Histone Demethylase KDM4C Stimulates the Proliferation of Prostate Cancer Cells via Activation of AKT and c-Myc
Abstract
:1. Introduction
2. Results
2.1. KDM4C Interferes with Prostate Acini Morphogenesis
2.2. Knockdown of KDM4C Suppresses the Proliferation of Prostate Cancer Cells
2.3. Knockdown of KDM4C Reduces Expression of Proteins Involved in AKT Signaling and c-Myc
2.4. Prostate Tumors Express Higher Protein Levels of KDM4C, AKT, and c-Myc
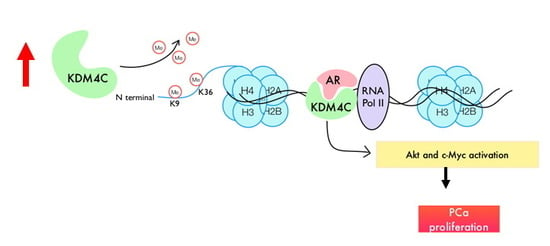
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. The 3D Cell Culture
4.3. Immuno-Fluorescent Staining and Confocal Image
4.4. Cell Proliferation Assay
4.5. Knockdown of KDM4C with siRNA
4.6. Western Blotting Analysis
4.7. Quantitative Real-Time PCR
4.8. AR Transcriptional Activity Assay
4.9. Micro-Western Arrays (MWAs)
4.10. Tissue Array
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rotili, D.; Mai, A. Targeting histone demethylases: A new avenue for the fight against cancer. Genes Cancer 2011, 2, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Nottke, A.; Colaiacovo, M.P.; Shi, Y. Developmental roles of the histone lysine demethylases. Development 2009, 136, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A. Histone demethylation mediated by the nuclear amine oxidase homolog lsd1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Mosammaparast, N.; Shi, Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010, 79, 155–179. [Google Scholar] [CrossRef] [PubMed]
- Coffey, K.; Rogerson, L.; Ryan-Munden, C.; Alkharaif, D.; Stockley, J.; Heer, R.; Sahadevan, K.; O’Neill, D.; Jones, D.; Darby, S.; et al. The lysine demethylase, kdm4b, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 2013, 41, 4433–4446. [Google Scholar] [CrossRef]
- Shin, S.; Janknecht, R. Activation of androgen receptor by histone demethylases jmjd2a and jmjd2d. Biochem. Biophys. Res. Commun. 2007, 359, 742–746. [Google Scholar] [CrossRef]
- Suikki, H.E.; Kujala, P.M.; Tammela, T.L.; van Weerden, W.M.; Vessella, R.L.; Visakorpi, T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate 2010, 70, 889–898. [Google Scholar] [CrossRef]
- Liu, G.; Bollig-Fischer, A.; Kreike, B.; van de Vijver, M.J.; Abrams, J.; Ethier, S.P.; Yang, Z.Q. Genomic amplification and oncogenic properties of the gasc1 histone demethylase gene in breast cancer. Oncogene 2009, 28, 4491–4500. [Google Scholar] [CrossRef]
- Luo, W.; Chang, R.; Zhong, J.; Pandey, A.; Semenza, G.L. Histone demethylase jmjd2c is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc. Natl. Acad. Sci. USA 2012, 109, E3367–E3376. [Google Scholar] [CrossRef]
- Yamamoto, S.; Tateishi, K.; Kudo, Y.; Yamamoto, K.; Isagawa, T.; Nagae, G.; Nakatsuka, T.; Asaoka, Y.; Ijichi, H.; Hirata, Y.; et al. Histone demethylase kdm4c regulates sphere formation by mediating the cross talk between wnt and notch pathways in colonic cancer cells. Carcinogenesis 2013, 34, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Ponnaluri, V.K.; Vavilala, D.T.; Putty, S.; Gutheil, W.G.; Mukherji, M. Identification of non-histone substrates for jmjd2a-c histone demethylases. Biochem. Biophys. Res. Commun. 2009, 390, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Katoh, M. Comparative integromics on jmjd2a, jmjd2b and jmjd2c: Preferential expression of jmjd2c in undifferentiated es cells. Int. J. Mol. Med. 2007, 20, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, M.; Zhang, Y.; Kou, Z.; Han, Z.; Chen, D.Y.; Sun, Q.Y.; Gao, S. The histone demethylase jmjd2c is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol. Reprod. 2010, 82, 105–111. [Google Scholar] [CrossRef]
