- 1Department of Colorectal Surgery, Changhai Hospital, Shanghai, China
- 2Department of Nephrology, Changhai Hospital, Shanghai, China
- 3Department of Pathology, Changhai Hospital, Shanghai, China
Background: Next generation sequencing (NGS)-based multi-gene panel tests have been performed to predict the treatment response and prognosis in patients with colorectal cancer (CRC). Whether the multi-gene mutation results of formalin-fixed paraffin-embedded (FFPE) tissues are identical to those of fresh frozen tissues remains unknown.
Methods: A 22-gene panel with 103 hotspots was used to detect mutations in paired fresh frozen tissue and FFPE tissue from 118 patients with CRC.
Results: In our study, 117 patients (99.2%) had one or more variants, with 226 variants in FFPE tissue and 221 in fresh frozen tissue. Of the 129 variants identified in this study, 96 variants were present in both FFPE and fresh frozen tissues; 27 variants were found in FFPE tissues only; 6 variants were found only in fresh frozen tissues. The mutation results demonstrated >94.0% concordance in all variants, with Kappa coefficient >0.500 in 64.3% (83/129) of variants. At the gene level, concordance ranged from 73.8 to 100.0%, with Kappa coefficient >0.500 in 81.3% (13/16) of genes.
Conclusions: The results of mutation analysis performed with a multi-gene panel and FFPE and fresh frozen tissue were highly concordant in patients with CRC, at both the variant and gene levels. There were, however, some important differences in mutation results between the two tissue types. Therefore, fresh frozen tissue should not routinely be replaced with FFPE tissue for mutation analysis with a multi-gene panel. Rather, FFPE tissue is a reasonable alternative for fresh frozen tissue when the latter is unavailable.
Introduction
The survival of patients with colorectal cancer (CRC) has improved greatly in recent decades, because of advancements in surgical technique, chemoradiotherapy, and targeted therapy (1, 2). However, CRC remains the third most common cancer and the fourth cause of cancer-related death worldwide (3). Molecular biomarkers have been reported to play a vital role in the early diagnosis of CRC, and individualized treatment for metastatic CRC (4, 5). In the past decade, somatic gene mutations of KRAS (KRAS proto-oncogene, GTPase), NRAS (NRAS proto-oncogene, GTPase), and BRAF (B-Raf proto-oncogene, serine/threonine kinase) have been used to predict the outcomes of EGFR (epidermal growth factor receptor)—targeted therapy for metastatic CRC (4). Research studies of targeted therapy and individualized medicine have identified additional genes associated with the development and treatment of CRC (6). The classification of CRC based on multiple gene detection may help to explain inter-individual differences in treatment response and long-term outcomes. For example, the UGT1A (UDP glucuronosyltransferase family 1 member A complex locus) polymorphism has been reported to predict drug toxicity (delayed severe diarrhea) in patients with CRC who receive irinotecan-based chemotherapy (7, 8). Somatic mutations of MLH1 (mutL homolog 1), MSH2 (mutS homolog 2), MSH6 (mutS homolog 6), and PMS2 (PMS1 homolog 2, mismatch repair system component) in CRC tissue could be used to screen for Lynch syndrome (9, 10). Moreover, the detection of multiple gene mutations could offer more options for new targeted treatment efforts in drug-resistant patients (11). Hence, the need for combined detection of multiple gene mutations has acquired increasing urgency.
Next generation sequencing (NGS) is an efficient and rapid tool for the detection of single-nucleotide mutations and small-fragment insertion/deletions. This approach has become the standard technique for multiple gene detection (12). Compared with the single-gene mutation detection, multiple gene detection with NGS is a timely and cost-effective technique that requires a small amount of DNA (13, 14). In our previous study (15), we established a 22-gene panel, which included 103 amplicons targeting the variants found to be most common in CRC. Those 22 genes included KRAS, BRAF, TP53 (tumor protein p53), EGFR, CTNNB1 (catenin beta 1), DDR2 (discoidin domain receptor tyrosine kinase 2), ERBB2 (erb-b2 receptor tyrosine kinase 2), ERBB4 (erb-b2 receptor tyrosine kinase 4), FBXW7 (F-box and WD repeat domain containing 7), FGFR1 (fibroblast growth factor receptor 1), FGFR2 (fibroblast growth factor receptor 2), FGFR3 (fibroblast growth factor receptor 3), AKT1 (AKT serine/threonine kinase 1), ALK (ALK receptor tyrosine kinase), MAP2K1 (mitogen-activated protein kinase kinase 1), MET (MET proto-oncogene, receptor tyrosine kinase), NOTCH1 (notch receptor 1), NRAS, PIK3CA (phosphatidylinositol-4,5- bisphosphate 3-kinase catalytic subunit alpha), PTEN (phosphatase and tensin homolog), SMAD4 (SMAD family member 4), and STK11 (serine/threonine kinase 11). Use of this panel requires only 10 ng of DNA and a single-tube multiplex polymerase chain reaction (PCR).
