- 1Department of Neurosurgery, West China Hospital of Sichuan University, Chengdu, China
- 2West China Brain Research Center, West China Hospital of Sichuan University, Chengdu, China
The urokinase-type plasminogen activator(PLAU) and its receptor PLAUR participate in a series of cell physiological activities on the extracellular surface. Abnormal expression of PLAU and PLAUR is associated with tumorigenesis. This study aims to evaluate the prognostic value of PLAU/PLAUR transcription expression in glioma and to explore how they affect the generation and progression of glioma. In this study, online databases are applied, such as Oncomine, GEPIA, CGGA, cBioPortal, and LinkedOmics. Overexpression of PLAU/PLAUR was found to be significantly associated with clinical variables including age, tumor type, WHO grade, histology, IDH-1 mutation, and 1p19q status. PLAU and PLAUR had a high correlation in transcriptional expression levels. High expression of PLAU and PLAUR predicted a poor prognosis in primary glioma and recurrent glioma patients, especially in lower grade gliomas. Cox regression analysis indicated that high expression of PLAU and PLAUR were independent prognostic factors for shorter overall survival in glioma patients. In gene co-expression network analysis PLAU and PLAUR and their co-expression genes were found to be involved in inflammatory activities and tumor-related signaling pathways. In conclusion, PLAU and PLAUR could be promising prognostic biomarkers and potential therapeutic targets of glioma patients.
Introduction
Glioma, a broad category of brain tumors with high fatality rate, is the most common type among primary malignant brain tumors in adults, though accounting for less than 1% of all newly diagnosed tumors (1). Among all kinds of diffuse glioma, 70-75% of them are glioblastoma(GBM), which is the most fatal one, with a median overall survival less than 2 years after standard chemoradiotherapy. Molecular therapy that targets epigenetic alterations is under evaluation and is supposed to lead to an important breakthrough in the treatment of malignancies like GBM (2). An increasing number of evidences suggest that the integrated histological-molecular classification may be superior to the traditional histological classification (3). Biomarkers like isocitrate dehydrogenase 1(IDH-1) and O6-methylguanine-DNA methyltransferase (MGMT) not only play a crucial role in diagnosis and prognosis but also work as potential therapeutic targets. A deeper research of these biomarkers provides an essential framework to treat particular glioma subtypes (4).
PLAU (plasminogen activator, urokinase), also known as UPA, encodes a selected serine protease that converts plasminogen to plasmin. PLAUR, the receptor of PLAU, is bound to cell membranes by a glycosyl phosphatidy linositol anchor, and plays an important role in localizing and promoting plasmin formation. The PLAU-PLAUR system participates in a variety of physiological and pathological processes of the non-malignant cells during embryogenesis, wound healing and post-lactational involution; and it is also involved in tumorigenesis such as angiogenesis and metastasis (5, 6). In recent years, more and more studies have discovered that PLAU/PLAUR system is both involved in chronic diseases like rheumatoid arthritis and Quebec platelet disorder, and in malignant diseases such as breast cancer, colorectal cancer, ovarian cancer, lung cancer, and melanoma, etc. (7–12). In addition to the abnormal expression during tumor progression, PLAU and PLAUR are also found involved in complicated tumor invasion and cell migration. These features result in biological more aggressive tumors and poor prognosis of patients (13, 14).
Previous studies about the relationship between glioma and PLAU/PLAUR have indicated that high expression of PLAU/PLAUR promotes glioma cell invasion, tumor growth, and angiogenesis (15, 16). Moreover, their mechanisms and pathways have been explored and reported these years, such as GRB2/AKT/BAD pathway, PI3k/AKT pathway, and other relevant mechanisms (17–20). Although the number of publications about PLAU/PLAUR in glioma has grown each year, less attention is paid to clinicopathological features and prognosis. In the mid-90s, Hsu et al. reported a correlation between high expression of PLAU and bad prognoses of glioma patients (21). And approximately 20 years ago, Zhang et al. came to the same results using Northern blot hybridization and immunohistochemical detection (22). Nowadays, as large datasets with molecular-genetic data from modern platforms are available, the mentioned survival analytic results with outdated methods and small samples are not representative any more.
Different kinds of online databases, tools and integrate data were applied in this study. First, the transcription expression level of PLAU/PLAUR among glioma patients were investigated. Then, their relations with clinical parameters were analyzed, while the prognostic factors were also analyzed. Furthermore, potential gene functions and pathways were predicted through data mining.
Materials and Methods
Ethics Statement
This study was approved by the Academic Committee of Sichuan University, and conducted according to the principles expressed in the Declaration of Helsinki. All the datasets were retrieved from the published literature, so it was confirmed that all written informed consent was obtained.
Oncomine Database
Oncomine database (http://www.oncomine.org) is a cancer microarray database and integrated data-mining platform, consisting of 715 independent databases and 86733 tumor samples (23). In the current study, the Oncomine database was retrieved for the transcription expression of PLAU and PLAUR in different glioma tissues and adjacent normal brain tissues, and the differences in transcription expression were analyzed with students’ t-test. Cut-off of p-value and fold change were set as follows: p-value: 0.05, fold change: 2, gene rank: top 10%.
GEPIA Database
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) is a newly developed interactive web server for analyzing the RNA sequencing and expression data of 9736 tumors and 8587 normal samples from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx), using a standard processing pipeline (24). Transcription expressions between gliomas and normal brain tissues were compared with GEPIA in this study, and a survival analysis in glioma patients with different WHO grades was also conducted.
CGGA Database
Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn) is a web application for data storage and analysis (25). It explores brain tumors datasets of over 2,000 samples from Chinese cohorts. This database includes the whole-exome sequencing, DNA methylation, mRNA sequencing, mRNA microarray, and microRNA microarray and matched clinical data. Clinicopathological data was downloaded from the CGGA database. A survival analysis was performed on the selected glioma patients. And a Pearson correlation analysis was also carried out.
cBioPortal
The cBio Cancer Genomics Portal (cBioPortal) (https://www.cbioportal.org) is a web for exploring, visualizing, and analyzing multidimensional cancer genomics data (26). Genomic data types integrated by cBioPortal include somatic mutations, DNA copy-number alterations, mRNA and microRNA expression, DNA methylation, protein abundance, and phosphoprotein abundance. The cBioPortal online tool was used for detecting the frequency of gene alterations and for survival analysis between the gene-altered group and unaltered group.
LinkedOmics
LinkedOmics (http://www.linkedomics.org) is a publicly available portal that includes multi-omics data from all 32 TCGA Cancer types (27). This web application is consist of three analytical modules: LinkFinder, LinkInterpreter and LinkCompare. With LinkFinder, attributes relevant to a query attribute would be searched out, for example, mRNA or protein expression signatures of genomic alterations, candidate biomarkers of clinical attributes, and candidate target genes of transcriptional factors, microRNAs, or protein kinases. To get biological interpretation from the association results, an enrichment analysis is performed with the LinkInterpreter module based on Gene Ontology, biological pathways, network modules, among other functional categories. In the current study, LinkFinder and LinkInterpreter were used to explore the potential gene regulation network.
Statistical Analysis
Statistic analyses were carried out using SPSS software(version 22.0, IBM Corp, New York, USA). Overall survival (OS) was defined from the time of surgery to death or last follow-up. Variables with normal distribution were analyzed with students’ t-test, otherwise by the Mann-Whitney U test. One-way analysis of variance was used in the case of more than 2 data sets. Cox regression analysis was used to assess the prognostic values of clinical factors based on the data from CGGA. Statistical significance was defined as two-sided p<0.05.
Results
PLAU and PLAUR Transcription Levels in Gliomas
Oncomine database was applied to investigate whether there were differences between glioma and normal brain tissue in the expression of the PLAU gene. As shown in Table 1, mRNA expressions of PLAU were significantly upregulated in 7 databases respectively, where 5 databases indicated overexpression of PLAU in GBM compared to normal brain tissue, the fold change ranged from 2.7 to 9.1 (28–31). In addition, PLAU mRNA overexpression in other kind of cells was also reported. The dataset of Gutman suggested PLAU mRNA overexpression in pilocytic astrocytoma with a fold change of 4.3 (32). And the Rickman’s dataset suggested it in astrocytoma with a fold change of 17.2 (33). With respect to the transcription expression of PLAUR, 3 databases were retrieved and indicated overexpression of PLAUR in GBM compared to normal brain tissue with fold changes of 2.4, 2.5 and 7.0 respectively (28–30).
