Induced Pluripotent Stem Cells and Their Use in Cardiac and Neural Regenerative Medicine
Abstract
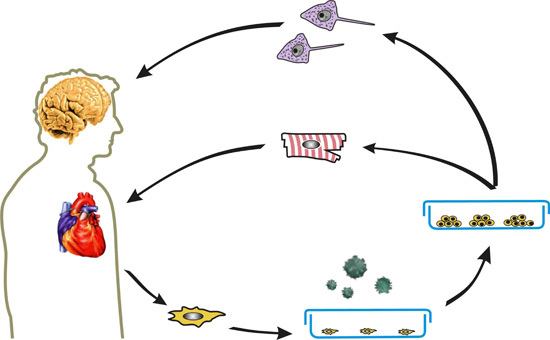
:1. Introduction
| Year | Event |
|---|---|
| 1908 | The term stem cell was associated with haemopoiesis [2] |
| 1961 | Existence of stem cells in mouse bone marrow was demonstrated [3] |
| 1981 | Embryonic stem cell isolation from inner cell mass of mouse blastocyst [4] |
| 1995 | Embryonic stem cells isolation from rhesus monkey [8] |
| 1998 | Isolation of first human ES cells [5] |
| 2006 | Induced pluripotent stem cells from adult mouse fibroblast cells [6] |
| 2007 | Induced pluripotent stem cells from human fibroblasts [7] |
2. Cellular Reprogramming and Induced Pluripotent Stem Cells
| Species | Germ Layer | Cell Type | Reprogramming Factors | Reference |
|---|---|---|---|---|
| mouse | mesoderm | mouse embryonic fibroblasts | O, K, S, M | [6,31] |
| O, K, S | [32] | |||
| adipose-derived stem cells | O, K, S, M | [33] | ||
| B lymphocytes | O, K, S, M | [34] | ||
| endoderm | hepatocytes | O, K, S, M | [35] | |
| O, K, S | [35] | |||
| pancreatic β cells | O, K, S, M | [36] | ||
| gastric epithelial cells | O, K, S, M | [35] | ||
| ectoderm | neural stem cells | O, K, S, M | [37] | |
| O, K, M | [38] | |||
| O, K | [37] | |||
| O, M | [37] | |||
| O | [39] | |||
| human | endoderm | hepatocytes | O, K, S, M | [40] |
| mesoderm | fibroblast | O, K, S, M | [6] | |
| O, L, S, N | [24] | |||
| O, K, S | [41] | |||
| mobilized peripheral blood | O, K, S, M | [42] | ||
| peripheral blood and bone marrow mononuclear cells | O, K, S, M | [43] | ||
| bone marrow stem cells | O, K, S, M | [42] | ||
| circulating T lymphocytes | O, K, S, M | [43] | ||
| umbilical endothelial cells | O, L, S, N | [44] | ||
| cord blood stem cells | O, K, S, M | [42] | ||
| O, S | [45] | |||
| adipose-derived stem cells | O, K, S, M | [33] | ||
| adipose stem cells | O, K, S | [46] | ||
| mesenchymal stromal cells | O, K, S | [47] | ||
| mesenchymal cells | O, K, S, M | [48] | ||
| ectoderm | keratinocytes | O, K, S, M | [49] | |
| O, K, S | [49] | |||
| neural stem cells | O | [39] | ||
| melanocytes | O, K, M | [50] |
3. iPSC Differentiation into Three Germ Layers
4. iPSC Differentiation into Cardiomyocytes
| Modifier | Name | Mechanism |
|---|---|---|
| Chemicals | Ascorbic acid | Enhances proliferation of CPCs via the MEK-ERK1/2 [72] |
| Cardiogenol C | Activation of the Wnt signaling pathway and modified expression of several key chromatin remodeling proteins [73] | |
| Retinoic acid | Effects to growth factor stimulation pathway(s) [74] | |
| Szh-1 | Unknown [69] | |
| Small molecules | Pluripotin (SC1) | ERK1/Ras-GAP inhibition [75] |
| RepSox | TGF-β receptor signaling inhibition [30] | |
| BIX01294 | Histone methyltransferase inhibitor [76] | |
| Bay K 8644 | Ca2+ channel agonist [77] | |
| RG108 | DNA methyltransferase inhibitor [78] | |
| 5-azacytidine | Inhibitors of DNA methyltransferases [79] | |
| Valproic acid | Histone deacetylase inhibitor | |
| SB431542 | TGF-β superfamily type I activin receptor inhibition | |
| KY02112 | Wnt inhibitor [80] | |
| DMSO | Decreases phosphorylation and increases levels of β-catenin [81] |
5. iPSCs in Cardiac Disease Modeling and Regenerative Medicine

6. iPSC Differentiation into Neurons
7. Direct Differentiation into Neurons with Small Molecules and Their Properties
| Name | Mechanism |
|---|---|
| Retinoic acid | Morphogen/agonist of the Sonic Hedgehog pathway [102,112] |
| Epidermal growth factor (EGF) | Mitogen [113] |
| Fibroblast growth factor (FGF-2, FGF-8, FGF-4) | Regulation of neural stem cells proliferation and self-renewal [113] |
| Platelet-derived growth factor (PDGF) | Neural induction factor [113] |
| Sonic hedgehog (SHH) | Morphogen, induction factor [112] |
| Noggin | BMP antagonist [113] |
| SB431542 | Inhibition of the TGFβ/Activin/Nodal pathway/inhibition of SMAD [114,115] |
| Dorsomorphin | Inhibition of BMP pathway/inhibition of SMAD [114] |
| LDN193189 | Inhibition of BMP pathway [116] |
| Purmorphamine | Activation of the Hedgehog pathway [117] |