Fresh frozen tissue is the preferred sample to detect gene mutation due to its superiority in preserving DNA. However, fresh frozen tissue is often not available in clinical practice, as the associated protocol requires that resected tissue be snap-frozen in liquid nitrogen 30–60 min after surgical resection. Moreover, the cost of preserving fresh frozen tissue is relatively high, as it requires the maintenance of a constant ultralow temperature. Compared with fresh frozen tissue, formalin-fixed paraffin-embedded (FFPE) tissue has several advantages: (1) preservation of the cellular and architectural morphology; (2) the possibility of storage at room temperature for several years; (3) easy availability, as FFPE blocks are routinely prepared in the pathology departments of most centers. Therefore, FFPE tissue has become the sample type used most commonly for molecular testing (11). However, the fixation and archiving process in FFPE often leads to the cross-linking, degradation, and fragmentation of DNA molecules. These alterations inevitably affect the quality and quantity of DNA extracted from FFPE tissue, which in turn would adversely impact the accuracy with which gene mutations are detected (16–19). Nevertheless, the detection of EGFR and KRAS mutations in DNA extracted from FFPE tissue has proven to be highly accurate (20). The detection of gene mutations using NGS may also be performed with DNA from FFPE samples (13, 14, 16, 21).
In our previous study, we showed the utility of this 22-gene panel when used with NGS to detect gene mutations in FFPE tissue from 207 patients with CRC (15). However, whether the gene mutation results of this multi-gene panel are concordant between FFPE and fresh frozen tissue remains unknown. In this study, gene mutations were detected in paired FFPE and fresh frozen primary tumor tissue from patients with CRC using this 22-gene panel. The results obtained were compared between tissue types at the variant and gene levels.
Materials and Methods
Inclusion Criteria
Patients who satisfied all of the following criteria were included: (1) pathologically proven primary CRC adenocarcinoma; (2) history of radical surgery for a primary tumor at Changhai Hospital (Shanghai, China) during the period from March 2015 to November 2016; (3) availability of FFPE and fresh frozen primary tumor tissue samples.
Exclusion Criteria
Patients were excluded if they met any one of the following criteria: (1) history of preoperative radiotherapy; (2) insufficient FFPE or fresh frozen tumor tissue for the extraction of DNA; (3) history of local tumor excision; (4) recurrent CRC; (5) personal history of other tumors; (6) hereditary CRC.
Patients
All patients with CRC satisfying the above-mentioned criteria were enrolled in the study. All the relevant clinicopathological information was prospectively maintained in an electronic CRC database. The study was approved by the ethical committee at Changhai Hospital. All included patients provided their written informed consent. All patients were followed up after surgery every 3 months for the first 2 years, every 6 months for the next 3 years, and every year from that point onward.
Processing of Tumor Tissue
All tumor tissue samples were obtained from surgically resected primary CRC tumor specimens. The resected tissue was divided into two parts. One part was frozen in liquid nitrogen <30 min after surgery and stored at −80°C until the time of DNA extraction; the other part was fixed in 4% formalin for 24–72 h and embedded in paraffin, and then stored at room temperature. The blocks containing FFPE tissues were divided to obtain ten consecutive sections with thickness of 10 μm. Hematoxylin and eosin staining was performed on the first section. Two pathologists examined each stained section individually and estimated the percentage of neoplastic cells. Sections with ≥ 40% neoplastic cells were considered as eligible. The remaining nine sections were pooled into a 1.5-mL tube.
DNA Isolation
The QIAamp DNA Mini Kit(Qiagen) and GeneRead DNA FFPE kit (Qiagen) were used to extract DNA from fresh frozen and FFPE tissues, respectively, according to the manufacturer's protocols. The detailed methods used for DNA extraction from FFPE tissue samples were reported previously by our group (15). Briefly, FFPE tissue samples were dewaxed with deparaffinization solution, then incubated with lysis buffer for 1 h. After incubation at 90°C to remove cross-links, Uracil-N-Glycosilase was added for the specific removal of deaminated cytosine residues from the DNA. For fresh frozen tissues, 25 mg of tumor tissues were used for DNA extraction. After being homogenized with the Bioprep-24 homogenizer, the tissues were incubated in lysis buffer at 56°C until the tissues were completely lysed. RNase A was used to digest the RNA molecules from the genomic DNA. For both FFPE and fresh frozen tissue, DNA was eluted and quantified with a Qubit 3.0 fluorometer and dsDNA HS assay kit (Life Technologies).