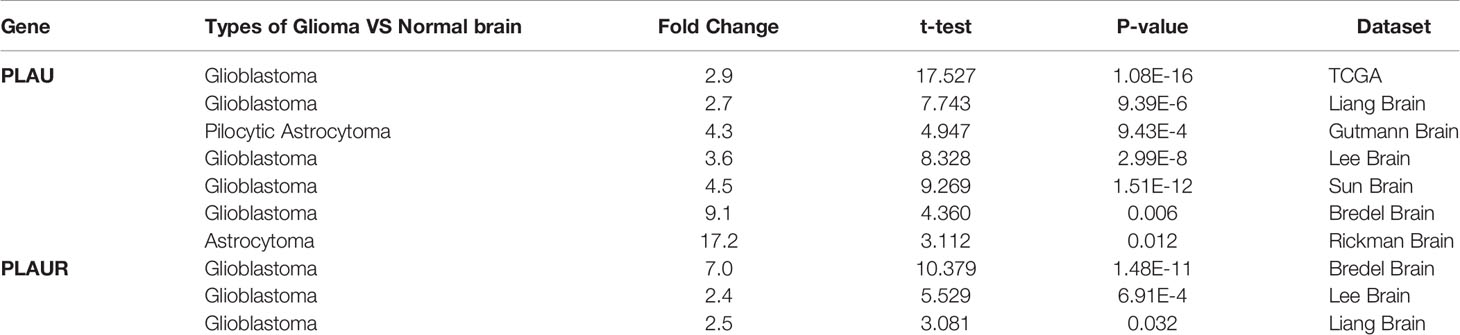
Table 1 Significant changes of PLAU and PLAUR expression in transcription level between glioma and normal brain tissues (ONCOMINE).
A similar analysis was performed in GEPIA. As shown in Figure 1, transcription levels of PLAU and PLAUR were evidently higher in GBM and lower-grade glioma (LGG) than in normal brain tissues. Figure 1 also indicated that the expression levels of two genes in GBM were higher than LGG.

Figure 1 Transcriptional expression of PLAU (A) and PLAUR (B) in gliomas and normal brain tissues(GEPIA). *p < 0.01.
Association of mRNA Expression of PLAU and PLAUR With Clinicopathological Parameters in Glioma Patients
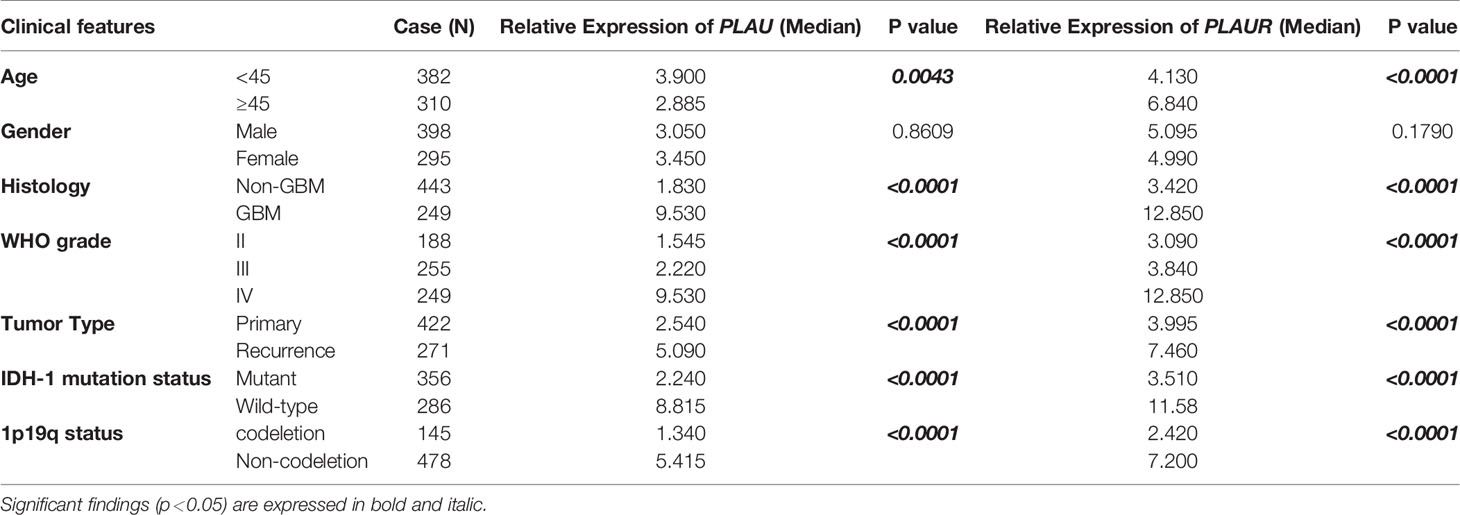
CGGA databases were retrieved for the clinical data of glioma patients, and one of the databases was acquired for further research (Supplementary Tables 1, 2). The database recorded 693 cases of glioma patients (Table 2), including 398 males and 295 females. Among them, 422 cases were primary gliomas while 271 cases were recurrent gliomas. Except for one case without information about the WHO grade, 249 cases were diagnosed as GBM, and 443 cases were diagnosed as LGG. There was no WHO grade I case in the dataset. IDH mutation status was found in 642 cases while 1p19q status was available in 623 cases. Results of stratification and statistic analysis were shown in Table 2. The mRNA expressions of PLAU and PLAUR varied remarkably among different ages, tumor histology, WHO grades and tumor types, IDH-1 mutation, and 1p19q status and other clinical characteristics. The expressions of PLAU and PLAUR were evidently higher in higher WHO grade, recurrent gliomas, wild type IDH-1 and non-codeletion of 1p19q than their counterparts (p<0.0001). With regard to age, older patients had higher PLAU(p=0.0043) and PLAUR(p<0.0001) mRNA level. There was no difference between men and women in PLAU and PLAUR expression (p=0.8609, p=0.1790, respectively).
Prognostic Value of PLAU/PLAUR Transcription Expression in Gliomas
Prognostic statistics were retrieved from GEPIA and CGGA (Figure 2). In the GEPEIA database, shown in Figures 2A, B, high expression of PLAU in LGG was associated with a shorter OS(p=2.2e-5) and DFS(p=0.0076), whereas no difference was found between high and low expression of PLAU in OS and DFS(p=0.19, p=0.15 respectively) (Figures 2C, D) in GBM. In the CGGA database, high expression of PLAU indicated poor prognosis in WHO grade II (p=0.03) and III patients(p=0.00094) (Figures S1A, B). Similarly, the overall survival was unchanged with the change of PLAU expression level(p=0.7) (Figure S1C). High expression of PLAU predicted poor prognosis in regard of all gliomas. (p<0.0001) (Figure 3A).
