Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
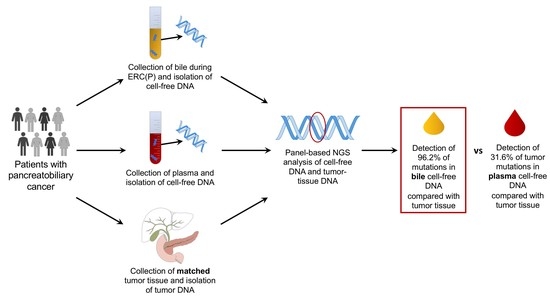
2.1. Detection of Pathogenic Tumor Mutations in Bile Samples
2.2. Detection of Pathogenic Tumor Mutations in the Plasma Samples
2.3. Bile cfDNA Sequencing Performs Better than Plasma cfDNA Sequencing
2.4. CA19-9 Displays Limitations as Serum Biomarker in Our Collective
2.5. Comparison of NGS and ddPCR
3. Discussion
4. Materials and Methods
4.1. Patients’ Collective
4.2. Blood and Bile Samples and cfDNA Isolation
4.3. DNA Isolation from Tumor Tissue
4.4. DNA Next Generation Sequencing (NGS)
4.5. Digital Droplet PCR (ddPCR)
4.6. Sensitivity and Specificity
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Emadossadaty, S.; Ladep, N.G.; Thomas, H.C.; Elliott, P.; Taylor-Robinson, S.D.; Toledano, M.B. Rising trends in cholangiocarcinoma: Is the ICD classification system misleading us? J. Hepatol. 2012, 56, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Groot, V.P.; van Santvoort, H.C.; Rombouts, S.J.E.; Hagendoorn, J.; Borel Rinkes, I.H.M.; van Vulpen, M.; Herman, J.M.; Wolfgang, C.L.; Besselink, M.G.; Molenaar, I.Q. Systematic review on the treatment of isolated local recurrence of pancreatic cancer after surgery; re-resection, chemoradiotherapy and SBRT. HPB 2017, 19, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, I.; Gonen, M.; Yopp, A.C.; Dalal, K.M.; Zhou, Q.; Klimstra, D.; D’Angelica, M.; DeMatteo, R.P.; Fong, Y.; Schwartz, L.; et al. Intrahepatic cholangiocarcinoma: Rising frequency, improved survival, and determinants of outcome after resection. Ann. Surg. 2008, 248, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-M.; Ni, J.-M.; Zhang, Z.-Y.; Zhang, L.; Li, B.; Jiang, C.-J. Presurgical Evaluation of Pancreatic Cancer: A Comprehensive Imaging Comparison of CT Versus MRI. AJR Am. J. Roentgenol. 2016, 206, 526–535. [Google Scholar] [CrossRef]
- Chu, L.C.; Goggins, M.G.; Fishman, E.K. Diagnosis and Detection of Pancreatic Cancer. Cancer J. 2017, 23, 333–342. [Google Scholar] [CrossRef]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goere, D.; Seufferlein, T.; Haustermans, K.; van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. S5), v56–v68. [Google Scholar] [CrossRef]
- Lewis, A.R.; Valle, J.W.; McNamara, M.G. Pancreatic cancer: Are “liquid biopsies” ready for prime-time? World J. Gastroenterol. 2016, 22, 7175–7185. [Google Scholar] [CrossRef]
- Levy, C.; Lymp, J.; Angulo, P.; Gores, G.J.; Larusso, N.; Lindor, K.D. The value of serum CA 19-9 in predicting cholangiocarcinomas in patients with primary sclerosing cholangitis. Dig. Dis. Sci. 2005, 50, 1734–1740. [Google Scholar] [CrossRef]
- Winter, J.M.; Cameron, J.L.; Campbell, K.A.; Arnold, M.A.; Chang, D.C.; Coleman, J.; Hodgin, M.B.; Sauter, P.K.; Hruban, R.H.; Riall, T.S.; et al. 1423 pancreaticoduodenectomies for pancreatic cancer: A single-institution experience. J. Gastrointest. Surg. 2006, 10, 1199–1210. [Google Scholar] [CrossRef]