DNA Amplification and Sequencing
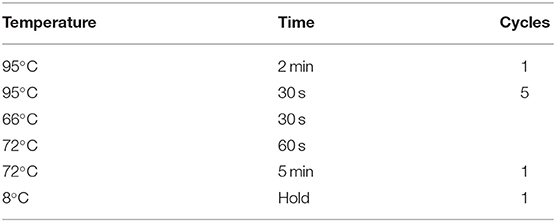
For sequencing, 20 ng DNA extracted from each FFPE or fresh frozen sample was used for library construction. In brief, gene-specific PCR was used to amplify 103 regions in the first round, followed by purification via size selection. The details of the first round of PCR were showed in Table 1. Subsequently, the second round of PCR (“indexing PCR”) was conducted (Table 2). The 22-gene panel was purchased from Pillar Biosciences, USA (ONCO/Reveal Lung & Colon Cancer Panel). The details of the 22-gene panel were same as described previously (14, 15). This process involves the addition of Illumina index adaptors to purify the products for sample tracking and sequencing. Lastly, the libraries were eluted in 22 μL nuclease-free water. The final libraries were quantified using a Qubit 3.0 fluorometer and dsDNA HS assay kit (Life Technologies), as per the manufacturer's protocol. The MiSeq was used for sequencing libraries according to the manufacturer's protocol. Each gene library was normalized to 4 nM and combined at equal volume (4 μL). The mixed library was then denatured with 0.2 N NaOH and diluted up to the concentration of 15 pM for sequencing with MiSeq Reagent Kit v2 at 300 cycles (or 20 pM for v3). Sample quality of FFPE tissues was determined by the amount of amplifiable DNA using qPCR and sequencing libraries were evaluated using Bioanalyzer.
Variant Analysis
Subsequently, they were annotated with the 1000 genomes (https://www.internationalgenome.org/home), which is one of the most frequently used databases in genetic research. Variants were selected for known single nucleotide polymorphisms (SNPs) and synonymous mutations. All non-coding, silent, synonymous, unknown and common germline variants were filtered out, as well as all variants present in 1,000 G data (22). Moreover, all variants at a locus with coverage of <200, or variants with a variant frequency <0.05 were excluded. The remaining mutations were assessed using the Catalog of Somatic Mutations in Cancer (COSMIC) database (14, 23). The SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) online databases were used to analyze the clinical significance of these variants.
Statistical Analysis
All data were analyzed using the Statistical Package for Social Sciences (SPSS 22.0, Chicago, IL). Categorical parameters were recorded as frequency and percentage; continuous parameters were described as mean and standard deviation, or median and interquartile range (IQR), as appropriate. Concordance rate and Kappa coefficient were used to compare FFPE and fresh frozen tissue in terms of mutation results, at both the variant and gene levels. P < 0.05 (two-sided) was considered as statistically significant.
Results
Clinicopathological Features of Patients Included in the Study
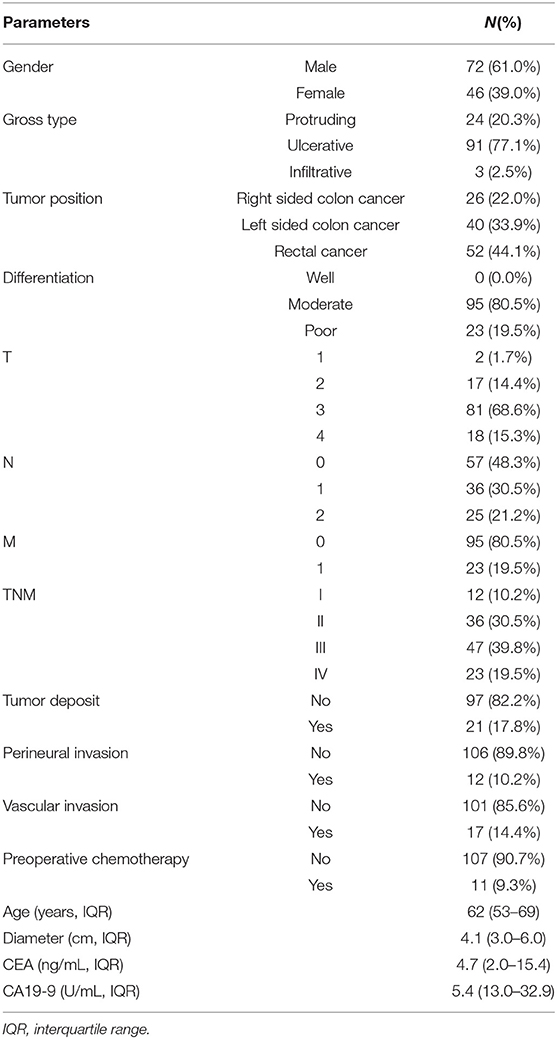
Of the 118 patients with CRC included in this study, 72 (61%) were men. The median age (IQR) was 62 (53–69) years. Most of the patients had TNM stage II (n = 36) or III (n = 47) disease. Eleven (9.3%) patients received preoperative chemotherapy (Table 3). The storage period of FFPE tissues ranged from 3 to 24 months, with a median period of 10 months.