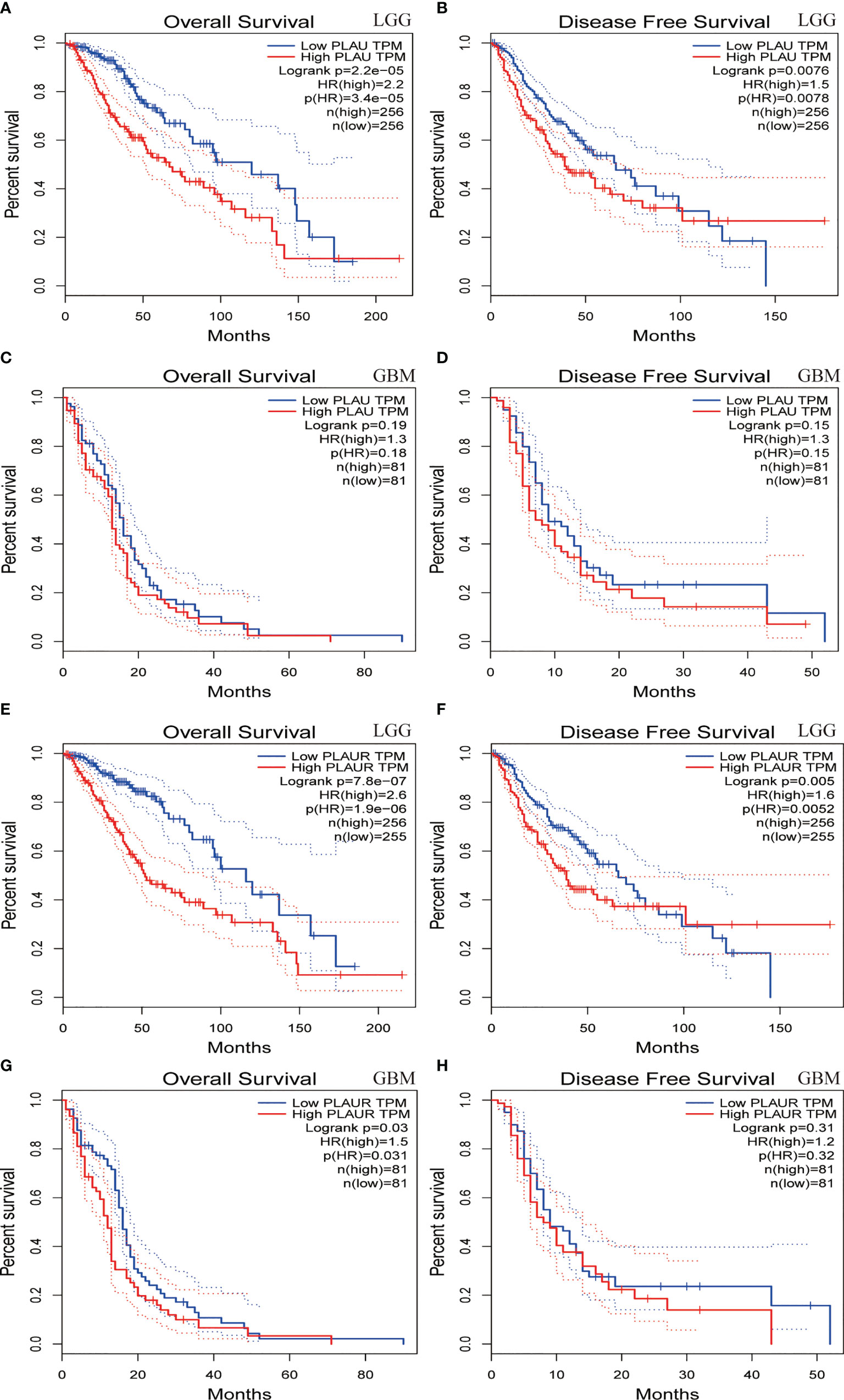
Figure 2 Overall survival and disease-free survival analyses based on PLAU and PLAUR expression in lower grade gliomas(LGG) (A, B, E, F) and GBM (C, D, G, H). (GEPIA from TCGA).
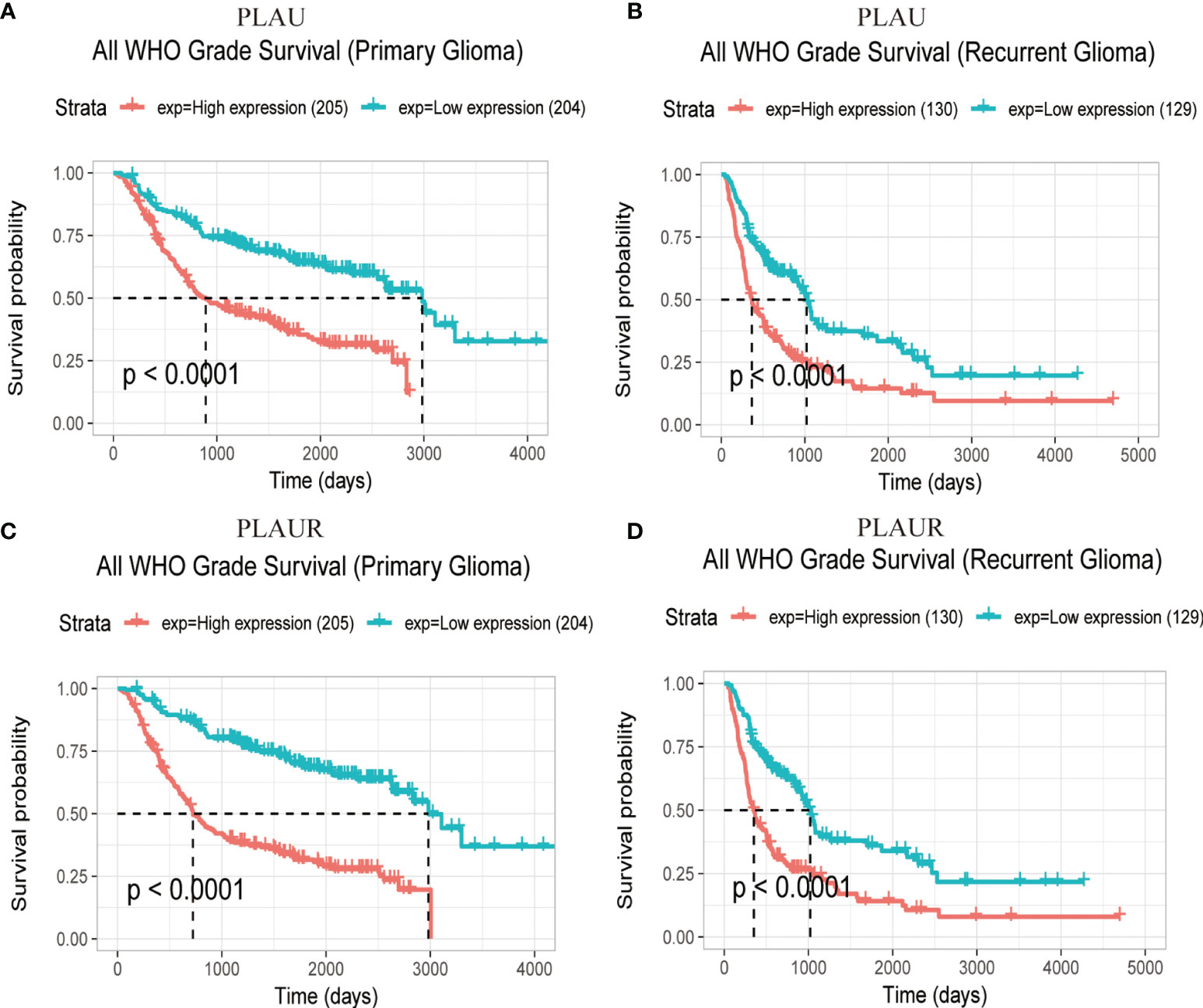
Figure 3 Prognostic significance of PLAU and PLAUR expression in primary gliomas (A, C) and recurrent gliomas (B, D). (CGGA).
Figures 2E, F demonstrated that among LGG patients, high expression of PLAUR resulted in shorter OS and DFS(p=7.8e-7, p=0.005 respectively), while in GBM patients, high expression of PLAUR predicted a shorter OS(p=0.03) (Figure 2G), but no significant correlation was found between high expression of PLAUR with shorter DFS(p=0.31) (Figure 2H). In the CGGA database, overall survival difference was detected between PLAUR high expression and PLAUR low expression group in WHO grade II (p=0.021) and III patients(p=0.0028) (Figure S1G, H), but not in GBM(p=0.2) (Figure S1I). Likewise, high expression of PLAUR indicated poor prognosis in all gliomas (p<0.0001) (Figure 3C).
With regard to recurrent glioma, complete data was achieved from CGGA. Among patients of WHO grade II and IV, expression of PLAU (p=0.84, p=0.2 respectively) (Figures S1D, F) and PLAUR (p=0.14, p=0.16 respectively) (Figures S1J, L) were of no significance in OS. However, high expression of PLAU and PLAUR indicated shorter OS in patients with current gliomas of WHO grade III (p=0.0041, p=0.00079 respectively) (Figures S1E, K). And in all recurrent patients, high expression of PLAU and PLAUR predicted poor prognosis (p<0.0001, p<0.0001 respectively) (Figures 3B, D).