- Wan, M.; Liang, J.; Xiong, Y.; Shi, F.; Zhang, Y.; Lu, W.; He, Q.; Yang, D.; Chen, R.; Liu, D.; et al. The trithorax group protein ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J. Biol. Chem. 2013, 288, 5039–5048. [Google Scholar] [CrossRef]
- Webber, M.M.; Bello, D.; Kleinman, H.K.; Hoffman, M.P. Acinar differentiation by non-malignant immortalized human prostatic epithelial cells and its loss by malignant cells. Carcinogenesis 1997, 18, 1225–1231. [Google Scholar] [CrossRef]
- Li, C.R.; Su, J.J.; Wang, W.Y.; Lee, M.T.; Wang, T.Y.; Jiang, K.Y.; Li, C.F.; Hsu, J.M.; Chen, C.K.; Chen, M.; et al. Molecular profiling of prostatic acinar morphogenesis identifies pdcd4 and klf6 as tissue architecture-specific prognostic markers in prostate cancer. Am. J. Pathol. 2013, 182, 363–374. [Google Scholar] [CrossRef]
- Ciaccio, M.F.; Wagner, J.P.; Chuu, C.P.; Lauffenburger, D.A.; Jones, R.B. Systems analysis of egf receptor signaling dynamics with microwestern arrays. Nat. Methods 2010, 7, 148–155. [Google Scholar] [CrossRef]
- True, L.; Coleman, I.; Hawley, S.; Huang, C.Y.; Gifford, D.; Coleman, R.; Beer, T.M.; Gelmann, E.; Datta, M.; Mostaghel, E.; et al. A molecular correlate to the gleason grading system for prostate adenocarcinoma. Proc. Natl. Acad. Sci. USA 2006, 103, 10991–10996. [Google Scholar] [CrossRef]
- Zhang, X.; Fournier, M.V.; Ware, J.L.; Bissell, M.J.; Yacoub, A.; Zehner, Z.E. Inhibition of vimentin or beta1 integrin reverts morphology of prostate tumor cells grown in laminin-rich extracellular matrix gels and reduces tumor growth in vivo. Mol. Cancer Ther. 2009, 8, 499–508. [Google Scholar] [CrossRef]
- Wissmann, M.; Yin, N.; Muller, J.M.; Greschik, H.; Fodor, B.D.; Jenuwein, T.; Vogler, C.; Schneider, R.; Gunther, T.; Buettner, R.; et al. Cooperative demethylation by jmjd2c and lsd1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007, 9, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Harma, V.; Knuuttila, M.; Virtanen, J.; Mirtti, T.; Kohonen, P.; Kovanen, P.; Happonen, A.; Kaewphan, S.; Ahonen, I.; Kallioniemi, O.; et al. Lysophosphatidic acid and sphingosine-1-phosphate promote morphogenesis and block invasion of prostate cancer cells in three-dimensional organotypic models. Oncogene 2012, 31, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.A.; Ortiz-Urda, S.; Kretz, M.; Garcia, J.; Scholl, F.A.; Pasmooij, A.M.; Cassarino, D.; Chang, H.Y.; Khavari, P.A. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell 2009, 15, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C.; Neel, B.G. New insights into tumor suppression: Pten suppresses tumor formation by restraining the phosphoinositide 3-kinase/akt pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 4240–4245. [Google Scholar] [CrossRef] [PubMed]
- Bedolla, R.; Prihoda, T.J.; Kreisberg, J.I.; Malik, S.N.; Krishnegowda, N.K.; Troyer, D.A.; Ghosh, P.M. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of pten expression and akt activation. Clin. Cancer Res. 2007, 13, 3860–3867. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yen, C.; Liaw, D.; Podsypanina, K.; Bose, S.; Wang, S.I.; Puc, J.; Miliaresis, C.; Rodgers, L.; McCombie, R.; et al. Pten, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Sarker, D.; Reid, A.H.; Yap, T.A.; de Bono, J.S. Targeting the pi3k/akt pathway for the treatment of prostate cancer. Clin. Cancer Res. 2009, 15, 4799–4805. [Google Scholar] [CrossRef]