Comparisons of Gene Mutation Characteristics Between FFPE and Fresh Frozen Tissue
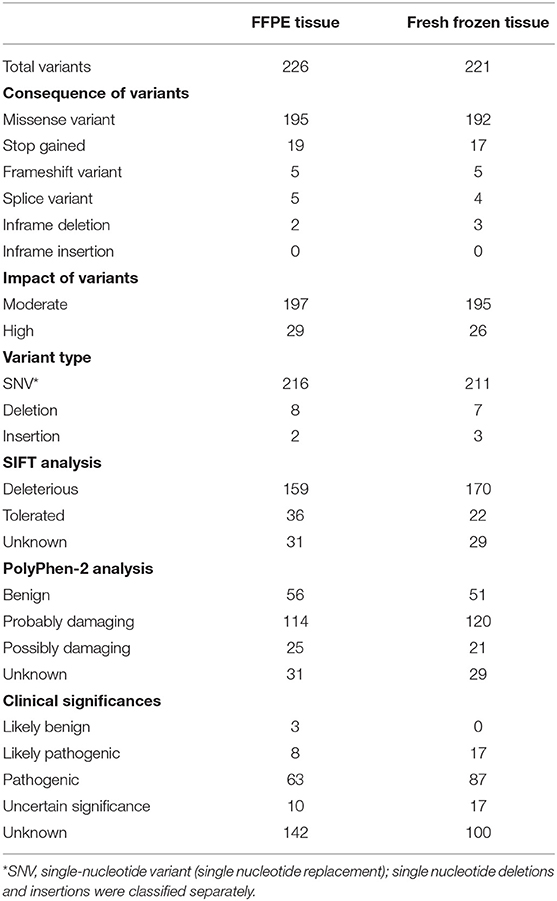
The median and interquartile of total coverage across all genes were 3739 (2148–5866) reads for fresh frozen tissues, which were significantly higher than those of FFPE tissues [2,814 (1,784–3,936) reads] (P < 0.001). In our study, 117 patients (99.2%) had one or more variants, with 226 variants in FFPE tissues and 221 in fresh frozen tissues. All variants identified in the FFPE and fresh frozen tissues are showed in Supplemental Table 1.
FFPE tissue analysis revealed at least one variant in 112 patients (94.9%), yielding a total of 226 variants. Among 112 patients, 44, 39, 19, 7, 2, and 1 had one, two, three, four, six and seven variants, respectively. The genes mutated most frequently were TP53 (54.2%), KRAS (47.5%), PIK3CA (21.2%), and FBXW7 (15.3%). No mutations were identified in ALK, FGFR1, FGFR3, MET, NOTCH1, or STK11 (Figure 1).
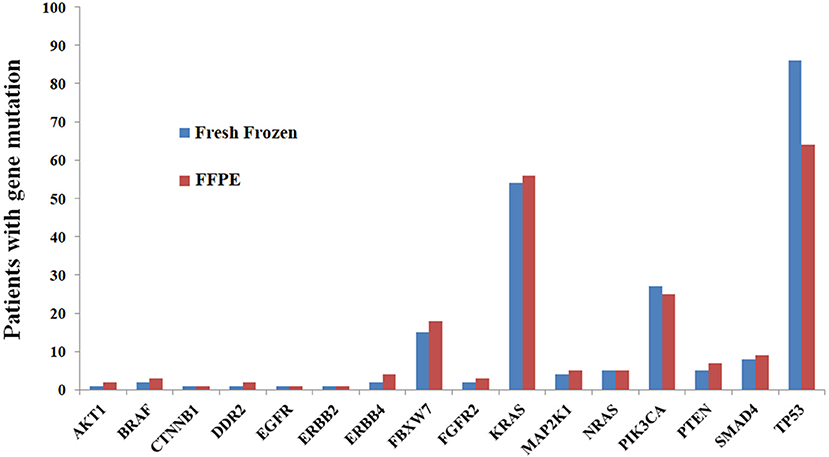
Figure 1. Comparisons of mutation results between FFPE and fresh frozen tissue at the gene level, showing high concordance.
