and Ki67 IHC staining from xenograft tumor tissues subcutaneously implanted with A549-vector and A549-DACH1 cells.
High CXCL1 and Low DACH1 Expression Predicted Poor Survival in ADC
To further confirm the clinical significance of CXCL1 and DACH1expressions, we first investigated the prognostic value of CXCL1 and DACH1 for patients with ADC on kmplot.com, respectively. The Kaplan–Meier curves from univariate Cox regression showed that patients with higher mRNA expression of CXCL1 tended to have shorter median OS (HR:1.31, 95%CI:1.04–1.66, P = 0.022) (Figure 8A). Nevertheless, this tendency was not observed in PFS (HR:1.07, 95%CI:0.79–1.46, P = 0.66) (Figure 8B). As expected, higher mRNA expression of DACH1 was significantly associated with better OS (HR:0.76, 95%CI:0.60–0.96, P = 0.023) (Figure 8C) and PFS (HR:0.60, 95%CI:0.44–0.83, P = 0.0015) (Figure 8D). In order to obtain more precise prognostic information for ADC patients, we performed combination analysis for the expression of CXCL1 and DACH1 in published database GSE31210. Apparently, patients characterized by high DACH1 with low CXCL1 achieved longest median OS (Figure 8E) and PFS (Figure 8F). The results proved that DACH1 was a potential prognostic factor for better clinical outcomes, while CXCL1 was an adverse biomarker for ADC patients. Combined detection of CXCL1 and DCCH1 provided more precise information for prognosis.
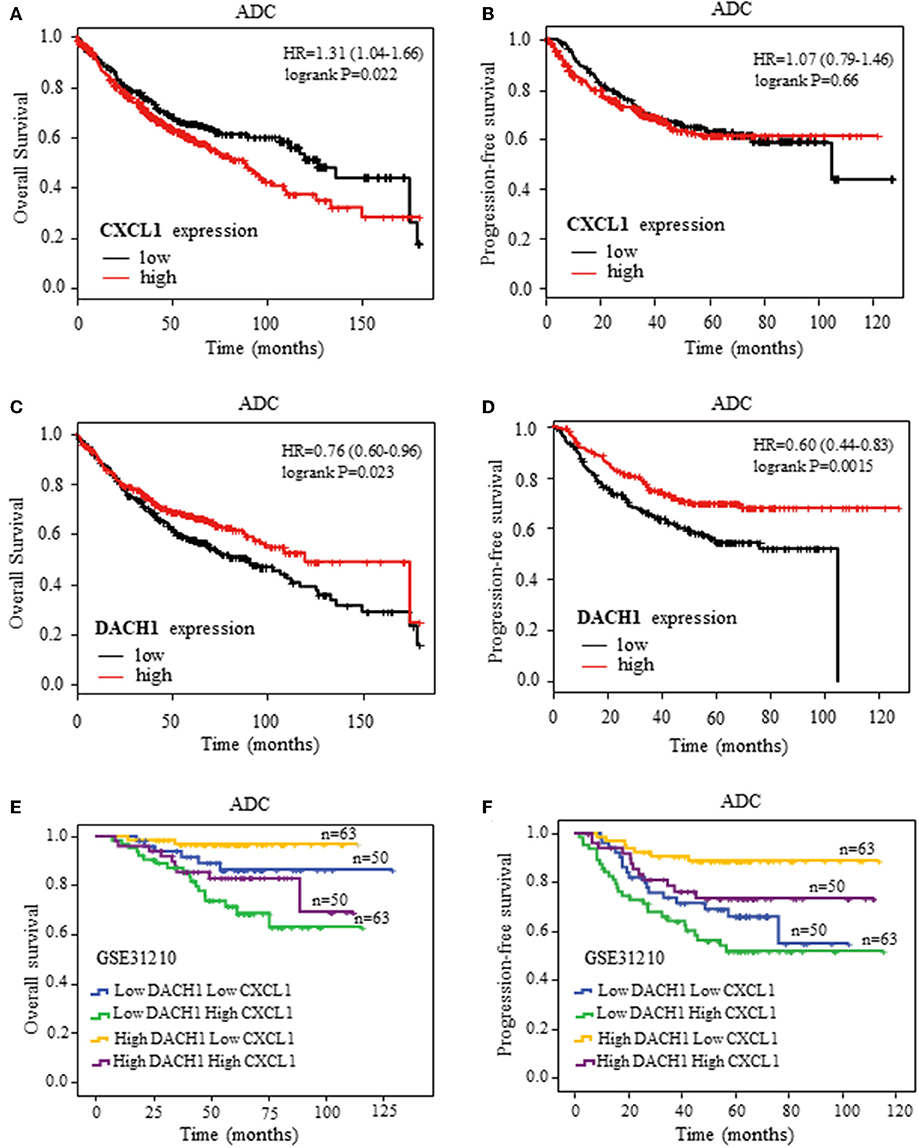
Figure 8. Kaplan–Meier survival curves. Survival curves of CXCL1 with OS (A) and PFS (B) for patients with ADC. Survival curves of DACH1 with OS (C) and PFS (D) for patients with ADC. The blend Kaplan–Meier survival curves of CXCL1 and DACH1 with OS (E) and PFS (F) for patients with ADC in GSE31210.