8. iPSCs in Neuronal Disease Modeling
9. iPSCs in Neuronal Diseases
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stadtfeld, M.; Hochedlinger, K. Induced pluripotency: History, mechanisms, and applications. Genes Dev. 2010, 24, 2239–2263. [Google Scholar] [CrossRef] [PubMed]
- Maximow, A. Der lymphozyt als gemeinsame stammzelle der verschiedenen blutelemente in der embryonalen entwicklung und im postfetalen leben der saugetiere. Fol. Haematol. 1909, 8, 125–134. [Google Scholar]
- Till, J.E.; McCulloch, E. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 1961, 14, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.; Kaufman, M. Establishment in culture of pluripotent cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, J.A.; Kalishman, J.; Golos, T.G.; Durning, M.; Harris, C.P.; Becker, R.A.; Hearn, J.P. Isolation of a primate embryonic stem cell line. Proc. Natl. Acad. Sci. USA 1995, 92, 7844–7848. [Google Scholar] [CrossRef] [PubMed]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef]
- Briggs, R.; King, T.J. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc. Natl. Acad. Sci. USA 1952, 38, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Gurdon, J.B.; Laskey, R.A.; Reeves, O.R. The developmental capacity of nuclei transplanted from keratinized skin dcells of adult frogs. J. Embryol. Exp. Morphol. 1975, 34, 93–112. [Google Scholar] [PubMed]
- Stevens, L.C.; Little, C.C. Spontaneous testicular teratomas in an inbred strain of mice. Proc. Natl. Acad. Sci. USA 1954, 40, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Guenther, M.G.; Frampton, G.M.; Soldner, F.; Hockemeyer, D.; Mitalipova, M.; Jaenisch, R.; Young, R.A. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 2010, 7, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar] [CrossRef]
- Loh, Y.H.; Wu, Q.; Chew, J.L.; Vega, V.B.; Zhang, W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Tomlinson, S.R. The transcriptional foundation of pluripotency. Development 2009, 136, 2311–2322. [Google Scholar] [CrossRef] [PubMed]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on Sox2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ai, W. Function of Klf4 in stem cell biology. In Pluripotent Stem Cells; Bhartiya, D., Lenka, N., Eds.; InTech: Winchester, UK, 2013; Chapter 15. [Google Scholar]
- Boxer, L.M.; Dang, C.V. Translocations involving c-Myc and c-Myc function. Oncogene 2001, 20, 5595–5610. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Clavel, C.; Kim, S.; Ang, Y.S.; Grisanti, L.; Lee, D.F.; Kelley, K.; Rendl, M. Oct4 and Klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells 2010, 28, 221–228. [Google Scholar] [PubMed]
- Feng, C.; Jia, Y.D.; Zhao, X.Y. Pluripotency of induced pluripotent stem cells. Genomics Proteomics Bioinform. 2013, 11, 299–303. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Do, J.T.; Desponts, C.; Hahm, H.S.; Scholer, H.R.; Ding, S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2008, 2, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H. Wnt: What’s needed to maintain pluripotency? Nat. Cell Biol. 2011, 13, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhou, H.; Abujarour, R.; Zhu, S.; Young Joo, J.; Lin, T.; Hao, E.; Scholer, H.R.; Hayek, A.; Ding, S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 2009, 27, 2992–3000. [Google Scholar] [PubMed]
- Miyazono, K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Blanchard, J.; Lam, K.; Son, E.Y.; Chung, J.E.; Egli, D.; Loh, K.M.; Carter, A.C.; di Giorgio, F.P.; Koszka, K.; et al. A small-molecule inhibitor of TGF-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Cassady, J.P.; Jaenisch, R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008, 2, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Sugii, S.; Kida, Y.; Kawamura, T.; Suzuki, J.; Vassena, R.; Yin, Y.Q.; Lutz, M.K.; Berggren, W.T.; Izpisua Belmonte, J.C.; Evans, R.M. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Markoulaki, S.; Schorderet, P.; Carey, B.W.; Beard, C.; Wernig, M.; Creyghton, M.P.; Steine, E.J.; Cassady, J.P.; Foreman, R.; et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 2008, 133, 250–264. [Google Scholar] [CrossRef]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Brennand, K.; Hochedlinger, K. Reprogramming of pancreatic β cells into induced pluripotent stem cells. Curr. Biol. 2008, 18, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Zaehres, H.; Wu, G.; Gentile, L.; Ko, K.; Sebastiano, V.; Arauzo-Bravo, M.J.; Ruau, D.; Han, D.W.; Zenke, M.; et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 2008, 454, 646–650. [Google Scholar] [CrossRef]