The analysis of fresh frozen tissue revealed the presence of at least one variant in 105 patients (89.0%), yielding a total of 221 variants. Among the 105 patients, 39, 36, 18, 5, 6, and 1 patients had one, two, three, four, five and six variants, respectively. The list of most frequently mutated genes was similar to that developed for FFPE tissues: TP53 (72.9%), KRAS (45.8%), PIK3CA (22.9%), and FBXW7 (12.7%). No mutations were identified in ALK, FGFR1, FGFR3, MET, NOTCH1, or STK11 (Figure 1).
A comparison between FFPE and fresh frozen tissue in terms of variant characteristics and their clinical significance was shown in Table 4. The impact of all variants were evaluated using the Variant Impact Predictor Database (VIPdb). No obvious difference in variant impact was found between the two samples (Table 4).
Comparison of Gene Mutations Between FFPE and Fresh Frozen Tissue at the Variant Level
A total of 129 variants were identified in this study. Among these variants, 96 were present in both FFPE and fresh frozen tissue; 27 variants were present in FFPE tissues alone; 6 variants were present in fresh frozen tissue alone. Of the 27 variants that existed in FFPE only, 59.3% (16/27) were C>T/G>A or A>G/T>C transitions, 14.8%(4/27) were G>T/T>G transversions, 7.4%(2/27) were A>C transversions, 7.4% (2/27) were A>T/T>A transversions, 3.7%(1/27) was C>G transversion and 7.4% (2/27) were deletion. Among the 6 variants that existed in fresh frozen tissue only, 83.3% (5/6) were C>T/G>A or A>G/T>C transitions, and 16.7% (1/6) was A>C transversion. Comparison of the gene mutations at the variant level revealed that concordance rates were 100.0% for 38.0% (49/129) variants, and concordance rates were higher than 94.0% for all variants (Supplemental Table 2). Kappa coefficients were higher than 0.500 in 64.3% (83/129) of variants (Supplemental Table 2).
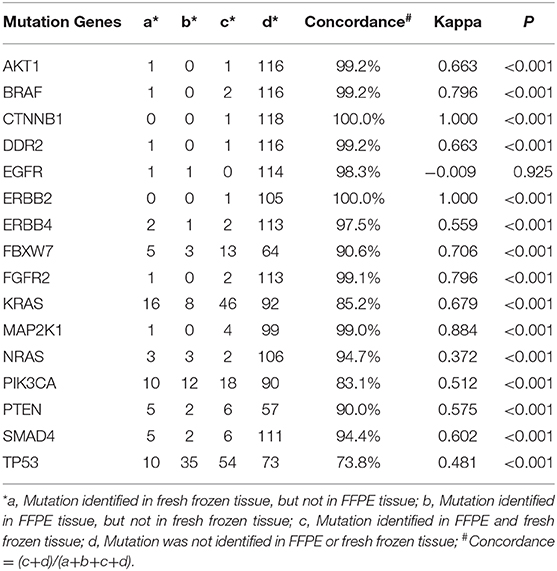
Comparison of Gene Mutations Between FFPE and Fresh Frozen Tissue at the Gene Level
The concordance rates of gene mutations ranged from 73.8 to 100.0% for all genes (Table 5, Figure 1). Kappa coefficients were >0.500 in 81.3% (13/16) of genes (Table 5).
Discussion
In the present study, we found that 117 patients (99.2%) had one or more variants, with 226 variants in FFPE tissues and 221 in fresh frozen tissues. The concordance rates of gene mutation results in FFPE vs. fresh frozen tissue were higher than 94% for all variants, with Kappa coefficient >0.500 in 64.3% (83/129) variants. At the gene level, concordance ranged from 73.8 to 100.0%, with Kappa coefficient >0.500 in 81.3% (13/16) of genes. Our results indicated that the mutation results for FFPE and fresh frozen tissue were highly concordant at the variant and gene levels, but there were still some important differences in mutation results between the two tissue types.