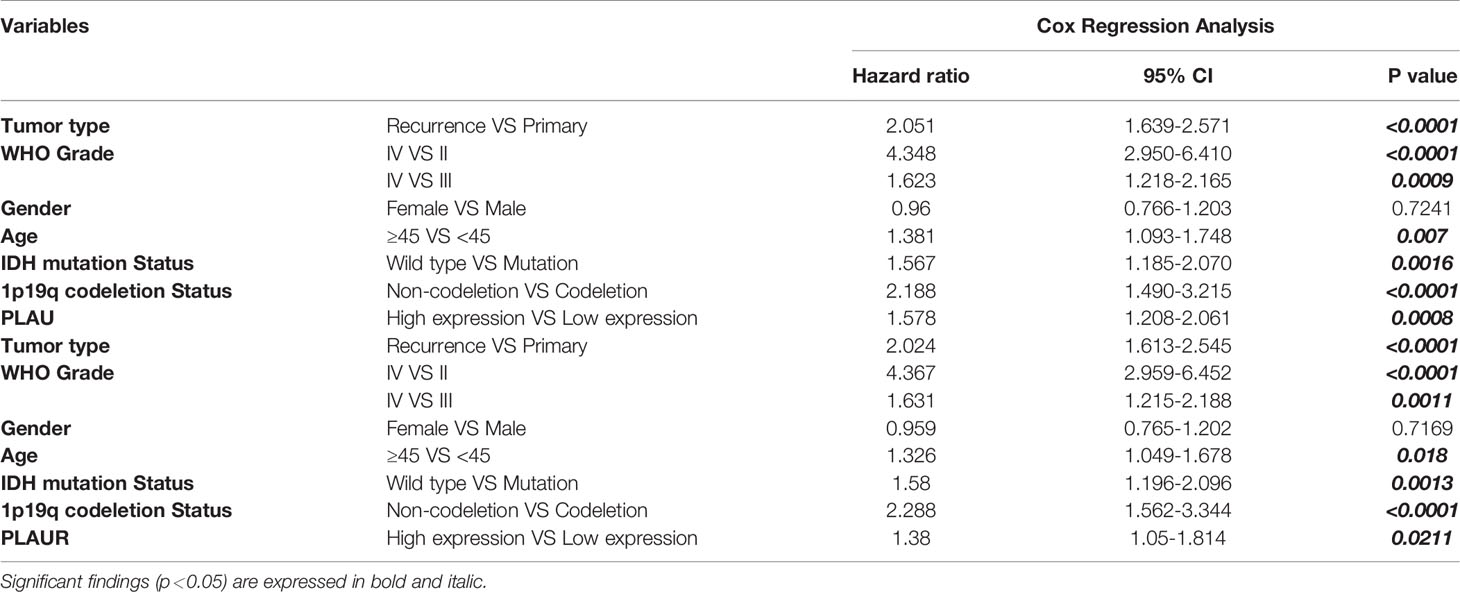
A cox regression analysis was conducted to further confirm the independent prognostic significance of PLAU/PLAUR mRNA expression. As shown in Table 3, high mRNA expression of PLAU (HR=1.578, 95%CI 1.208-2.061, p=0.0008) and PLAUR(HR=1.38, 95%CI 1.05-1.814, p=0.0211) were independently associated with significantly shorter OS of glioma patients. At the same time, clinical variables including tumor type, WHO grade, age, IDH mutation, and 1p19q status were regarded as independent prognostic factors.
Genetic Alterations in PLAU/PLAUR and Their Association With OS of Glioma Patients
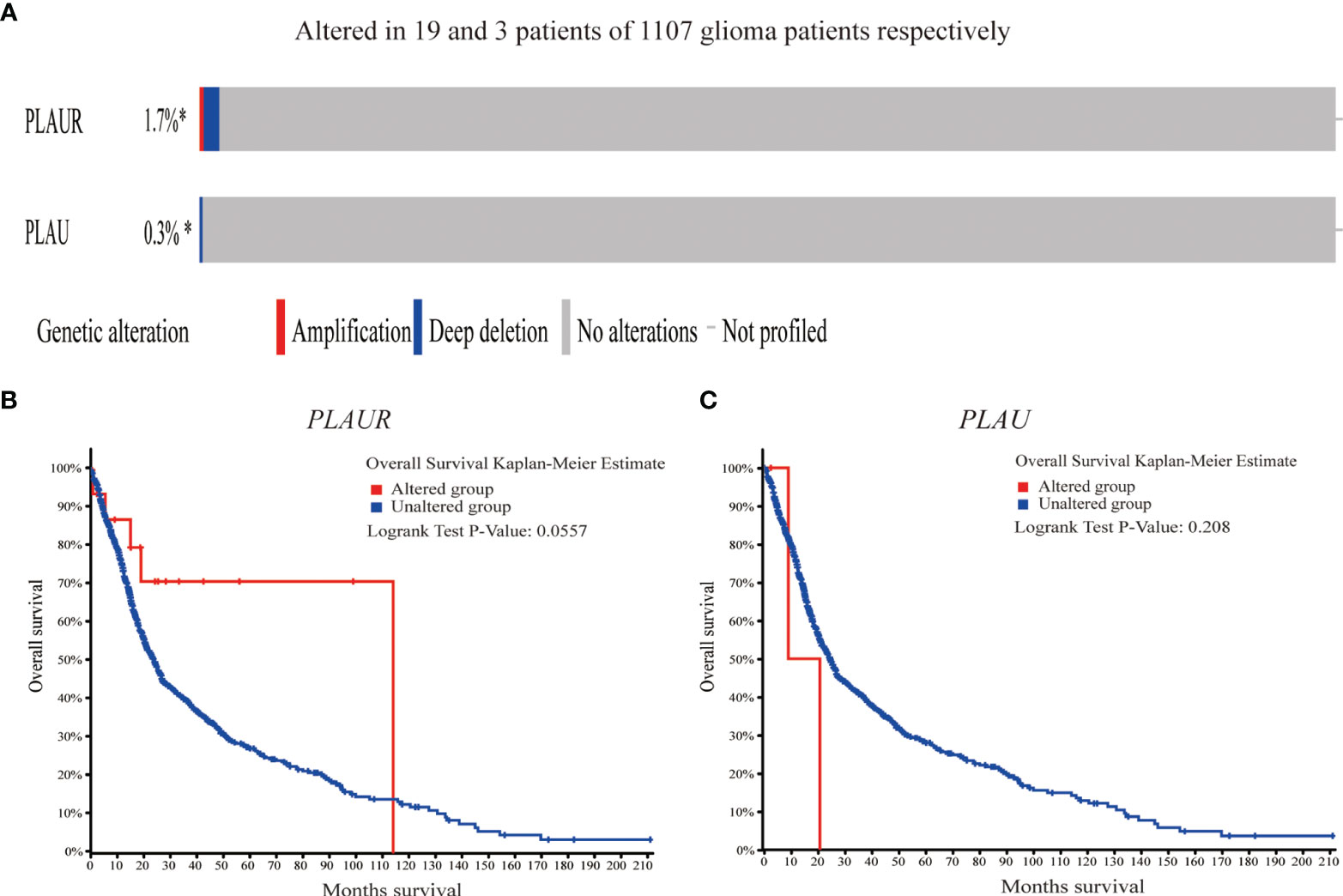
As shown in Figure 4, genetic alterations (amplification and deep deletion) were analyzed in 1107 sequenced glioma samples, with alteration rates of 0.3% and 1.7% in PLAU and PLAUR respectively. Furthermore, survival analysis indicated that there was no significant difference between the altered group and the unaltered group in OS, considering either PLAU (log-rank test p=0.208) or PLAUR(log-rank test p=0.0557).