- Ayala, G.; Thompson, T.; Yang, G.; Frolov, A.; Li, R.; Scardino, P.; Ohori, M.; Wheeler, T.; Harper, W. High levels of phosphorylated form of akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin. Cancer Res. 2004, 10, 6572–6578. [Google Scholar] [CrossRef]
- Kreisberg, J.I.; Malik, S.N.; Prihoda, T.J.; Bedolla, R.G.; Troyer, D.A.; Kreisberg, S.; Ghosh, P.M. Phosphorylation of akt (ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004, 64, 5232–5236. [Google Scholar] [CrossRef]
- McCall, P.; Gemmell, L.K.; Mukherjee, R.; Bartlett, J.M.; Edwards, J. Phosphorylation of the androgen receptor is associated with reduced survival in hormone-refractory prostate cancer patients. Br. J. Cancer 2008, 98, 1094–1101. [Google Scholar] [CrossRef]
- Sircar, K.; Yoshimoto, M.; Monzon, F.A.; Koumakpayi, I.H.; Katz, R.L.; Khanna, A.; Alvarez, K.; Chen, G.; Darnel, A.D.; Aprikian, A.G.; et al. Pten genomic deletion is associated with p-akt and ar signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 2009, 218, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Hammarsten, P.; Cipriano, M.; Josefsson, A.; Stattin, P.; Egevad, L.; Granfors, T.; Fowler, C.J. Phospho-akt immunoreactivity in prostate cancer: Relationship to disease severity and outcome, ki67 and phosphorylated egfr expression. PLoS ONE 2012, 7, e47994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebello, R.J.; Pearson, R.B.; Hannan, R.D.; Furic, L. Therapeutic approaches targeting myc-driven prostate cancer. Genes 2017, 8, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, W.H.; Hamel, A.; MacDonald, R.; Ramsey, E.; Pettigrew, N.M.; Johnston, B.; Dodd, J.G.; Matusik, R.J. Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer Res. 1986, 46, 1535–1538. [Google Scholar]
- Gurel, B.; Iwata, T.; Koh, C.M.; Jenkins, R.B.; Lan, F.; van Dang, C.; Hicks, J.L.; Morgan, J.; Cornish, T.C.; Sutcliffe, S.; et al. Nuclear myc protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 2008, 21, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. C-myc binds to human ribosomal DNA and stimulates transcription of rrna genes by rna polymerase i. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef]
- Grewal, S.S.; Li, L.; Orian, A.; Eisenman, R.N.; Edgar, B.A. Myc-dependent regulation of ribosomal rna synthesis during drosophila development. Nat. Cell Biol. 2005, 7, 295–302. [Google Scholar] [CrossRef]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlen, S.; Hydbring, P.; Soderberg, O.; Grummt, I.; et al. C-myc associates with ribosomal DNA and activates rna polymerase i transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef]
- Wang, H.J.; Pochampalli, M.; Wang, L.Y.; Zou, J.X.; Li, P.S.; Hsu, S.C.; Wang, B.J.; Huang, S.H.; Yang, P.; Yang, J.C.; et al. Kdm8/jmjd5 as a dual coactivator of ar and pkm2 integrates ar/ezh2 network and tumor metabolism in crpc. Oncogene 2019, 38, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Hamada, S.; Suzuki, T.; Mino, K.; Koseki, K.; Oehme, F.; Flamme, I.; Ozasa, H.; Itoh, Y.; Ogasawara, D.; Komaarashi, H.; et al. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J. Med. Chem. 2010, 53, 5629–5638. [Google Scholar] [CrossRef]