- Eminli, S.; Utikal, J.; Arnold, K.; Jaenisch, R.; Hochedlinger, K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 2008, 26, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Greber, B.; Arauzo-Bravo, M.J.; Meyer, J.; Park, K.I.; Zaehres, H.; Scholer, H.R. Direct reprogramming of human neural stem cells by Oct4. Nature 2009, 461, 649–643. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ye, Z.; Kim, Y.; Sharkis, S.; Jang, Y.Y. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology 2010, 51, 1810–1819. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhan, H.; Mali, P.; Dowey, S.; Williams, D.M.; Jang, Y.Y.; Dang, C.V.; Spivak, J.L.; Moliterno, A.R.; Cheng, L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 2009, 114, 5473–5480. [Google Scholar] [CrossRef] [PubMed]
- Kunisato, A.; Wakatsuki, M.; Shinba, H.; Ota, T.; Ishida, I.; Nagao, K. Direct generation of induced pluripotent stem cells from human nonmobilized blood. Stem Cells Dev. 2011, 20, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.; Olmer, R.; Schwanke, K.; Wunderlich, S.; Merkert, S.; Hess, C.; Zweigerdt, R.; Gruh, I.; Meyer, J.; Wagner, S.; et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 2009, 5, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Giorgetti, A.; Montserrat, N.; Rodriguez-Piza, I.; Azqueta, C.; Veiga, A.; Izpisua Belmonte, J.C. Generation of induced pluripotent stem cells from human cord blood cells with only two factors: Oct4 and Sox2. Nat. Protoc. 2010, 5, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Ohnishi, H.; Oda, Y.; Tadokoro, M.; Sasao, M.; Kato, H.; Hattori, K.; Ohgushi, H. Generation of induced pluripotent stem cells from human adipose-derived stem cells without c-Myc. Tissue Eng. Part A 2010, 16, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Yoshimura, Y.; Ohnishi, H.; Tadokoro, M.; Katsube, Y.; Sasao, M.; Kubo, Y.; Hattori, K.; Saito, S.; Horimoto, K.; et al. Induction of pluripotent stem cells from human third molar mesenchymal stromal cells. J. Biol. Chem. 2010, 285, 29270–29278. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-specific induced pluripotent stem cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilic, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Utikal, J.; Maherali, N.; Kulalert, W.; Hochedlinger, K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009, 122, 3502–3510. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Badylak, S.F.; Nerem, R.M. Progress in tissue engineering and regenerative medicine. Proc. Natl. Acad. Sci. USA 2010, 107, 3285–3286. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007, 2, 3081–3089. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.H.; Hannan, N.R.; Docherty, F.M.; Docherty, H.M.; Joao Lima, M.; Trotter, M.W.; Docherty, K.; Vallier, L. Inhibition of activin/nodal signalling is necessary for pancreatic differentiation of human pluripotent stem cells. Diabetologia 2012, 55, 3284–3295. [Google Scholar] [CrossRef] [PubMed]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Maehr, R.; Chen, S.; Chen, A.E.; Tang, W.; Fox, J.L.; Schreiber, S.L.; Melton, D.A. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 2009, 4, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.P.; Bear, C.E.; Chin, S.; Pasceri, P.; Thompson, T.O.; Huan, L.J.; Ratjen, F.; Ellis, J.; Rossant, J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012, 30, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.P.; Rossant, J. Generation of lung epithelium from pluripotent stem cells. Curr. Pathobiol. Rep. 2013, 1, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.; D’Arcy, S.; Barry, F.P.; Murphy, J.M.; Coleman, C.M. Mesenchymal chondroprogenitor cell origin and therapeutic potential. Stem Cell Res. Ther. 