Gene Set Enrichment Analysis of PLAU/PALUR Functional Networks in Gliomas
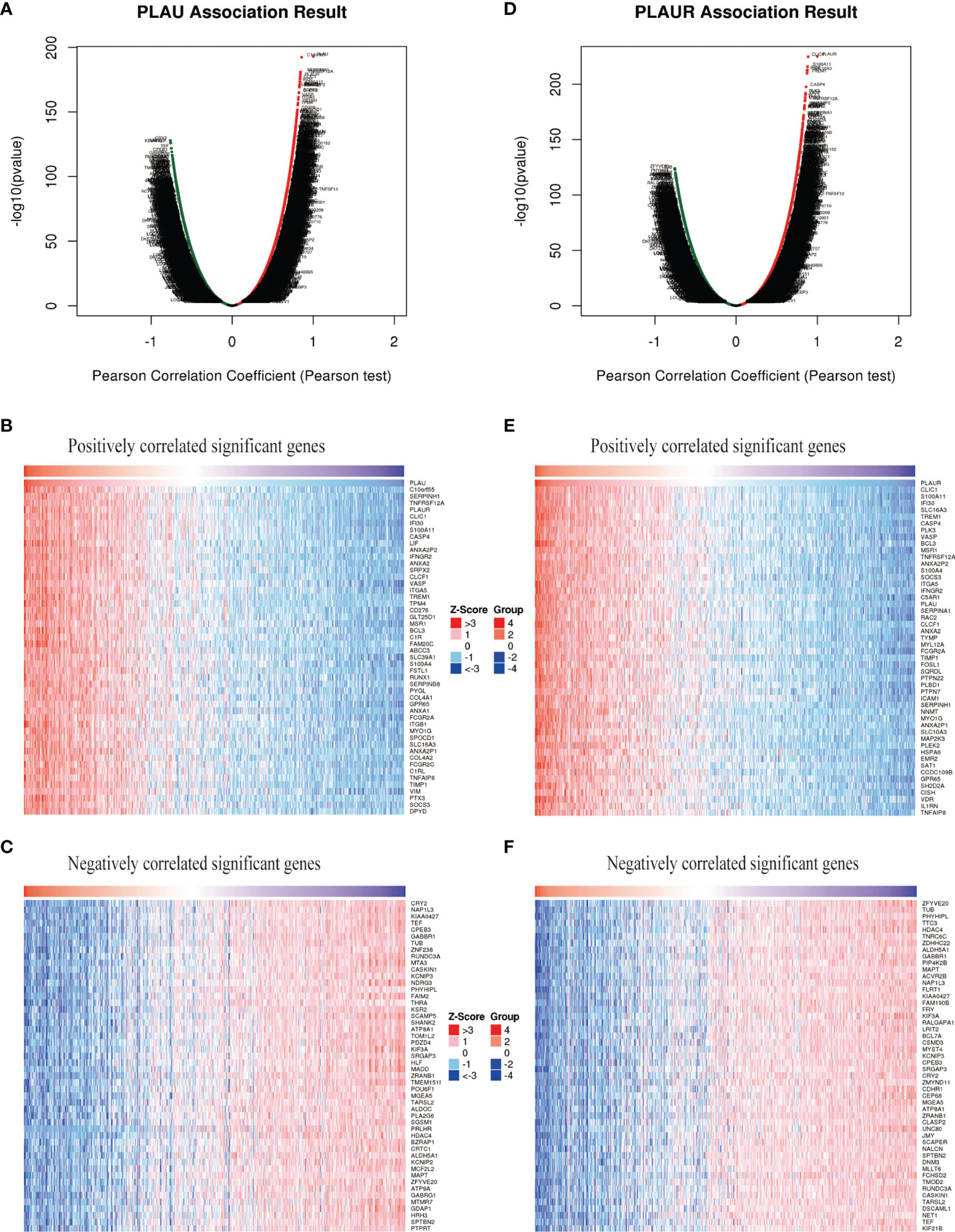
To further explore the biological meaning of PLAU and PLAUR in glioma patients, the function module of LinkedOmics was applied to analyze mRNA sequencing data from the 669 glioma patients. As shown in Figures 5A, D, genes that were positively or negatively correlated with PLAU and PLAUR(false discovery rate, |FDR|<0.01) were represented by dark red dots and dark green pots respectively. And the top 50 positively and negatively correlated significant genes were shown in the heat map (Figures 5B, C, E, F). The above-mentioned co-expressed genes were detailedly described in Supplementary Tables 3, 4. A positively significant correlation of PLAU and PLAUR was found in the gene list through Pearson correlation analysis (Pearson correlation coefficient=0.8386) (Figure 6A). Same analysis was carried out on the previous 693 patients of the CGGA database to verify the accuracy of the result. Pearson correlation analysis indicated a high positive correlation between PLAU and PLAUR transcription expression in primary gliomas (Pearson correlation coefficient=0.83) and recurrent gliomas (Pearson correlation coefficient=0.81) (Figures 6B, C).
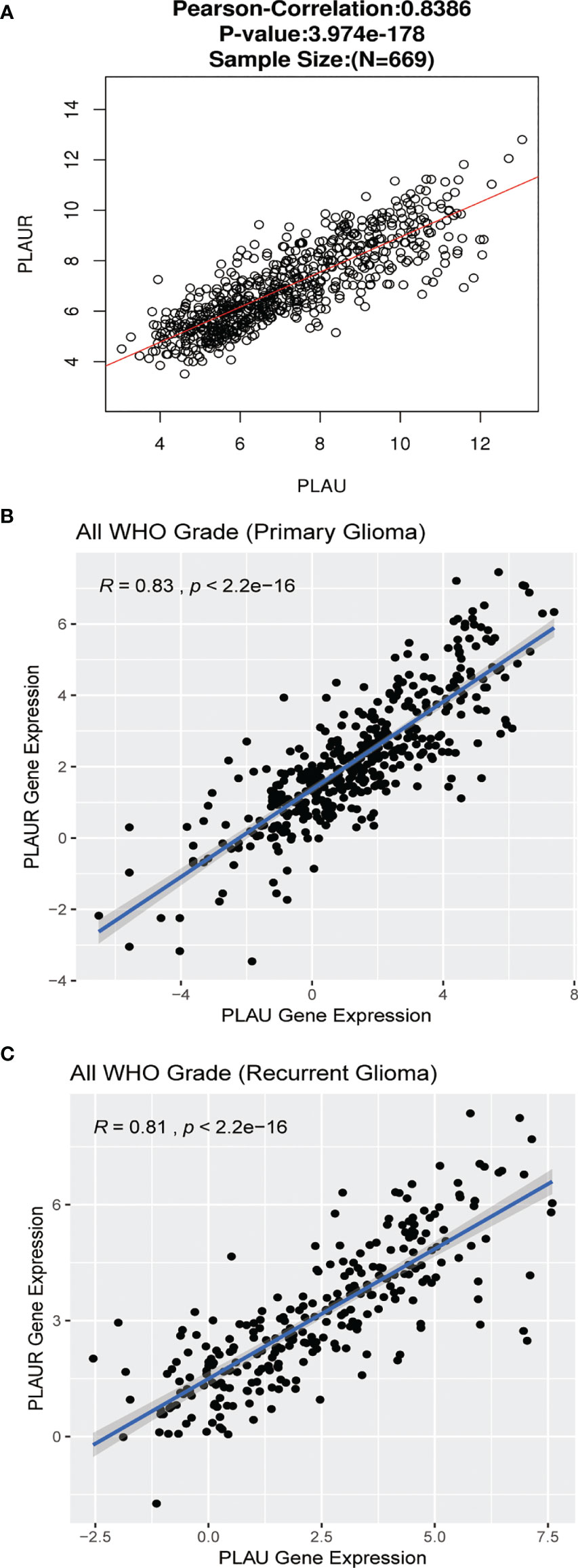
Figure 6 Genetic correlation of PLAU and PLAUR with Pearson correlation analysis. (A) Linkedomics. (B, C) CGGA database.
Gene set enrichment analysis(GSEA) included Gene Ontology(GO) analysis and Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis. GO analysis indicated that PLAU co-expressed genes were mainly involved in the biological processing of neutrophil and T cell, and participated in the construction of the nerve-related structure and cell-substrate junction(Figures 7A, B). And these genes were linked to the activities of transmembrane transporter and channels (Figure 7C). Likewise, PLAUR and its co-expressed genes not only had a close relationship with immune cells but also played a crucial role in the neuronal structure, cell-substrate junction, and transmembrane activity (Figures 7E–G).