- Okamura, R.; Hasegawa, S.; Hida, K.; Hoshino, N.; Kawada, K.; Sugihara, K.; Sakai, Y. The role of periodic serum CA19-9 test in surveillance after colorectal cancer surgery. Int. J. Clin. Oncol. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Fujii, L.L.; Levy, M.J. Basic techniques in endoscopic ultrasound-guided fine needle aspiration for solid lesions: Adverse events and avoiding them. Endosc. Ultrasound 2014, 3, 35–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimbas, M.; Deaconu, M.; Croitoru, A.; Haidar, A. Sudden appearance of free fluid during endoscopic ultrasound-guided fine-needle aspiration. Endosc. Ultrasound 2016, 5, 55–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Z.-H.; Xu, H.-X.; Zhang, S.-R.; Xu, J.-Z.; Li, S.; Gao, H.-L.; Jin, W.; Wang, W.-Q.; Wu, C.-T.; Ni, Q.-X.; et al. The Significance of Liquid Biopsy in Pancreatic Cancer. J. Cancer 2018, 9, 3417–3426. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Gormally, E.; Caboux, E.; Vineis, P.; Hainaut, P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. Mutat. Res. 2007, 635, 105–117. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Esposito, A.; Criscitiello, C.; Locatelli, M.; Milano, M.; Curigliano, G. Liquid biopsies for solid tumors: Understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol. Ther. 2016, 157, 120–124. [Google Scholar] [CrossRef]
- Ilié, M.; Hofman, P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016, 5, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef] [Green Version]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Tu, H.; Meng, Z.Q.; Chen, Z.; Wang, P.; Liu, L.M. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur. J. Surg. Oncol. 2010, 36, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Macarulla Mercade, T.; Javle, M.; Kelley, R.K.; Lubner, S.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.; Borad, M.J.; Bridgewater, J.A.; et al. LBA10_PRClarIDHy: A global, phase III, randomized, double-blind study of ivosidenib (IVO) vs placebo in patients with advanced cholangiocarcinoma (CC) with an isocitrate dehydrogenase 1 (IDH1) mutation. Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef]
- Vogel, A.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. LBA40FIGHT-202: A phase II study of pemigatinib in patients (pts) with previously treated locally advanced or metastatic cholangiocarcinoma (CCA). Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.W.; Lee, K.; Lee, S.H.; Kim, H.; You, M.S.; Hwang, Y.J.; Paik, W.H.; Ryu, J.K.; Kim, Y.-T. Comparison of liquid-based cytology with conventional smear cytology for EUS-guided FNA of solid pancreatic masses: A prospective randomized noninferiority study. Gastrointest. Endosc. 2020, 91, 837–846.e1. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-Y.; Liu, B.; Wang, Y.-Z.; Wang, W.-J.; Wang, W.; Li, D.; Li, Y.-L. Percutaneous transhepatic cholangiography versus endoscopic retrograde cholangiography for the pathological diagnosis of suspected malignant bile duct strictures. Medicine 2020, 99, e19545. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Ako, S.; Dohi, C.; Matsushita, H.; Matsumoto, K.; Kato, H.; Okada, H. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol. Ther. 2018, 19, 934–938. [Google Scholar] [CrossRef]