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.D.; Yu, J.; Smuga-Otto, K.; Salvagiotto, G.; Rehrauer, W.; Vodyanik, M.; Thomson, J.; Slukvin, I. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 2009, 27, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, R.M.; Gibson, J.; Xu, R.H.; Lee, F.Y.; Drissi, H. Efficient differentiation of human iPSC-derived mesenchymal stem cells to chondroprogenitor cells. J. Cell. Biochem. 2013, 114, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Gumenyuk, M.; Kang, H.; Vodyanik, M.; Yu, J.; Thomson, J.A.; Slukvin, I.I. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011, 20, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Shaikh Qureshi, W.M.; Latif, M.L.; Parker, T.L.; Pratten, M.K. Lithium carbonate teratogenic effects in chick cardiomyocyte micromass system and mouse embryonic stem cell derived cardiomyocyte—Possible protective role of myo-inositol. Reprod. Toxicol. 2014, 46, 106–114. [Google Scholar]
- Kattman, S.J.; Witty, A.D.; Gagliardi, M.; Dubois, N.C.; Niapour, M.; Hotta, A.; Ellis, J.; Keller, G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011, 8, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Ema, M.; Takahashi, S.; Rossant, J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 2006, 107, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Kawamata, S.; Murakami, Y.; Inoue, K.; Nagahashi, A.; Tosaka, M.; Yoshimura, N.; Miyamoto, Y.; Iwasaki, H.; Asahara, T.; et al. Coexpression of platelet-derived growth factor receptor α and fetal liver kinase 1 enhances cardiogenic potential in embryonic stem cell differentiation in vitro. J. Biosci. Bioeng. 2007, 103, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Uosaki, H.; Yamashita, J.K. Chemicals regulating cardiomyocyte differentiation. In Embryonic Stem Cells: The Hormonal Regulation of Pluripotency and Embryogenesis; Atwood, C., Ed.; InTech: Winchester, UK, Chapter 24.
- Terami, H.; Hidaka, K.; Katsumata, T.; Iio, A.; Morisaki, T. Wnt11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem. Biophys. Res. Commun. 2004, 325, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.; Laurent, T.; Ding, S. Small molecules, big roles—The chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 2012, 125, 5609–5620. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.; Liu, Z.; Chen, Z.; Wang, J.; Chen, T.; Zhao, X.; Ma, Y.; Qin, L.; Kang, J.; Wei, B.; et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012, 22, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Yau, W.W.; Tang, M.K.; Chen, E.; Yao, Y.; Wong, I.W.C.; Lee, H.S.S.; Lee, K.K.H. Cardiogenol C can induce mouse hair bulge progenitor cells to transdifferentiate into cardiomyocyte-like cells. Proteome Sci. 2011, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Dolle, P.; Ryckebusch, L.; Noseda, M.; Zaffran, S.; Schneider, M.D.; Niederreither, K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 9234–9239. [Google Scholar] [CrossRef] [PubMed]
- Osada, H. Protein Targeting with Small Molecules: Chemical Biology Techniques and Applications; Wiley: Hoboken, NJ, USA, 2009; p. 297. [Google Scholar]
- Mezentseva, N.V.; Yang, J.; Kaur, K.; Iaffaldano, G.; Remond, M.C.; Eisenberg, C.A.; Eisenberg, L.M. The histone methyltransferase inhibitor BIX01294 enhances the cardiac potential of bone marrow cells. Stem Cells Dev. 2013, 22, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.; Chung, M.; Cohen, C.J. A dihydropyridine (Bay k 8644) that enhances calcium currents in guinea pig and calf myocardial cells. A new type of positive inotropic agent. Circ. Res. 1985, 56, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Savickiene, J.; Treigyte, G.; Jazdauskaite, A.; Borutinskaite, V.V.; Navakauskiene, R. DNA methyltransferase inhibitor RG108 and histone deacetylase inhibitors cooperate to enhance NB4 cell differentiation and E-cadherin re-expression by chromatin remodelling. Cell Biol. Int. 2012, 36, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Qian, H.; Zhang, X.; Zhu, W.; Yan, Y.; Ye, S.; Peng, X.; Li, W.; Xu, Z.; Sun, L.; et al. 5-Azacytidine induces cardiac differentiation of human umbilical cord-derived mesenchymal stem cells by activating extracellular regulated kinase. Stem Cells Dev. 2012, 21, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Minami, I.; Yamada, K.; Otsuji, T.G.; Yamamoto, T.; Shen, Y.; Otsuka, S.; Kadota, S.; Morone, N.; Barve, M.; Asai, Y.; et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012, 2, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Sano, M.; Songyang, Z.; Schneider, M.D. A Wnt- and β-catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 5834–5839. [Google Scholar] [CrossRef] [PubMed]
- Xu, C. Turning cardiac fibroblasts into cardiomyocytes in vivo. Trends Mol. Med. 2012, 18, 575–576. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, P.; Sdek, P.; MacLellan, W.R. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol. Rev. 2007, 87, 521–544. [Google Scholar] [CrossRef] [PubMed]
- Guyette, J.P.; Moser, P.T.; Charest, J.M.; Ott, H.C. Generation of functional human myocardium from native human heart matrix and human induced pluripotent stem (iPS) derived cardiomyocytes. J. Am. Coll. Surg. 2014, 219, e49. [Google Scholar] [CrossRef]
- Becker, A.; Rubart, M.; Field, L. Inducing embryonic stem cells to become cardiomyocytes. In Regenerating the Heart; Cohen, I.S., Gaudette, G.R., Eds.; Humana Press: New York, NY, USA, 2011; pp. 7–24. [Google Scholar]
- Freund, C.; Mummery, C.L. Prospects for pluripotent stem cell-derived cardiomyocytes in cardiac cell therapy and as disease models. J. Cell. Biochem. 2009, 107, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Segers, V.F.M.; Lee, R.T. Stem-cell therapy for cardiac disease. Nature 2008, 451, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, W.H.; Melnychenko, I.; Wasmeier, G.; Didie, M.; Naito, H.; Nixdorff, U.; Hess, A.; Budinsky, L.; Brune, K.; Michaelis, B.; et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat. Med. 2006, 12, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, J.; Liu, Z.; Wang, C. Injectable cardiac tissue engineering for the treatment of myocardial infarction. J. Cell. Mol. Med. 2010, 14, 1044–1055. [Google Scholar] [PubMed]
- Liu, Z.; Tang, Y.; Lu, S.; Zhou, J.; Du, Z.; Duan, C.; Li, Z.; Wang, C. The tumourigenicity of iPS cells and their differentiated derivates. J. Cell. Mol. Med. 2013, 17, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Vincent, G.M. The long QT syndrome. Ind. Pac. Electrophysiol. J. 2002, 2, 127–142. [Google Scholar]
- Sheng, C.C.; Hong, C.C. Pluripotent stem cells to model human cardiac diseases. In Pluripotent Stem Cells; Bhartiya, D., Lenka, N., Eds.; InTech: Winchester, UK, 2013; Chapter 20. [Google Scholar]
- Yazawa, M.; Dolmetsch, R.E. Modeling Timothy syndrome with iPS cells. J. Cardiovasc. Transl. Res. 2013, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, M.; Hsueh, B.; Jia, X.; Pasca, A.M.; Bernstein, J.A.; Hallmayer, J.; Dolmetsch, R.E. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011, 471, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Towbin, J.A.; Thiene, G.; Antzelevitch, C.; Corrado, D.; Arnett, D.; Moss, A.J.; Seidman, C.E.; Young, J.B. Contemporary definitions and classification of the cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006, 113, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Willott, R.H.; Gomes, A.V.; Chang, A.N.; Parvatiyar, M.S.; Pinto, J.R.; Potter, J.D. Mutations in Troponin that cause HCM, DCM and RCM: What can we learn about thin filament function? J. Mol. Cell. Cardiol. 2010, 48, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yazawa, M.; Liu, J.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.; Wang, L.; Lee, A.; et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012, 4, 130–147. [Google Scholar] [CrossRef]