Discussion
Accumulating evidence has proved that CXCL1 plays an important role in the development of various malignant tumors. CXCL1, as an inflammatory factor, participates in the recruitment of granulocytes (63). Tumor-derived CXCL1 contributes to tumor-associated neutrophils infiltration in lung cancer which promotes tumor growth (31). Colorectal CSCs secrete CXCL1 and CXCL2 to attract neutrophils, which promoted tumorigenesis of colorectal cancer cells via interleukin-1β (64). CXCL1/2 recruit CD11b(+)Gr1(+) myeloid cells into the tumor, which produce chemokines including S100A8/9 that enhance cancer cell survival, chemoresistance and metastasis (28). Feedback loop between NF-κB and CXCL1/-2 enhanced metastasis formation in vivo (65). In ER-negative breast cancer, CXCL1 is highly expressed and stimulates cancer cell migration and invasion via activating the ERK-MMP2/9 signaling pathway (66).
Through immunohistochemical analysis, we found that CXCL1 protein expressed in ADC was higher than in other kinds of lung cancer. Therefore, we further evaluated the expression level of CXCL1 in ADC and its relationship with clinicopathological characteristics and prognosis. The analysis showed that the expression of CXCL1 in ADC was positively related to the TNM stage, tumor size and lymph node metastasis. CXCL1 was also positively associated with tumor grade, though it did not reach statistical significance. The survival curve plotted with survival information exhibited that the higher CXCL1 level accompanied with the shorter survival time. Previous study reported that circulating CXCL1 combining CCL18 could be as tumor markers for the differential diagnosis between ovarian cancer and benign ovarian masses (67). Therefore, except for tumor tissues, we also collected the peripheral blood of ADC patients and normal donors to detect the amount of circulating CXCL1 by ELISA. The results demonstrated that the serum CXCL1 of ADC patients obviously higher than that of normal donors and correlated with TNM stage though there is no statistical significance, possibly because of the limited samples. Our results were in agreement with previous study that NSCLC patients with metastasis had high levels of CXCL1 than patients in stage IA-IIB (33). From the above results, we speculate that CXCL1 could be used as an indicator for monitoring cancer progression. In particular, detection of circulating CXCL1 would be a more convenient and acceptable method.
The analysis results of 20 databases showed that mRNA CXCL1 positively associated with OS, PFS, TNM stage and lymph node metastasis for the ADC patients. But for the SQC patients, higher mRNA CXCL1 only related with shorter OS. Therefore, although the mRNA level of CXCL1 in SQC is higher than that of in ADC, the functions of CXCL1 were more obvious in ADC, suggesting that CXCL1 protein abundance and functions were also regulated by other factors.
As previous studies proved that DACH1 was considered as a tumor suppressor in several kinds of cancers including lung cancer and regulated multiple cytokine expression (8, 11, 14). Therefore, we performed the correlation analysis between CXCL1 and DACH1 in lung cancer cell lines and cancer tissues. The results suggested that CXCL1 negatively correlated with DACH1 at mRNA level. In vitro study confirmed that secretion of CXCL1 protein was significantly suppressed by DACH1 in lung cancer cell lines A549 and SKLU-1. Our previous study has demonstrated that DACH1 overexpression remarkably inhibited the growth of lung cancer in nude mice (11). The tumor tissues isolated from those nude mice were further immunized with four antibodies: DACH1, CXCL1, cyclin D1, Ki67. The results showed that DACH1 significantly inhibited the expression of CXCL1, cyclin D1, and Ki67 in vivo. Previous finding demonstrated that tumor derived CXCL1 promoted the growth of lung cancer (31), our study suggested that the tumor suppress function of DACH1 might be through inhibiting CXCL1. CXCL1 was regulated by several pathways, including JNK, p38 MAPK, and PI-3K/Akt signaling pathways (24, 25). Our previous study indicated DACH1 suppressed CXCL8 through AP-1 and NF-κB sites of CXCL8 promoter depending on the DS domain of DACH1 in NSCLC (11). And other study showed that DACH1 repressed CXCL6 and CXCL8 in a dose-dependent manner, and repression required the DS domain in prostate cancer (21). CXCL1, CXCL6, and CXCL8 all belong to CXC chemokine family. Therefore, it is rationally assumed that CXCL1 was suppressed by DACH1 in a similar way.