The total coverage of fresh frozen tissues was significantly higher than that of FFPE tissues. We hypothesized that this could be due to the smaller DNA fragment size in FFPE tissues as some were below the detection limit. In our study, 226 variants were identified in FFPE tissues and 221 variants were identified in fresh frozen tissues. The more variants detected in FFPE samples might be caused by DNA damage during the formalin fixation process (e.g., fragmentation, degradation, crosslinking). Of the 27 variants identified only in FFPE tissues, 16 (59.3%) were C>T/G>A or A>G/T>C transitions. These 16 transitions may be artifacts secondary to postmortem deamination of cytosine or adenine to uracil or hypoxanthine residues (20). We took special care to decrease the rate of false-negative results by including only sections with >40% tumor cells, because adjacent normal cells usually have no mutations (20). In a study by Gallegos et al., the authors compared FFPE samples with fresh frozen tissue samples from 47 lung cancer patients in terms of EGFR and KRAS mutations. The authors showed that the success rate of PCR amplification was only 50% in FFPE tissues, with a false-positive rate up to 50% (19). The high false-positive rate may be related to tissue type and fixation method (18). The fixation and archiving processes required for FFPE often lead to the degradation and fragmentation of DNA. In our study, all PCR primers were designed to make sure that the PCR products were <200 bp, which may have contributed to the high concordance observed.
A few small studies previously compared paired FFPE and fresh frozen tissue from cancer patients in terms of microRNA expression, gene expression, and DNA methylation (24–27). De et al. compared the results of whole-exome sequencing in ten matched FFPE and fresh frozen tissue samples from patients with melanoma (28), and found that the average concordance rate was 43.2% over a total of 1,299 variants for the chosen 27 genes (28). The low concordance rate may be related to the length of PCR product. Spencer et al. compared the variants of 27 cancer-related genes between 16 pairs of fresh frozen and FFPE tissues from patients with lung carcinoma (29), and found that the concordance rate was up to 96.8% in the single-nucleotide variants. Solassol et al. compared the mutation status of KRAS in 33 patients with metastatic CRC, using paired fresh frozen and FFPE tumor tissues. The findings obtained revealed a concordance rate of only 81.9% (20). Betge et al. studied gene mutations in paired FFPE and fresh frozen tissue samples from hepatic metastases from 10 patients with CRC. The results revealed a high concordance between samples, with 21 identical variants and 2 different variants (22). All of the above-mentioned studies used NGS to detect mutation, but the testing method were different. The various concordance rates may be related to the various factors such as primer, length of PCR product, tumor type, and testing methods. These results proved that remarkable differences existed between results of FFPE tissues and fresh frozen tissues, which were consistent with our finding. To the best of our knowledge, this is the first study of its kind to systematically compare the rate of mutation in paired fresh frozen and FFPE CRC tissue samples, at the gene and variant levels. Furthermore, our study had a relatively large sample size (118 patients), compared with previous relevant studies.
Although fresh frozen tissue is the gold standard for molecular analyses, its use in clinical practice is impractical because of its high cost and technical difficulty (20). Based on the results of this study, we suggest that archived tissues from pathology departments may be used for mutation detection with the 22-gene panel. The feasibility of using FFPE tissues for mutation detection will facilitate future studies of CRC. However, because the accuracy of mutation detection in FFPE tissues is influenced by multiple factors, it is important to standardize the procedure in order to minimize variability. Standardized protocols should be elaborated for sample preparation, storage room requirements, library preparation, evaluating the quality of extracted DNA, and the exclusion of poor-quality samples (22).
This study had some limitations. First, it was a retrospective study of stored FFPE tissues. In clinical practice, the fixation and embedding of specimens (which includes tissue thickness, fixative volume, and fixation time of 24–72 h) was not strictly controlled. Variation in the above factors may have affected the quality of preserved DNA, leading to inaccurate results (20). Second, the processes of fixation and embedding may result in deamination, leading to artifactual mutations or false-negative results. These factors may have had variable impacts at different tumor sites (20). In this study, we only took 10 consecutive sections of FFPE tumor tissues. We were therefore unable to rule out the possibility of intra-tumor heterogeneity in terms of DNA deamination. Third, although it is the largest study of its kind, larger prospective studies are required to validate our results. The number of variants will increase with sample size, leading to a more accurate evaluation of concordance, which is especially important for rare variants. Fourth, our study included FFPE specimens that had been stored in the pathology department for <2 years. The degradation and fragmentation of DNA in FFPE tissues may increase with time. Therefore, our observations cannot be extrapolated to all FFPE specimens.