- Yuan, T.; Liao, W.; Feng, N.H.; Lou, Y.L.; Niu, X.; Zhang, A.J.; Wang, Y.; Deng, Z.F. Human induced pluripotent stem cell-derived neural stem cells survive, migrate, differentiate, improve neurologic function in a rat model of middle cerebral artery occlusion. Stem Cell. Res. Ther. 2013, 4, 73. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; An, M.C.; Montoro, D.; Ellerby, L.M. Characterization of human huntington’s disease cell model from induced pluripotent stem cells. PLoS Curr. 2010, 2, RRN1193. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.A.; McCaffery, P.J.; Drager, U.C.; de Luca, L.M. Retinoids in embryonal development. Physiol. Rev. 2000, 80, 1021–1054. [Google Scholar] [PubMed]
- Okada, Y.; Shimazaki, T.; Sobue, G.; Okano, H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 2004, 275, 124–142. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.M.; Ahn, J.I.; Lee, K.H.; Lee, Y.S. Ascorbic acid responsive genes during neuronal differentiation of embryonic stem cells. Neuroreport 2004, 15, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- Rebec, G.V.; Pierce, R.C. A vitamin as neuromodulator: Ascorbate release into the extracellular fluid of the brain regulates dopaminergic and glutamatergic transmission. Prog. Neurobiol. 1994, 43, 537–565. [Google Scholar] [CrossRef] [PubMed]
- Bagga, V.; Dunnett, S.B.; Fricker-Gates, R.A. Ascorbic acid increases the number of dopamine neurons in vitro and in transplants to the 6-OHDA-lesioned rat brain. Cell Transplant. 2008, 17, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Kalir, H.H.; Mytilineou, C. Ascorbic acid in mesencephalic cultures: Effects on dopaminergic neuron development. J. Neurochem. 1991, 57, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Hartfield, E.M.; Yamasaki-Mann, M.; Ribeiro Fernandes, H.J.; Vowles, J.; James, W.S.; Cowley, S.A.; Wade-Martins, R. Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS One 2014, 9, e87388. [Google Scholar] [CrossRef] [PubMed]
- Tornero, D.; Wattananit, S.; Gronning Madsen, M.; Koch, P.; Wood, J.; Tatarishvili, J.; Mine, Y.; Ge, R.; Monni, E.; Devaraju, K.; et al. Human induced pluripotent stem cell-derived cortical neurons integrate in stroke-injured cortex and improve functional recovery. Brain J. Neurol. 2013, 136, 3561–3577. [Google Scholar] [CrossRef]
- Young, W.; D’Souza, S.L.; Lemischka, I.R.; Schaniel, C. Patient-specific induced pluripotent stem cells as a platform for disease modeling, drug discovery and precision personalized medicine. J. Stem Cell Res. Ther. 2012, 2, S10. [Google Scholar]
- Menendez, L.; Yatskievych, T.A.; Antin, P.B.; Dalton, S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. USA 2011, 108, 19240–19245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pak, C.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Boulting, G.L.; Kiskinis, E.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; Wainger, B.J.; Williams, D.J.; Kahler, D.J.; Yamaki, M.; Davidow, L.; et al. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, M.; Yan, Y.; Yang, S.T. Neural differentiation from pluripotent stem cells: The role of natural and synthetic extracellular matrix. World J. Stem Cells 2014, 6, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Su, P.; Li, D.; Tsang, S.; Duan, E.; Wang, F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells 2010, 28, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Mak, S.K.; Huang, Y.A.; Iranmanesh, S.; Vangipuram, M.; Sundararajan, R.; Nguyen, L.; Langston, J.W.; Schule, B. Small molecules greatly improve conversion of human-induced pluripotent stem cells to the neuronal lineage. Stem Cells Int. 2012, 2012, 140427. [Google Scholar] [CrossRef] [PubMed]
- Kreitzer, F.R.; Salomonis, N.; Sheehan, A.; Huang, M.; Park, J.S.; Spindler, M.J.; Lizarraga, P.; Weiss, W.A.; So, P.L.; Conklin, B.R. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am. J. Stem Cells 2013, 2, 119–131. [Google Scholar] [PubMed]