Our previous study showed that DACH1 antagonized the function of CXCL5 and revealed opposite effect on prognosis (22). To explore the whether CXCL1 and DACH1 had similar role, we plotted the survival curves of CXCL1 and DACH1 for ADC patients. The results showed that high CXCL1 associated with short survival time, while as the high DACH1 predicted long OS and PFS as previously reported (22). To explore the combined effects of CXCL1 and DACH1 on prognosis, the blend curves was performed on public database GSE31210 which includes 226 samples from ADC patients. The results suggested that high CXCL1 with low DACH1 indicated worse OS and PFS. Therefore, CXCL1 combining DACH1 could more precisely stratify patients with lung ADC in term of prognosis.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of the Tongji Hospital of Huazhong University of Science and Technology. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SY performed the experiments and drafted the manuscript. LX, MY, and SQ prepared the figures and tables. AL and KW designed the study and revised the manuscript. All authors contributed to this manuscript, read, and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81874120, 81572608) and Wuhan Science and Technology Bureau (No. 2017060201010170).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer; ADC, adenocarcinoma; SQC, squamous cell carcinoma; EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; CXCL1, chemokine (C-X-C motif) ligand 1; CXCR2, CXC receptor 2; VEGF, vascular endothelial growth factor; TNF, tumor necrosis factor; MPE, malignant pleural effusion; DACH1, Dachshund homolog 1; RDGN, retinal determination gene network; EMT, epithelial-mesenchymal transition; ELISA, enzyme-linked immunosorbent assay; CSC, cancer stem cell; TMA, tissue microarray; OD, optical densities; GEO, Gene Expression Omnibus; OS, overall survival; PFS, progression-free survival; HR, hazard ratio.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
2. Postmus PE, Kerr KM, Oudkerk M, Senan S, Waller DA, Vansteenkiste J, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2017) 28(Suppl. 4):iv1–21. doi: 10.1093/annonc/mdx222
3. Liu Q, Yu S, Zhao W, Qin S, Chu Q, Wu K. EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol Cancer. (2018) 17:53. doi: 10.1186/s12943-018-0793-1
4. Wu AJ, Garay E, Foster A, Hsu M, Zhang Z, Chaft JE, et al. Definitive radiotherapy for local recurrence of NSCLC after surgery. Clin Lung Cancer. (2017) 18:e161–8. doi: 10.1016/j.cllc.2017.01.014
5. Zhao H, Huang Y, Shi J, Dai Y, Wu L, Zhou H. ABCC10 plays a significant role in the transport of gefitinib and contributes to acquired resistance to gefitinib in NSCLC. Front Pharmacol. (2018) 9:1312. doi: 10.3389/fphar.2018.01312
6. Schmid S, Gautschi O, Rothschild S, Mark M, Froesch P, Klingbiel D, et al. Clinical outcome of ALK-positive non-small cell lung cancer (NSCLC) patients with de novo EGFR or KRAS co-mutations receiving tyrosine kinase inhibitors (TKIs). J Thorac Oncol. (2017) 12:681–8. doi: 10.1016/j.jtho.2016.12.003
7. Bylicki O, Paleiron N, Margery J, Guisier F, Vergnenegre A, Robinet G, et al. Targeting the PD-1/PD-L1 immune checkpoint in EGFR-mutated or ALK-translocated non-small-cell lung cancer. Target Oncol. (2017) 12:563–9. doi: 10.1007/s11523-017-0510-9
8. Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. (2012) 150:1121–34. doi: 10.1016/j.cell.2012.08.024
9. Popov VM, Wu K, Zhou J, Powell MJ, Mardon G, Wang C, et al. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol Metab. (2010) 21:41–9. doi: 10.1016/j.tem.2009.08.002
10. Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu Rev Biochem. (2007) 76:513–38. doi: 10.1146/annurev.biochem.76.052705.164916
11. Liu Q, Li A, Yu S, Qin S, Han N, Pestell RG, et al. DACH1 antagonizes CXCL8 to repress tumorigenesis of lung adenocarcinoma and improve prognosis. J Hematol Oncol. (2018) 11:53. doi: 10.1186/s13045-018-0597-1
12. Chen K, Wu K, Cai S, Zhang W, Zhou J, Wang J, et al. Dachshund binds p53 to block the growth of lung adenocarcinoma cells. Cancer Res. (2013) 73:3262–74. doi: 10.1158/0008-5472.CAN-12-3191
13. Zhu H, Wu K, Yan W, Hu L, Yuan J, Dong Y, et al. Epigenetic silencing of DACH1 induces loss of transforming growth factor-beta1 antiproliferative response in human hepatocellular carcinoma. Hepatology. (2013) 58:2012–22. doi: 10.1002/hep.26587
14. Watanabe A, Ogiwara H, Ehata S, Mukasa A, Ishikawa S, Maeda D, et al. Homozygously deleted gene DACH1 regulates tumor-initiating activity of glioma cells. Proc Natl Acad Sci USA. (2011) 108:12384–9. doi: 10.1073/pnas.0906930108
15. Xu H, Yu S, Yuan X, Xiong J, Kuang D, Pestell RG, et al. DACH1 suppresses breast cancer as a negative regulator of CD44. Sci Rep. (2017) 7:4361. doi: 10.1038/s41598-017-04709-2
16. Yan W, Wu K, Herman JG, Brock MV, Fuks F, Yang L, et al. Epigenetic regulation of DACH1, a novel Wnt signaling component in colorectal cancer. Epigenetics. (2013) 8:1373–83. doi: 10.4161/epi.26781
17. Chu Q, Han N, Yuan X, Nie X, Wu H, Chen Y, et al. DACH1 inhibits cyclin D1 expression, cellular proliferation and tumor growth of renal cancer cells. J Hematol Oncol. (2014) 7:73. doi: 10.1186/s13045-014-0073-5
18. Wu K, Chen K, Wang C, Jiao X, Wang L, Zhou J, et al. Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. (2014) 74:829–39. doi: 10.1158/0008-5472.CAN-13-2466
19. Wu K, Jiao X, Li Z, Katiyar S, Casimiro MC, Yang W, et al. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J Biol Chem. (2011) 286:2132–42. doi: 10.1074/jbc.M110.148395
20. Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu H, et al. The DACH-EYA-SIX gene network and its role in tumor initiation and progression. Int J Cancer. (2016) 138:1067–75. doi: 10.1002/ijc.29560
21. Chen K, Wu K, Jiao X, Wang L, Ju X, Wang M, et al. The endogenous cell-fate factor dachshund restrains prostate epithelial cell migration via repression of cytokine secretion via a cxcl signaling module. Cancer Res. (2015) 75:1992–2004. doi: 10.1158/0008-5472.CAN-14-0611
22. Han N, Yuan X, Wu H, Xu H, Chu Q, Guo M, et al. DACH1 inhibits lung adenocarcinoma invasion and tumor growth by repressing CXCL5 signaling. Oncotarget. (2015) 6:5877–88. doi: 10.18632/oncotarget.3463
23. Zheng X, Liu Q, Yi M, Qin S, Wu K. The regulation of cytokine signaling by retinal determination gene network pathway in cancer. Oncotargets Ther. (2018) 11:6479–87. doi: 10.2147/OTT.S176113
24. Lo HM, Shieh JM, Chen CL, Tsou CJ, Wu WB. Vascular endothelial growth factor induces CXCL1 chemokine release via JNK and PI-3K-dependent pathways in human lung carcinoma epithelial cells. Int J Mol Sci. (2013) 14:10090–106. doi: 10.3390/ijms140510090
25. Shieh JM, Tsai YJ, Tsou CJ, Wu WB. CXCL1 regulation in human pulmonary epithelial cells by tumor necrosis factor. Cell Physiol Biochem. (2014) 34:1373–84. doi: 10.1159/000366344
26. Cabrero-de Las Heras S, Martinez-Balibrea E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J Gastroenterol. (2018) 24:4738–49. doi: 10.3748/wjg.v24.i42.4738
27. Miyake M, Furuya H, Onishi S, Hokutan K, Anai S, Chan O, et al. Monoclonal antibody against CXCL1 (HL2401) as a novel agent in suppressing IL6 expression and tumoral growth. Theranostics. (2019) 9:853–67. doi: 10.7150/thno.29553