- Liang, Y.; Vogel, J.L.; Arbuckle, J.H.; Rai, G.; Jadhav, A.; Simeonov, A.; Maloney, D.J.; Kristie, T.M. Targeting the jmjd2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 2013, 5, 167ra5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigotti, N.A.; Pasternak, R.C. Cigarette smoking and coronary heart disease: Risks and management. Cardiol. Clin. 1996, 14, 51–68. [Google Scholar] [CrossRef]
- Chu, C.H.; Wang, L.Y.; Hsu, K.C.; Chen, C.C.; Cheng, H.H.; Wang, S.M.; Wu, C.M.; Chen, T.J.; Li, L.T.; Liu, R.; et al. Kdm4b as a target for prostate cancer: Structural analysis and selective inhibition by a novel inhibitor. J. Med. Chem. 2014, 57, 5975–5985. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.K.; Bonaldi, T.; Cuomo, A.; Del Rosario, J.R.; Hosfield, D.J.; Kanouni, T.; Kao, S.C.; Lai, C.; Lobo, N.A.; Matuszkiewicz, J.; et al. Design of kdm4 inhibitors with antiproliferative effects in cancer models. ACS Med. Chem. Lett. 2017, 8, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Leurs, U.; Clausen, R.P.; Kristensen, J.L.; Lohse, B. Inhibitor scaffold for the histone lysine demethylase kdm4c (jmjd2c). Bioorg. Med. Chem. Lett. 2012, 22, 5811–5813. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Hsu, C.C.; Wang, T.Y.; Li, C.R.; Hou, Y.C.; Chu, J.M.; Lee, C.T.; Liu, M.S.; Su, J.J.; Jian, K.Y.; et al. A gene expression signature of epithelial tubulogenesis and a role for aspm in pancreatic tumor progression. Gastroenterology 2013, 145, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Bello, D.; Webber, M.M.; Kleinman, H.K.; Wartinger, D.D.; Rhim, J.S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis 1997, 18, 1215–1223. [Google Scholar] [CrossRef]
- Weaver, V.M.; Petersen, O.W.; Wang, F.; Larabell, C.A.; Briand, P.; Damsky, C.; Bissell, M.J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997, 137, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Chuu, C.P.; Kokontis, J.M.; Hiipakka, R.A.; Fukuchi, J.; Lin, H.P.; Lin, C.Y.; Huo, C.; Su, L.C.; Liao, S. Androgen suppresses proliferation of castration-resistant lncap 104-r2 prostate cancer cells through androgen receptor, skp2, and c-myc. Cancer Sci. 2011, 102, 2022–2028. [Google Scholar] [CrossRef] [Green Version]
- Chuu, C.P.; Lin, H.P.; Ciaccio, M.F.; Kokontis, J.M.; Hause, R.J., Jr.; Hiipakka, R.A.; Liao, S.; Jones, R.B. Caffeic acid phenethyl ester suppresses the proliferation of human prostate cancer cells through inhibition of p70s6k and akt signaling networks. Cancer Prev. Res. (Phila) 2012, 5, 788–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Wang, B.-J.; Chen, B.-C.; Tseng, J.-C.; Jiang, S.S.; Tsai, K.K.; Shen, Y.-Y.; Yuh, C.H.; Sie, Z.-L.; Wang, W.-C.; et al. Histone Demethylase KDM4C Stimulates the Proliferation of Prostate Cancer Cells via Activation of AKT and c-Myc. Cancers 2019, 11, 1785. https://doi.org/10.3390/cancers11111785
Lin C-Y, Wang B-J, Chen B-C, Tseng J-C, Jiang SS, Tsai KK, Shen Y-Y, Yuh CH, Sie Z-L, Wang W-C, et al. Histone Demethylase KDM4C Stimulates the Proliferation of Prostate Cancer Cells via Activation of AKT and c-Myc. Cancers. 2019; 11(11):1785. https://doi.org/10.3390/cancers11111785
Chicago/Turabian StyleLin, Ching-Yu, Bi-Juan Wang, Bo-Chih Chen, Jen-Chih Tseng, Shih Sheng Jiang, Kelvin K. Tsai, Ying-Ying Shen, Chiou Hwa Yuh, Zong-Lin Sie, Wen-Ching Wang, and et al. 2019. "Histone Demethylase KDM4C Stimulates the Proliferation of Prostate Cancer Cells via Activation of AKT and c-Myc" Cancers 11, no. 11: 1785. https://doi.org/10.3390/cancers11111785