- Karumbayaram, S.; Novitch, B.G.; Patterson, M.; Umbach, J.A.; Richter, L.; Lindgren, A.; Conway, A.E.; Clark, A.T.; Goldman, S.A.; Plath, K.; et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells 2009, 27, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.; Brennand, K.J.; Boyer, L.F.; Gage, F.H. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: Progress and promises. Hum. Mol. Genet. 2011, 20, R109–R115. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.D.; Yu, J.; Rose, F.F., Jr.; Mattis, V.B.; Lorson, C.L.; Thomson, J.A.; Svendsen, C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009, 457, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.X.; Marchetto, M.C.; Gage, F.H. Therapeutic translation of iPSCs for treating neurological disease. Cell Stem Cell 2013, 12, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Nakajo, S.; Tu, P.H.; Tomita, T.; Nakaya, K.; Lee, V.M.; Trojanowski, J.Q.; Iwatsubo, T. Aggregation of α-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 1998, 152, 879–884. [Google Scholar] [PubMed]
- Nguyen, H.N.; Byers, B.; Cord, B.; Shcheglovitov, A.; Byrne, J.; Gujar, P.; Kee, K.; Schule, B.; Dolmetsch, R.E.; Langston, W.; et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 2011, 8, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Devine, M.J.; Ryten, M.; Vodicka, P.; Thomson, A.J.; Burdon, T.; Houlden, H.; Cavaleri, F.; Nagano, M.; Drummond, N.J.; Taanman, J.W.; et al. Parkinson’s disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat. Commun. 2011, 2, 440. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ren, Y.; Yuen, E.Y.; Zhong, P.; Ghaedi, M.; Hu, Z.; Azabdaftari, G.; Nakaso, K.; Yan, Z.; Feng, J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat. Commun. 2012, 3, 668. [Google Scholar] [CrossRef] [PubMed]
- Woodard, C.M.; Campos, B.A.; Kuo, S.-H.; Nirenberg, M.J.; Nestor, M.W.; Zimmer, M.; Mosharov, E.V.; Sulzer, D.; Zhou, H.; Paull, D.; et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s Disease. Cell Rep. 2014, 9, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Ito, D.; Okada, Y.; Akamatsu, W.; Nihei, Y.; Yoshizaki, T.; Yamanaka, S.; Okano, H.; Suzuki, N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 4530–4539. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 2012, 482, 216–220. [Google Scholar] [PubMed]
- Duan, L.; Bhattacharyya, B.J.; Belmadani, A.; Pan, L.; Miller, R.J.; Kessler, J.A. Stem cell derived basal forebrain cholinergic neurons from Alzheimer’s disease patients are more susceptible to cell death. Mol. Neurodegener. 2014, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Asai, M.; Tsukita, K.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Imamura, K.; Egawa, N.; Yahata, N.; Okita, K.; et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell 2013, 12, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Nishitsuji, K.; Tomiyama, T.; Ishibashi, K.; Ito, K.; Teraoka, R.; Lambert, M.P.; Klein, W.L.; Mori, H. The E693∆ mutation in amyloid precursor protein increases intracellular accumulation of amyloid β oligomers and causes endoplasmic reticulum stress-induced apoptosis in cultured cells. Am. J. Pathol. 2009, 174, 957–969. [Google Scholar] [CrossRef] [PubMed]
- The HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 2012, 11, 264–278. [Google Scholar]
- Carter, R.L.; Chen, Y.; Kunkanjanawan, T.; Xu, Y.; Moran, S.P.; Putkhao, K.; Yang, J.; Huang, A.H.C.; Parnpai, R.; Chan, A.W.S. Reversal of cellular phenotypes in neural cells derived from Huntington’s disease monkey-induced pluripotent stem cells. Stem Cell Rep. 2014, 3, 585–593. [Google Scholar] [CrossRef]
- Camnasio, S.; Delli Carri, A.; Lombardo, A.; Grad, I.; Mariotti, C.; Castucci, A.; Rozell, B.; lo Riso, P.; Castiglioni, V.; Zuccato, C.; et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012, 46, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.I.; Kim, D.W.; Lee, N.; Jeon, Y.J.; Jeon, I.; Kwon, J.; Kim, J.; Soh, Y.; Lee, D.S.; Seo, K.S.; et al. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington’s disease patient. Biochem. J. 2012, 446, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Koch, P.; Breuer, P.; Peitz, M.; Jungverdorben, J.; Kesavan, J.; Poppe, D.; Doerr, J.; Ladewig, J.; Mertens, J.; Tüting, T.; et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature 2011, 480, 543–546. [Google Scholar] [PubMed]