Conclusion
The gene mutation results of a 22-gene panel showed high concordance between paired FFPE and fresh frozen tissue samples, at both the variant and gene levels. This indicates that FFPE tissues stored for <2 years may be used as an alternative to fresh frozen tissue for detecting gene mutations in patients with CRC. However, any alteration in the preparation or detection process may affect the accuracy of results. Factors that may be affected include fixation time, duration of storage of FFPE specimens, DNA sample quality, and tools used to analyze the variants. This should be taken into consideration when interpreting the findings presented above. Furthermore, the mutation results still showed some differences between tissues. Therefore, fresh frozen tissue should not routinely be replaced with FFPE for mutation analysis with a multi-gene panel; instead, FFPE is a reasonable alternative for fresh frozen tissue when the latter is unavailable. In addition, if the clinical response of EGFR-targeted therapy were not consistent with the mutation results based on FFPE tissue, gene mutation test might be performed again with fresh frozen tissue.
Data Availability Statement
The raw data supporting the conclusions of this article can be found in the supplementary files, and are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethical committee at Changhai Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LL, CB, and WZ: conceptualization. XG, JL, and HG: data curation and writing—original draft. XG and JL: formal analysis. XG: funding acquisition. HG, GY, and PL: investigation. LH: methodology. CB: resources. LL and WZ: supervision. GY: validation. GY, PL, LH, LL, CB, and WZ: writing—review and editing.
Funding
This work was sponsored by Shanghai Pujiang Program (#2019PJD052) and the National Natural Science Foundation of China (#81572332 and #81572358).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00310/full#supplementary-material
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
2. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. (2019) doi: 10.3322/caac.21565
3. Stewart B, Wild, CP. (Eds.). World Cancer Report 2014. Lyon: International Agency for Research on Cancer (IARC) (2014).
4. Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. (2017) 18:197–236. doi: 10.3390/ijms18010197
5. Garde Noguera J, Jantus-Lewintre E, Gil-Raga M, Evgenyeva E, Macia Escalante S, Llombart-Cussac A, et al. Role of RAS mutation status as a prognostic factor for patients with advanced colorectal cancer treated with first-line chemotherapy based on fluoropyrimidines and oxaliplatin, with or without bevavizumab: a retrospective analysis. Mol Clin Oncol. (2017) 6:403–8. doi: 10.3892/mco.2017.1149
6. Goel G. Molecular characterization and biomarker identification in colorectal cancer: toward realization of the precision medicine dream. Cancer Manag Res. (2018) 10:5895–908. doi: 10.2147/CMAR.S162967
7. Liu D, Li J, Gao J, Li Y, Yang R, Shen L. Examination of multiple UGT1A and DPYD polymorphisms has limited ability to predict the toxicity and efficacy of metastatic colorectal cancer treated with irinotecan-based chemotherapy: a retrospective analysis. BMC Cancer. (2017) 17:437. doi: 10.1186/s12885-017-3406-2
8. Liu XH, Lu J, Duan W, Dai ZM, Wang M, Lin S, et al. Predictive value of UGT1A1*28 polymorphism in irinotecan-based chemotherapy. J Cancer. (2017) 8:691–703. doi: 10.7150/jca.17210
9. Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, Goossens M, Ouchene H, Hendriks-Cornelissen SJ, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. (2014) 146:643–6e8. doi: 10.1053/j.gastro.2013.12.002
10. Ziada-Bouchaar H, Sifi K, Filali T, Hammada T, Satta D, Abadi N. First description of mutational analysis of MLH1, MSH2 and MSH6 in algerian families with suspected lynch syndrome. Fam Cancer. (2017) 16:57–66. doi: 10.1007/s10689-016-9917-1
11. Normanno N, Rachiglio AM, Roma C, Fenizia F, Esposito C, Pasquale R, et al. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. J Cell Biochem. (2013) 114:514–24. doi: 10.1002/jcb.24401
12. Rey JM, Ducros V, Pujol P, Wang Q, Buisine MP, Aissaoui H, et al. Improving mutation screening in patients with colorectal cancer predisposition using next-generation sequencing. J Mol Diagn. (2017) 19:589–601. doi: 10.1016/j.jmoldx.2017.04.005
13. Tops BB, Normanno N, Kurth H, Amato E, Mafficini A, Rieber N, et al. Development of a semi-conductor sequencing-based panel for genotyping of colon and lung cancer by the onconetwork consortium. BMC Cancer. (2015) 15:26. doi: 10.1186/s12885-015-1015-5
14. Dijkstra JR, Tops BB, Nagtegaal ID, van Krieken JH, Ligtenberg MJ. The homogeneous mutation status of a 22 gene panel justifies the use of serial sections of colorectal cancer tissue for external quality assessment. Virchows Arch. (2015) 467:273–8. doi: 10.1007/s00428-015-1789-5
15. Gao XH, Yu GY, Hong YG, Lian W, Chouhan H, Xu Y, et al. Clinical significance of multiple gene detection with a 22-gene panel in formalin-fixed paraffin-embedded specimens of 207 colorectal cancer patients. Int J Clin Oncol. (2019) 24:141–52. doi: 10.1007/s10147-018-1377-1
16. Esteve-Codina A, Arpi O, Martinez-Garcia M, Pineda E, Mallo M, Gut M, et al. A comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS ONE. (2017) 12:e0170632. doi: 10.1371/journal.pone.0170632
17. Suciu BA, Pap Z, Denes L, Brinzaniuc K, Copotoiu C, Pavai Z. Allele-specific PCR method for identification of EGFR mutations in non-small cell lung cancer: formalin-fixed paraffin-embedded tissue versus fresh tissue. Rom J Morphol Embryol. (2016) 57:495–500.
18. Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. (2001) 25:409–18. doi: 10.1006/meth.2001.1263
19. Gallegos Ruiz MI, Floor K, Rijmen F, Grunberg K, Rodriguez JA, Giaccone G. EGFR and K-ras mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. (2007) 29:257–64. doi: 10.1155/2007/568205
20. Solassol J, Ramos J, Crapez E, Saifi M, Mange A, Vianes E, et al. KRAS mutation detection in paired frozen and Formalin-Fixed Paraffin-Embedded (FFPE) colorectal cancer tissues. Int J Mol Sci. (2011) 12:3191–204. doi: 10.3390/ijms12053191
21. Hadd AG, Houghton J, Choudhary A, Sah S, Chen L, Marko AC, et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn. (2013) 15:234–47. doi: 10.1016/j.jmoldx.2012.11.006
22. Betge J, Kerr G, Miersch T, Leible S, Erdmann G, Galata CL, et al. Amplicon sequencing of colorectal cancer: variant calling in frozen and formalin-fixed samples. PLoS ONE. (2015) 10:e0127146. doi: 10.1371/journal.pone.0127146
23. Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. (2004) 91:355–8. doi: 10.1038/sj.bjc.6601894
24. Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. (2007) 13:1668–74. doi: 10.1261/rna.642907
25. Kalmar A, Wichmann B, Galamb O, Spisak S, Toth K, Leiszter K, et al. Gene expression analysis of normal and colorectal cancer tissue samples from fresh frozen and matched formalin-fixed, paraffin-embedded (FFPE) specimens after manual and automated RNA isolation. Methods. (2013) 59:S16–9. doi: 10.1016/j.ymeth.2012.09.011
26. Kalmar A, Peterfia B, Wichmann B, Patai AV, Bartak BK, Nagy ZB, et al. Comparison of automated and manual DNA isolation methods for DNA methylation analysis of biopsy, fresh frozen, and formalin-fixed, paraffin-embedded colorectal cancer samples. J Lab Autom. (2015) 20:642–51. doi: 10.1177/2211068214565903
27. Moran B, Das S, Smeets D, Peutman G, Klinger R, Fender B, et al. Assessment of concordance between fresh-frozen and formalin-fixed paraffin embedded tumor DNA methylation using a targeted sequencing approach. Oncotarget. (2017) 8:48126–37. doi: 10.18632/oncotarget.18296
28. De Paoli-Iseppi R, Johansson PA, Menzies AM, Dias KR, Pupo GM, Kakavand H, et al. Comparison of whole-exome sequencing of matched fresh and formalin fixed paraffin embedded melanoma tumours: implications for clinical decision making. Pathology. (2016) 48:261–6. doi: 10.1016/j.pathol.2016.01.001
Keywords: colorectal cancer, multi-gene panel, gene mutation, fresh frozen tissue, formalin-fixed paraffin-embedded tissue
Citation: Gao XH, Li J, Gong HF, Yu GY, Liu P, Hao LQ, Liu LJ, Bai CG and Zhang W (2020) Comparison of Fresh Frozen Tissue With Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients With Colorectal Cancer. Front. Oncol. 10:310. doi: 10.3389/fonc.2020.00310
Received: 03 June 2019; Accepted: 21 February 2020;
Published: 13 March 2020.
Edited by:
Xinmin Li, University of California, Los Angeles, United StatesReviewed by:
Pasquale Pisapia, University of Naples Federico II, ItalyManfred Dietel, Charité Medical University of Berlin, Berlin, Germany
Copyright © 2020 Gao, Li, Gong, Yu, Liu, Hao, Liu, Bai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lian Jie Liu, lianjieliu@yeah.net; Chen Guang Bai, bcg709@126.com; Wei Zhang, weizhang2000cn@163.com
†These authors have contributed equally to this work
 Xian Hua Gao
Xian Hua Gao Juan Li2†
Juan Li2† Guan Yu Yu
Guan Yu Yu Peng Liu
Peng Liu