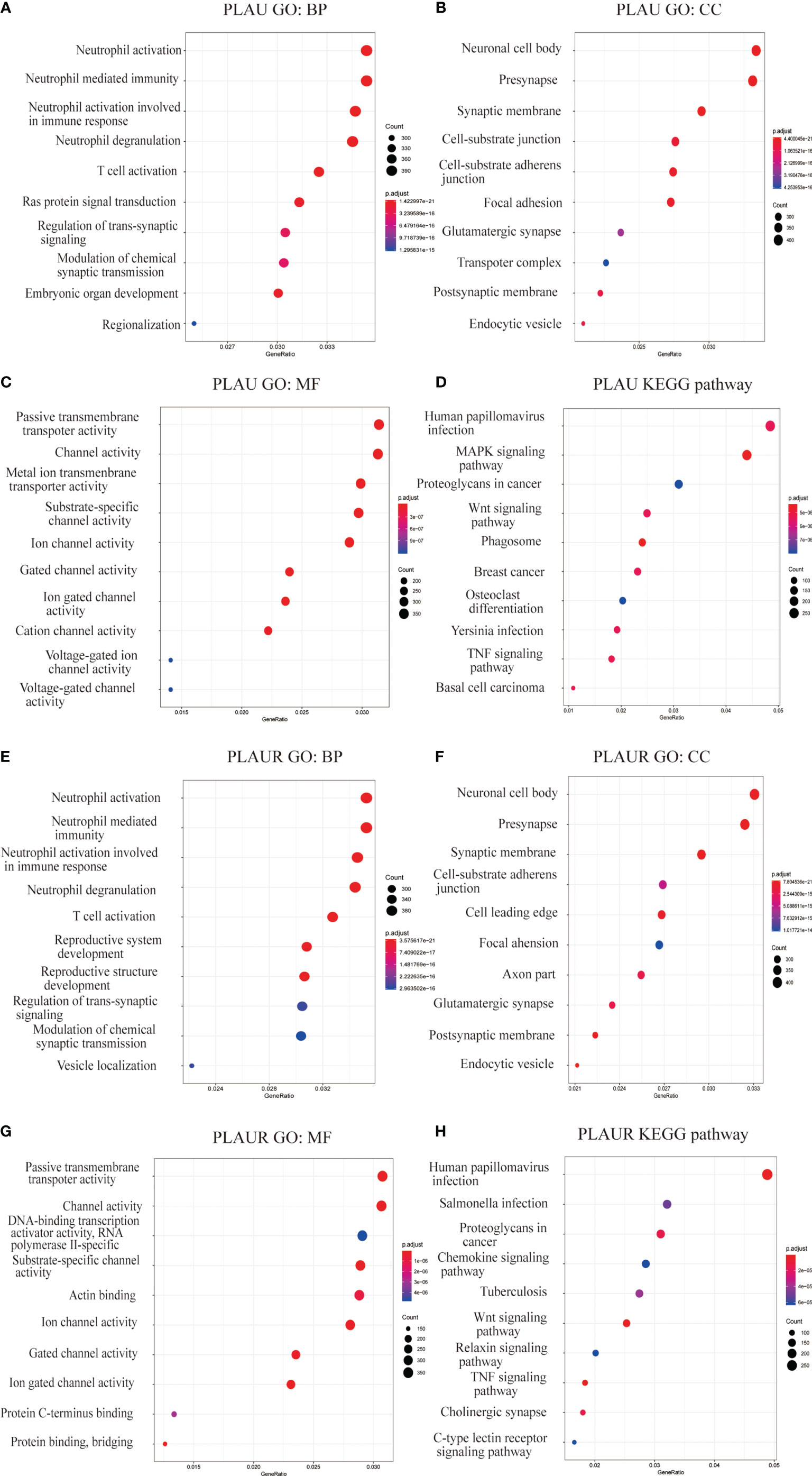
Figure 7 Gene set enrichment analyses of PLAU and PLAUR co-expression genes in gliomas. (A–C, E–G) Gene Ontology analyses including biological process (BP), cellular component (CC), molecular function (MF) of PLAU and PLAUR co-expression genes in gliomas. (D, H) KEGG pathway analyses of PLAU and PLAUR co-expression genes in gliomas.
KEGG pathway analysis showed PLAU and co-expressed genes enrichment in human papillomavirus infection, MAPK signaling pathway, Wnt signaling pathway, and proteoglycans in cancer, etc.(Figure 7D). Whereas, PLAUR and co-expressed genes enriched in human papillomavirus infection, salmonella infection, proteoglycans in cancer, and chemokine signaling pathway, etc. (Figure 7H).
Discussion
PLAU, by binding to PLAUR which is located on the extracellular surface, activates a cascade of extracellular proteases, which are involved in matrix remodeling and cell migration. As functional integrity, the PLAU-PLAUR system is supposed to not only plays a crucial role in mediating proteolysis during cancer invasion and metastasis but also participates into multiple stages of tumorigenesis (34). In recent years, high transcriptional levels of PLAU and PLAUR have been discovered in various tumors and have predicted a poor prognosis among the patients (12, 35–37). As a representative of malignancies, glioma, especially GBM, possesses common tumor characteristics including invasion, angiogenesis, epithelial-mesenchymal transition, cancer stem cell-like properties, and metastasis. Previous studies have demonstrated that these malignant features have a positive correlation with high expression of PLAU and PLAUR (16, 17, 38, 39). This study aims to evaluate the prognostic value of PLAU/PLAUR transcription expression in glioma and to explore how these pairs of genes affect the generation and progression of glioma.
Due to the small sample size and uncertainty of sample quality, we abandoned the traditional research methods and turned to data-rich online tools including Oncomine, GEPIA, and CGGA, etc. We can acquire a more authentic and comprehensive perspective using different online databases, which provide more information on tumors, both supplementing and verifying each other. The data of patients and gliomas are mainly from TCGA and CGGA; the former involves the Western region while CGGA only includes Chinese patients’ information. By the way, in consideration of the rarity of WHO I glioma, we usually regard LGG as WHO II and III glioma.
PLAU and PLAUR transcriptional expressions are found significantly higher in glioma compared with normal brain tissue, and are more obvious in high-grade gliomas. The result is coincident with previous studies (21, 22). The transcription expression even has a close relationship with IDH-1 mutation and 1p19q status. Higher expression occurred in wild-type IDH-1 and 1p-19q non-codeletion glioma. This demonstrates that high malignancy leads to high expression of PLAU/PLAUR since wild-type IDH-1 and 1p19q non-codeletion are features of malignancy (40). This can also explain why GBM has the highest expression. With regard to the survival analysis, there is high mRNA level of PLAU/PLAUR in primary and recurrent gliomas as a whole. After stratification of WHO grade, only LGG predicts poor prognosis. In GBM and part of recurrent gliomas, the prognostic value is questionable. There might be two possible explanations. On the one hand, high heterogeneity of malignant glioma leads to the diversity of OS; on the other hand, the sample size of GBMs and recurrent gliomas is relatively small. The results of Cox regression analysis further demonstrate the independent prognostic significance of PLAU/PLAUR in glioma. Interestingly, the two genes are similarly in the shape of the Kaplan-Mier plots, especially in the CGGA database. High Pearson correlation efficiency between PLAU and PLAUR indicates that the two genes share the same prognosis significance and could be integrated into one prognostic prediction model.
In view of the high incidence of genetic mutations in glioma, cBioPortal is used to evaluate the gene alteration frequency and whether the alteration affects overall survival (41). As shown in Figure 3, the gene alteration frequencies of 2 genes are negligible, without impact on overall survival. Another TCGA glioma cohort is retrieved in cBioPortal, and the results remain the same. It means PLUR/PLAUR and transcriptional expression are less affected by genetic mutations which usually cause phenotypic changes.
Extracellular matrix(ECM) breakdown is an important step for cell invasion and metastasis, and ECM proteinases such as PLAU/PLAUR system plays a key role in this process. Growing evidences indicate that down-regulation of PLAU and PLAUR attenuate the ability of tumor cells to invade and metastasize, but what triggers the regulation remains a critical question (6). DNA methylation and MicroRNAs are speculated to be the effective transcriptional regulatory ways (42).
PLAU/PLAUR co-expression network is constructed to further analyze their role in cell function and signaling pathway. PLAU and PLAUR share a lot of similarities in the gene set enrichment analysis. They both had a close relationship with immune cells like T cell and neutrophil during the biological process. The PLAU-deficient mice failed to develop T cells and macrophages and thus dying of bacterial infection was once reported (43). It is also indicated in the previous research that PLAU mediates regulatory T cells suppressor function through STAT5 and ERK signaling pathways, whereas regulatory T cells participates in complicated immunoreaction in glioma microenvironment (44, 45). That PLAUR plays a role in lymphocyte migration is also reported (46). At the same time, the close relationship between the PLAU/PLAUR expression and immune cells might be a key factor in the development of rheumatoid arthritis (7). Except for some inflammation-related signaling pathways, several tumor-related signaling pathways have a relationship with PLAU and PLAUR co-expression networks. KEGG pathway analysis suggests that PLAU and PLAUR are mainly involved in the proteoglycan-related cancer signaling pathway, Wnt signaling pathway, and TNF signaling pathway. Proteoglycan-related signaling pathway contributes to the biology of various types of cancer including proliferation, adhesion, angiogenesis, and metastasis, thus affecting tumor progress (47, 48). The Wnt signaling pathway and TNF signaling pathway are also involved in tumorigenesis of various tumors. Inhibition of expression of PLAU/PLAUR might blockade these crucial cancer-related pathways, and plays an anti-tumor role in gliomas. Gondi et al. revealed that bicistronic adenoviral construct targeting PLAU and PLAUR inhibited invasiveness and tumorigenicity in GBM cell lines (15). Other researches also indicated suppression of PLAU and PLAUR could attenuate the ability of glioma cells in vivo and vitro (16, 18, 20, 49).