28. Wang L, Zhang C, Xu J, Wu H, Peng J, Cai S, et al. CXCL1 gene silencing inhibits HGC803 cell migration and invasion and acts as an independent prognostic factor for poor survival in gastric cancer. Mol Med Rep. (2016) 14:4673–79. doi: 10.3892/mmr.2016.5843
29. Keshamouni VG, Arenberg DA, Reddy RC, Newstead MJ, Anthwal S, Standiford TJ. PPAR-gamma activation inhibits angiogenesis by blocking ELR+CXC chemokine production in non-small cell lung cancer. Neoplasia. (2005) 7:294–301. doi: 10.1593/neo.04601
30. Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. (2012) 150:165–78. doi: 10.1016/j.cell.2012.04.042
31. Yuan M, Zhu H, Xu J, Zheng Y, Cao X, Liu Q. Tumor-derived CXCL1 promotes lung cancer growth via recruitment of tumor-associated neutrophils. J Immunol Res. (2016) 2016:6530410. doi: 10.1155/2016/6530410
32. Yu PF, Huang Y, Han YY, Lin LY, Sun WH, Rabson AB, et al. TNFalpha-activated mesenchymal stromal cells promote breast cancer metastasis by recruiting CXCR2(+) neutrophils. Oncogene. (2017) 36:482–90. doi: 10.1038/onc.2016.217
33. Spaks A, Jaunalksne I, Spaka I, Chudasama D, Pirtnieks A, Krievins D. Diagnostic value of circulating CXC chemokines in non-small cell lung cancer. Anticancer Res. (2015) 35:6979–83.
34. Sharma B, Nawandar DM, Nannuru KC, Varney ML, Singh RK. Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis. Mol Cancer Ther. (2013) 12:799–808. doi: 10.1158/1535-7163.MCT-12-0529
35. Keane MP, Belperio JA, Xue YY, Burdick MD, Strieter RM. Depletion of CXCR2 inhibits tumor growth and angiogenesis in a murine model of lung cancer. J Immunol. (2004) 172:2853–60. doi: 10.4049/jimmunol.172.5.2853
36. Fujimori A, Okayasu R, Ishihara H, Yoshida S, Eguchi-Kasai K, Nojima K, et al. Extremely low dose ionizing radiation up-regulates CXC chemokines in normal human fibroblasts. Cancer Res. (2005) 65:10159–63. doi: 10.1158/0008-5472.CAN-05-2015
37. Chen G, Kim S, King A, Zhao L, Simpson R, Christensen P, et al. CYP24A1 is an independent prognostic marker of survival in patients with lung adenocarcinoma. Clin Cancer Res. (2011) 17:817–26. doi: 10.1158/1078-0432.CCR-10-1789
38. Wu K, Li A, Rao M, Liu M, Dailey V, Yang Y, et al. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol Cell Biol. (2006) 26:7116–29. doi: 10.1128/MCB.00268-06
39. Xu H, Wu K, Tian Y, Liu Q, Han N, Yuan X, et al. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol. (2016) 49:1343–1350. doi: 10.3892/ijo.2016.3639
40. Fromowitz FB, Viola MV, Chao S, Oravez S, Mishriki Y, Finkel G, et al. ras p21 expression in the progression of breast cancer. Hum Pathol. (1987) 18:1268–75. doi: 10.1016/S0046-8177(87)80412-4
41. Liu Q, Li A, Tian Y, Liu Y, Li T, Zhang C, et al. The expression profile and clinic significance of the SIX family in non-small cell lung cancer. J Hematol Oncol. (2016) 9:119. doi: 10.1186/s13045-016-0339-1
42. Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. (2006) 439:353–7. doi: 10.1038/nature04296
43. Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. (2006) 66:7466–72. doi: 10.1158/0008-5472.CAN-06-1191
44. Baty F, Facompre M, Kaiser S, Schumacher M, Pless M, Bubendorf L, et al. Gene profiling of clinical routine biopsies and prediction of survival in non-small cell lung cancer. Am J Respir Crit Care Med. (2010) 181:181–8. doi: 10.1164/rccm.200812-1807OC
45. Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. (2006) 24:1679–88. doi: 10.1200/JCO.2005.03.8224
46. Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. (2009) 27:2793–9. doi: 10.1200/JCO.2008.19.7053
47. Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. (2010) 5:e10312. doi: 10.1371/journal.pone.0010312
48. Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. (2011) 17:5705–14. doi: 10.1158/1078-0432.CCR-11-0196
49. Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. (2013) 19:194–204. doi: 10.1158/1078-0432.CCR-12-1139
50. Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. (2013) 19:1577–86. doi: 10.1158/1078-0432.CCR-12-2321
51. Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. (2014) 9:59–64. doi: 10.1097/JTO.0000000000000042
52. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. (2008) 14:822–27. doi: 10.1038/nm.1790
53. Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. (2002) 8:816–24. doi: 10.1038/nm733
54. Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. (2013) 5:186ra166. doi: 10.1126/scitranslmed.3005723
55. Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. (2008) 14:7397–404. doi: 10.1158/1078-0432.CCR-07-4937
56. Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. (2008) 3:e1651. doi: 10.1371/journal.pone.0001651
57. Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. (2010) 19:2590–97. doi: 10.1158/1055-9965.EPI-10-0332
58. Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. (2009) 63:32–8. doi: 10.1016/j.lungcan.2008.03.033
59. Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. (2010) 28:4417–24. doi: 10.1200/JCO.2009.26.4325
60. Tarca AL, Lauria M, Unger M, Bilal E, Boue S, Kumar Dey K, et al. Strengths and limitations of microarray-based phenotype prediction: lessons learned from the IMPROVER diagnostic signature challenge. Bioinformatics. (2013) 29:2892–9. doi: 10.1093/bioinformatics/btt492
61. Heiskanen M, Topal M, Hoadley K. caArray_EXP-576: TCGA (LUAD): Gene Expression Analysis of TCGA Samples Using Agilent Expression 244K Microarrays (UNC).
62. Wu K, Yu S, Liu Q, Bai X, Zheng X, Wu K. The clinical significance of CXCL5 in non-small cell lung cancer. Oncotargets Ther. (2017) 10:5561–73. doi: 10.2147/OTT.S148772
63. Drummond RA, Swamydas M, Oikonomou V, Zhai B, Dambuza IM, Schaefer BC, et al. CARD9(+) microglia promote antifungal immunity via IL-1beta- and CXCL1 mediated neutrophil recruitment. Nat Immunol. (2019) 20:559–70. doi: 10.1038/s41590-019-0377-2
64. Hwang WL, Lan HY, Cheng WC, Huang SC, Yang MH. Tumor stem-like cell-derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. J Hematol Oncol. (2019) 12:10. doi: 10.1186/s13045-019-0699-4
65. Killian PH, Kronski E, Michalik KM, Barbieri O, Astigiano S, Sommerhoff CP, et al. Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and−2. Carcinogenesis. (2012) 33:2507–19. doi: 10.1093/carcin/bgs312
66. Yang C, Yu H, Chen R, Tao K, Jian L, Peng M, et al. CXCL1 stimulates migration and invasion in ER-negative breast cancer cells via activation of the ERK/MMP2/9 signaling axis. Int J Oncol. (2019) 55:684–96. doi: 10.3892/ijo.2019.4840
Keywords: NSCLC, lung adenocarcinoma, CXCL1, DACH1, progression, prognosis
Citation: Yu S, Yi M, Xu L, Qin S, Li A and Wu K (2020) CXCL1 as an Unfavorable Prognosis Factor Negatively Regulated by DACH1 in Non-small Cell Lung Cancer. Front. Oncol. 9:1515. doi: 10.3389/fonc.2019.01515
Received: 30 August 2019; Accepted: 16 December 2019;
Published: 10 January 2020.
Edited by:
Jorge J. Nieva, Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, United StatesReviewed by:
Yangfu Jiang, Sichuan University, ChinaTianhong Li, Comprehensive Cancer Center, University of California, Davis, United States
Copyright © 2020 Yu, Yi, Xu, Qin, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anping Li, li_anpig@yahoo.com; Kongming Wu, kmwu@tjh.tjmu.edu.cn
 Shengnan Yu1
Shengnan Yu1 Ming Yi
Ming Yi Kongming Wu
Kongming Wu