- Shen, N.; Zhang, D.; Yin, L.; Qiu, Y.; Liu, J.; Yu, W.; Fu, X.; Zhu, B.; Xu, X.; Duan, A.; et al. Bile cellfree DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 2019, 42, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Nikiforova, M.N.; Chennat, J.; Papachristou, G.I.; Khalid, A.; Rabinovitz, M.; Das, R.; Sarkaria, S.; Ayasso, M.S.; Wald, A.I.; et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020, 69, 52–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef] [Green Version]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodriguez-Garrote, M.; Lisa, E.; Mendia, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef] [Green Version]
- Prat, F.; Chapat, O.; Ducot, B.; Ponchon, T.; Fritsch, J.; Choury, A.D.; Pelletier, G.; Buffet, C. Predictive factors for survival of patients with inoperable malignant distal biliary strictures: A practical management guideline. Gut 1998, 42, 76–80. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, Y.V.M.; Arvanitakis, M. Early Diagnosis and Management of Malignant Distal Biliary Obstruction: A Review On Current Recommendations And Guidelines. Clin. Exp. Gastroenterol. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Hruban, R.H.; Pitman, M.B.; Klimstra, D.S. Tumors of the Pancreas; American Registry of Pathology in Collaboration with the Armed Forces Institute of Pathology: Washington, DC, USA, 2007; ISBN 1933477024. [Google Scholar]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef]
- Lui, Y.Y.N.; Chik, K.-W.; Chiu, R.W.K.; Ho, C.-Y.; Lam, C.W.K.; Lo, Y.M.D. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, X.; Wang, J.; Zhou, S.; Wang, C.L.; Ye, M.Z.; Wang, X.Y.; Song, Y.; Wang, Y.Q.; Zhang, L.T.; et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann. Oncol. 2019, 30, 464–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, D.G.; Bland, J.M. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Diagnosis | N | Age (Mean) | Gender ♂:♀ | Bile Samples | Plasma Samples | Tissues | CA19-9 (U/mL) |
|---|---|---|---|---|---|---|---|
| Localized disease | 16 | 66.5 | 8:8 | 9 | 14 | 13 | 856.1 |
| CCA | 6 | 62.5 | 4:2 | 2 | 5 | 5 | 112.0 |
| PDAC | 10 | 70.8 | 4:6 | 6 | 8 | 8 | 1302.6 |
| Metastatic disease | 29 | 64.6 | 18:11 | 13 | 24 | 16 | 3725.1 |
| CCA | 4 | 58.0 | 2:2 | 2 | 3 | 4 | 4235.5 |
| PDAC | 25 | 65.6 | 16:9 | 11 | 21 | 12 | 3655.6 |
| Control | 35 | 63.7 | 20:15 | 23 | 14 | - | 93.5 |
| CBD obstruction | 10 | 69.5 | 7:3 | 10 | 0 | - | 126.7 |
| Choledocholithiasis | 4 | 80.0 | 0:4 | 4 | 1 | - | - |
| Chronic pancreatitis | 7 | 50.1 | 5:2 | 3 | 4 | - | 53.1 |
| IPMN | 6 | 72.5 | 1:5 | 1 | 5 | - | 17.7 |
| PSC | 5 | 49.0 | 3:2 | 5 | 1 | - | 130.0 |
| Pseudocyst | 5 | 63.0 | 4:1 | 1 | 4 | - | 6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Driescher, C.; Fuchs, K.; Haeberle, L.; Goering, W.; Frohn, L.; Opitz, F.V.; Haeussinger, D.; Knoefel, W.T.; Keitel, V.; Esposito, I. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers 2021, 13, 39. https://doi.org/10.3390/cancers13010039
Driescher C, Fuchs K, Haeberle L, Goering W, Frohn L, Opitz FV, Haeussinger D, Knoefel WT, Keitel V, Esposito I. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers. 2021; 13(1):39. https://doi.org/10.3390/cancers13010039
Chicago/Turabian StyleDriescher, Caroline, Katharina Fuchs, Lena Haeberle, Wolfgang Goering, Lisa Frohn, Friederike V. Opitz, Dieter Haeussinger, Wolfram Trudo Knoefel, Verena Keitel, and Irene Esposito. 2021. "Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer" Cancers 13, no. 1: 39. https://doi.org/10.3390/cancers13010039