There are some limitations in this study. First, though some recurrent gliomas are found with high expression of PLAU and PLAUR that predict poor prognosis, it is unknown whether the two genes are involved in the recurrence of glioma. Second, the data are insufficient to access the potential diagnostic and therapeutic roles of PLAU/PLAUR in glioma. Third, more detailed clinic data including tumor size, location, the extent of resection, and even pathological and imaging information are needed to stratify and analyze in a more accurate way. Fourth, these analyses are almost limited to the mRNA level, lacking evidence from protein level and functional studies make it less persuasive. Fifth, it is hard to know how PLAU and PLAUR expression can be involved in the glioma formation, or their expression is a consequence of the glioma development. These limitations should be addressed in follow-up studies.
In conclusion, our current study indicated that overexpression of PLAU and PLAUR is associated with poor prognosis in primary and recurrent glioma patients, especially in LGG. Overexpression of PLAU and PLAUR is regarded as independent prognostic factors for shorter OS of glioma patients through Cox regression analysis. Moreover, Gene co-expression network analysis enlightens us that immune therapy and specific cancer-related signaling pathway blocking by targeting PLAU/PLAUR might be a new idea for treating glioma.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Oncomine, GEPIA, LinkedOmics, cBioPortal, and CGGA databases.
Author Contributions
Study design: YL and JL. Database retrieve: JL and HF. Statistical Analysis: JL, YX, and HF. Result interpretation: YL, JL, and XZ. Manuscript preparation: JL and XZ. Text correction: YL and YX. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Key Research and Development Item from the Department of Science and Technology of Sichuan Province, China (No.2017SZ0006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.602321/full#supplementary-material
Supplementary Figure 1 | Prognostic significance of PLAU and PLAUR expression in gliomas with stratification of WHO grade(CGGA).
Supplementary Table 1 | Clinical characteristics with PLAU(CGGA).
Supplementary Table 2 | Clinical characteristics with PLAUR(CGGA).
Supplementary Table 3 | Co-expression genes of PLAU(LinkedOmics).
Supplementary Table 4 | Co-expression genes of PLAUR(LinkedOmics).
References
1. Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR. Genetic and Molecular Epidemiology of Adult Diffuse Glioma. Nat Rev Neurol (2019) 15(7):405–17. doi: 10.1038/s41582-019-0220-2
2. Gusyatiner O, Hegi ME. Glioma Epigenetics: From Subclassification to Novel Treatmsent Options. Semin Cancer Biol (2018) 51:50–8. doi: 10.1016/j.semcancer.2017.11.010
3. Wesseling P, Capper D. WHO 2016 Classification of Gliomas. Neuropathol Appl Neurobiol (2018) 44(2):139–50. doi: 10.1111/nan.12432
4. Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics (2017) 14(2):284–97. doi: 10.1007/s13311-017-0519-x
5. Andreasen PA, Egelund R, Petersen HH. The Plasminogen Activation System in Tumor Growth, Invasion, and Metastasis. Cell Mol Life Sci (2000) 57(1):25–40. doi: 10.1007/s000180050497
6. Irigoyen JP, Muñoz-Cánoves P, Montero L, Koziczak M, Nagamine Y. The Plasminogen Activator System: Biology and Regulation. Cell Mol Life Sci (1999) 56(1-2):104–32. doi: 10.1007/PL00000615
7. Dinesh P, Rasool M. uPA/uPAR Signaling in Rheumatoid Arthritis: Shedding Light on its Mechanism of Action. Pharmacol Res (2018) 134:31–9. doi: 10.1016/j.phrs.2018.05.016
8. Diamandis M, Paterson AD, Rommens JM, Veljkovic DK, Blavignac J, Bulman DE, et al. Quebec Platelet Disorder Is Linked to the Urokinase Plasminogen Activator Gene (PLAU) and Increases Expression of the Linked Allele in Megakaryocytes. Blood (2009) 113(7):1543–6. doi: 10.1182/blood-2008-08-175216
9. Hugdahl E, Bachmann IM, Schuster C, Ladstein RG, Akslen LA. Prognostic Value of uPAR Expression and Angiogenesis in Primary and Metastatic Melanoma. PloS One (2019) 14(1):e0210399–e. doi: 10.1371/journal.pone.0210399
10. Li D, Wei P, Peng Z, Huang C, Tang H, Jia Z, et al. The Critical Role of Dysregulated FOXM1-PLAUR Signaling in Human Colon Cancer Progression and Meastasis. Clin Cancer Res (2013) 19(1):62–72. doi: 10.1158/1078-0432.CCR-12-1588
11. Pappot H, Pfeiffer P, Grondahlhansen J, Skov B. Presence of Urokinase Plasminogen Activator, its Inhibitor and Receptor in Small Cell Lung Cancer and Non-Small Cell Lung Cancer. Int J Oncol (1997) 10(1):177–82. doi: 10.3892/ijo.10.1.177
12. Will C, Wilhelm O, Hohl S, Mobus V, Weidle U, Kreienberg R, et al. Expression of Urokinase-Type Plasminogen-Activator (Upa) and its Receptor (Upar) in Human Ovarian-Cancer Cells and in-Vitro Invasion Capacity. Int J Oncol (1994) 5(4):753–61. doi: 10.3892/ijo.5.4.753
13. Su S-C, Lin C-W, Yang W-E, Fan W-L, Yang S-F. The Urokinase-Type Plasminogen Activator (uPA) System as a Biomarker and Therapeutic Target in Human Malignancies. Expert Opin Ther Targets (2016) 20(5):551–66. doi: 10.1517/14728222.2016.1113260
14. Madunić J. The Urokinase Plasminogen Activator System in Human Cancers: An Overview of Its Prognostic and Predictive Role. Thromb Haemostasis (2018) 118(12):2020–36. doi: 10.1055/s-0038-1675399
15. Gondi CS, Lakka SS, Yanamandra N, Siddique K, Dinh DH, Olivero WC, et al. Expression of Antisense uPAR and Antisense uPA From a Bicistronic Adenoviral Construct Inhibits Glioma Cell Invasion, Tumor Growth, and Angiogenesis. Oncogene (2003) 22(38):5967–75. doi: 10.1038/sj.onc.1206535
16. Raghu H, Nalla AK, Gondi CS, Gujrati M, Dinh DH, Rao JS. uPA and uPAR shRNA Inhibit Angiogenesis via Enhanced Secretion of SVEGFR1 Independent of GM-CSF But Dependent on TIMP-1 in Endothelial and Glioblastoma Cells. Mol Oncol (2012) 6(1):33–47. doi: 10.1016/j.molonc.2011.11.008
17. Kunigal S, Gondi CS, Gujrati M, Lakka SS, Dinh DH, Olivero WC, et al. SPARC-Induced Migration of Glioblastoma Cell Lines via uPA-uPAR Signaling and Activation of Small GTPase RhoA. Int J Oncol (2006) 29(6):1349–57. doi: 10.3892/ijo.29.6.1349
18. Gondi CS, Kandhukuri N, Dinh DH, Gujrati M, Rao JS. Down-Regulation of uPAR and uPA Activates Caspase-Mediated Apoptosis and Inhibits the PI3K/AKT Pathway. Int J Oncol (2007) 31(1):19–27. doi: 10.3892/ijo.31.1.19
19. Le DM, Besson A, Fogg DK, Choi K-S, Waisman DM, Goodyer CG, et al. Exploitation of Astrocytes by Glioma Cells to Facilitate Invasiveness: A Mechanism Involving Matrix Metalloproteinase-2 and the Urokinase-Type Plasminogen Activator-Plasmin Cascade. J Neurosci (2003) 23(10):4034–43. doi: 10.1523/JNEUROSCI.23-10-04034.2003
20. Raghu H, Lakka SS, Gondi CS, Mohanam S, Dinh DH, Gujrati M, et al. Suppression of uPA and uPAR Attenuates Angiogenin Mediated Angiogenesis in Endothelial and Glioblastoma Cell Lines. PloS One (2010) 5(8):e12458–e. doi: 10.1371/journal.pone.0012458
21. Hsu DW, Efird JT, Hedley-Whyte ET. Prognostic Role of Urokinase-Type Plasminogen Activator in Human Gliomas. Am J Pathol (1995) 147(1):114–23.