- Nihei, Y.; Ito, D.; Okada, Y.; Akamatsu, W.; Yagi, T.; Yoshizaki, T.; Okano, H.; Suzuki, N. Enhanced aggregation of androgen receptor in induced pluripotent stem cell-derived neurons from spinal and bulbar muscular atrophy. J. Biol. Chem. 2013, 288, 8043–8052. [Google Scholar] [CrossRef] [PubMed]
- Hook, V.; Brennand, K.J.; Yongsung Kim, Y.; Toneff, T.; Funkelstein, L.; Lee, K.C.; Ziegler, M.; Gage, F.H. Human iPSC neurons display activity-dependent neurotransmitter secretion: Aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep. 2014, 3, 531–538. [Google Scholar] [CrossRef]
- Bird, M.J.; Needham, K.; Frazier, A.E.; van Rooijen, J.; Leung, J.; Hough, S.; Denham, M.; Thornton, M.E.; Parish, C.L.; Nayagam, B.A.; et al. Functional characterization of Friedreich ataxia iPS-derived neuronal progenitors and their integration in the adult brain. PLoS One 2014, 7, e101718. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, Z.N.; Rong, Z.; Xu, Y. Immunogenicity of induced pluripotent stem cells. Nature 2011, 474, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Guha, P.; Morgan, J.W.; Mostoslavsky, G.; Rodrigues, N.P.; Boyd, A.S. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 2013, 12, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Uda, M.; Hoki, Y.; Sunayama, M.; Nakamura, M.; Ando, S.; Sugiura, M.; Ideno, H.; Shimada, A.; Nifuji, A.; et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 2013, 494, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jiang, H.; Ma, D.; Nakaso, K.; Feng, J. Parkin degrades estrogen-related receptors to limit the expression of monoamine oxidases. Hum. Mol. Genet. 2011, 20, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Cooper, O.; Seo, H.; Andrabi, S.; Guardia-Laguarta, C.; Graziotto, J.; Sundberg, M.; McLean, J.R.; Carrillo-Reid, L.; Xie, Z.; Osborn, T.; et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012, 4, 141–190. [Google Scholar] [CrossRef]
- Yahata, N.; Asai, M.; Kitaoka, S.; Takahashi, K.; Asaka, I.; Hioki, H.; Kaneko, T.; Maruyama, K.; Saido, T.C.; Nakahata, T.; et al. Anti-Aβ drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer’s disease. PLoS One 2011, 6, e25788. [Google Scholar] [CrossRef] [PubMed]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Bjorklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef] [PubMed]
- An, M.C.; Zhang, N.; Scott, G.; Montoro, D.; Wittkop, T.; Mooney, S.; Melov, S.; Ellerby, L.M. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 2012, 11, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Suzuki, K.; Qu, J.; Sancho-Martinez, I.; Yi, F.; Li, M.; Kumar, S.; Nivet, E.; Kim, J.; Soligalla, R.D.; et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell 2011, 8, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Soldner, F.; Laganiere, J.; Cheng, A.W.; Hockemeyer, D.; Gao, Q.; Alagappan, R.; Khurana, V.; Golbe, L.I.; Myers, R.H.; Lindquist, S.; et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 2011, 146, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Durrani, S.; Pandey, R.; Jiang, S.; Ahmed, R.P. MicroRNA mediated proliferation of human iPS derived cardiomyocytes for cardiac repair. Circulation 2014, 130, A16332. [Google Scholar]
- Seyhan, A.A. RNAi: A potential new class of therapeutic for human genetic disease. Hum. Genet. 2011, 130, 583–605. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalova, S.; Svadlakova, T.; Qureshi, W.M.S.; Dev, K.; Mokry, J. Induced Pluripotent Stem Cells and Their Use in Cardiac and Neural Regenerative Medicine. Int. J. Mol. Sci. 2015, 16, 4043-4067. https://doi.org/10.3390/ijms16024043
Skalova S, Svadlakova T, Qureshi WMS, Dev K, Mokry J. Induced Pluripotent Stem Cells and Their Use in Cardiac and Neural Regenerative Medicine. International Journal of Molecular Sciences. 2015; 16(2):4043-4067. https://doi.org/10.3390/ijms16024043
Chicago/Turabian StyleSkalova, Stepanka, Tereza Svadlakova, Wasay Mohiuddin Shaikh Qureshi, Kapil Dev, and Jaroslav Mokry. 2015. "Induced Pluripotent Stem Cells and Their Use in Cardiac and Neural Regenerative Medicine" International Journal of Molecular Sciences 16, no. 2: 4043-4067. https://doi.org/10.3390/ijms16024043