22. Zhang X, Fei Z, Bu X, Zhen H, Zhang Z, Gu J, et al. Expression and Significance of Urokinase Type Plasminogen Activator Gene in Human Brain Gliomas. J Surg Oncol (2000) 74(2):90–4. doi: 10.1002/1096-9098(200006)74:2<90::AID-JSO2>3.0.CO;2-7
23. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia (New York NY) (2004) 6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2
24. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–W102. doi: 10.1093/nar/gkx247
25. Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource With Functional Genomic Data From Chinese Glioma Patients. Genomics Proteomics Bioinf (2021) 19(1):1–12. doi: 10.1016/j.gpb.2020.10.005
26. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6(269):pl1–pl. doi: 10.1126/scisignal.2004088
27. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46(D1):D956–D63. doi: 10.1093/nar/gkx1090
28. Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, et al. High-Resolution Genome-Wide Mapping of Genetic Alterations in Human Glial Brain Tumors. Cancer Res (2005) 65(10):4088–96. doi: 10.1158/0008-5472.CAN-04-4229
29. Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor Stem Cells Derived From Glioblastomas Cultured in bFGF and EGF More Closely Mirror the Phenotype and Genotype of Primary Tumors Than do Serum-Cultured Cell Lines. Cancer Cell (2006) 9(5):391–403. doi: 10.1016/j.ccr.2006.03.030
30. Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene Expression Profiling Reveals Molecularly and Clinically Distinct Subtypes of Glioblastoma Multiforme. Proc Natl Acad Sci USA (2005) 102(16):5814–9. doi: 10.1073/pnas.0402870102
31. Sun L, Hui A-M, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and Glioma-Derived Stem Cell Factor Induces Angiogenesis Within the Brain. Cancer Cell (2006) 9(4):287–300. doi: 10.1016/j.ccr.2006.03.003
32. Gutmann DH, Hedrick NM, Li J, Nagarajan R, Perry A, Watson MA. Comparative Gene Expression Profile Analysis of Neurofibromatosis 1-Associated and Sporadic Pilocytic Astrocytomas. Cancer Res (2002) 62(7):2085–91.
33. Rickman DS, Bobek MP, Misek DE, Kuick R, Blaivas M, Kurnit DM, et al. Distinctive Molecular Profiles of High-Grade and Low-Grade Gliomas Based on Oligonucleotide Microarray Analysis. Cancer Res (2001) 61(18):6885–91.
34. Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front Oncol (2018) 8:24. doi: 10.3389/fonc.2018.00024
35. Li Y, Cozzi PJ. Targeting uPA/uPAR in Prostate Cancer. Cancer Treat Rev (2007) 33(6):521–7. doi: 10.1016/j.ctrv.2007.06.003
36. Luo J, Sun X, Gao F, Zhao X, Zhong B, Wang H, et al. Effects of Ulinastatin and Docetaxel on Breast Cancer Invasion and Expression of uPA, uPAR and ERK. J Exp Clin Cancer Res CR (2011) 30(1):71. doi: 10.1186/1756-9966-30-71
37. Rubina KA, Sysoeva VY, Zagorujko EI, Tsokolaeva ZI, Kurdina MI, Parfyonova YV, et al. Increased Expression of uPA, uPAR, and PAI-1 in Psoriatic Skin and in Basal Cell Carcinomas. Arch Dermatol Res (2017) 309(6):433–42. doi: 10.1007/s00403-017-1738-z
38. Gupta R, Chetty C, Bhoopathi P, Lakka S, Mohanam S, Rao JS, et al. Downregulation of uPA/uPAR Inhibits Intermittent Hypoxia-Induced Epithelial-Mesenchymal Transition (EMT) in DAOY and D283 Medulloblastoma Cells. Int J Oncol (2011) 38(3):733–44.
39. Raghu H, Gondi CS, Dinh DH, Gujrati M, Rao JS. Specific Knockdown of uPA/uPAR Attenuates Invasion in Glioblastoma Cells and Xenografts by Inhibition of Cleavage and Trafficking of Notch -1 Receptor. Mol Cancer (2011) 10:130–. doi: 10.1186/1476-4598-10-130
40. Kawaguchi T, Sonoda Y, Shibahara I, Saito R, Kanamori M, Kumabe T, et al. Impact of Gross Total Resection in Patients With WHO Grade III Glioma Harboring the IDH 1/2 Mutation Without the 1p/19q Co-Deletion. J Neurooncol (2016) 129(3):505–14. doi: 10.1007/s11060-016-2201-2
41. Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med (2015) 372(26):2499–508. doi: 10.1056/NEJMoa1407279
42. Santibanez JF. Urokinase Type Plasminogen Activator and the Molecular Mechanisms of its Regulation in Cancer. Protein Pept Lett (2017) 24(10):936–46.
43. Gyetko MR, Chen GH, McDonald RA, Goodman R, Huffnagle GB, Wilkinson CC, et al. Urokinase is Required for the Pulmonary Inflammatory Response to Cryptococcus Neoformans. A Murine Transgenic Model. J Clin Invest (1996) 97(8):1818–26. doi: 10.1172/JCI118611
44. He F, Chen H, Probst-Kepper M, Geffers R, Eifes S, Del Sol A, et al. PLAU Inferred From a Correlation Network Is Critical for Suppressor Function of Regulatory T Cells. Mol Syst Biol (2012) 8:624. doi: 10.1038/msb.2012.56
45. Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. CCL2 Produced by the Glioma Microenvironment Is Essential for the Recruitment of Regulatory T Cells and Myeloid-Derived Suppressor Cells. Cancer Res (2016) 76(19):5671–82. doi: 10.1158/0008-5472.CAN-16-0144
46. Bianchi E, Ferrero E, Fazioli F, Mangili F, Wang J, Bender JR, et al. Integrin-Dependent Induction of Functional Urokinase Receptors in Primary T Lymphocytes. J Clin Invest (1996) 98(5):1133–41. doi: 10.1172/JCI118896
47. Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhão JG, et al. Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21. Sci Signal (2011) 4(199):ra75–ra. doi: 10.1126/scisignal.2001868
48. Iozzo RV, Sanderson RD. Proteoglycans in Cancer Biology, Tumour Microenvironment and Angiogenesis. J Cell Mol Med (2011) 15(5):1013–31. doi: 10.1111/j.1582-4934.2010.01236.x
49. Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Intraperitoneal Injection of a Hairpin RNA-Expressing Plasmid Targeting Urokinase-Type Plasminogen Activator (uPA) Receptor and uPA Retards Angiogenesis and Inhibits Intracranial Tumor Growth in Nude Mice. Clin Cancer Res an Off J Am Assoc Cancer Res (2007) 13(14):4051–60. doi: 10.1158/1078-0432.CCR-06-3032
Keywords: PLAU, PLAUR, glioma, prognosis, gene network
Citation: Li J, Fan H, Zhou X, Xiang Y and Liu Y (2022) Prognostic Significance and Gene Co-Expression Network of PLAU and PLAUR in Gliomas. Front. Oncol. 11:602321. doi: 10.3389/fonc.2021.602321
Received: 04 September 2020; Accepted: 16 December 2021;
Published: 11 January 2022.
Edited by:
Jürgen Schlegel, Technical University of Munich, GermanyReviewed by:
Carsten Friedrich, Klinikum Oldenburg AöR, GermanyPaola Merino, Emory University, United States
Juan Francisco Santibanez, University of Belgrade, Serbia
Copyright © 2022 Li, Fan, Zhou, Xiang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhui Liu, yhliu2001@163.com
 Junhong Li
Junhong Li Huanhuan Fan2
Huanhuan Fan2