Molecular Mechanism of the Anti-Inflammatory Action of Heparin
Abstract
:1. Introduction
2. Results
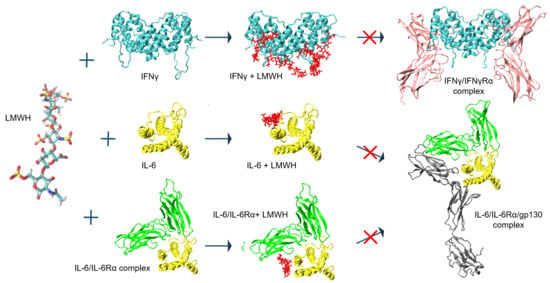
2.1. Simulation of Heparin Hexasaccharides’ Interaction with IFNγ
2.2. Effect of Heparin on the Activity of IFNγ
2.3. LMWH Binding to the IL-6 and IL-6/IL-6Rα Complex
3. Discussion
The COVID-19 Case
4. Materials and Methods
4.1. Explicit Solvent MD Simulations
4.2. Heparin Hexasaccharide Structure
4.3. Human Interferon Gamma
4.4. Human Interleukin 6
4.5. IL-6/IL-6Rα/gp130 Complex
4.6. Heparin–Protein Complexes Simulations
4.7. Reference Simulations
4.8. Complex-Interactions Analysis
4.9. Cell Culture
4.10. Inhibitory Antiproliferative Assay
- 100 ng/mL IFNγ purified as described in [13],
- IFNγ (100 ng/mL) supplemented with Fraxiparine (GlaxoSmithKline Pharmaceuticals S.A.) in concentrations varying from 1.5 to 150 anti-Xa IU/mL pre-incubated for 1 h at room temperature, and
- Fraxiparine in the same concentrations as in ii. Further, the antiproliferative activity of IFNγ was measured by a modified kynurenine bioassay as described earlier [12].
4.11. Cellular Localisation of Phosphorylated STAT1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Barrowcliffe, T. History of Heparin in Heparin—A Century of Progress; Lever, R., Mulloy, B., Page, C.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 3–22. [Google Scholar]
- Call, D.R.; Remick, D. Low molecular weight heparin is associated with greater cytokine production in a stimulated whole blood model. Shock 1998, 10, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Veraldi, N.; Hughes, A.J.; Rudd, T.R.; Thomas, H.; Edwards, S.W.; Hadfield, L.; Skidmore, M.; Siligardi, G.; Cosentino, C.; Shute, J.K.; et al. Heparin derivatives for the targeting of multiple activities in the inflammatory response. Carbohydr. Polym. 2015, 117, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Rudd, T.; Yates, E. New Applications of Heparin and Other Glycosaminoglycans. Molecules 2017, 22, 749. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Lortat-Jacob, H.; Kleinman, H.K.; A Grimaud, J. High-affinity binding of interferon-gamma to a basement membrane complex (matrigel). J. Clin. Investig. 1991, 87, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Lortat-Jacob, H.; Baltzer, F.; Grimaud, J.-A. Heparin Decreases the Blood Clearance of Interferon-γ and Increases Its Activity by Limiting the Processing of Its Carboxyl-terminal Sequence. J. Biol. Chem. 1996, 271, 16139–16143. [Google Scholar] [CrossRef] [Green Version]
- Vanhaverbeke, C.; Simorre, J.-P.; Sadir, R.; Gans, P.; Lortat-Jacob, H. NMR characterization of the interaction between the C-terminal domain of interferon-γ and heparin-derived oligosaccharides. Biochem. J. 2004, 384, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Mummery, R.S.; Rider, C. Characterization of the Heparin-Binding Properties of IL-6. J. Immunol. 2000, 165, 5671–5679. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.R.; Windsor, W.T.; Nagabhushan, T.L.; Lundell, D.J.; Lunn, C.A.; Zauodny, P.J.; Narula, S.K. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature 1995, 376, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Thiel, D.J.; le Du, M.-H.; Walter, R.L.; D’Arcy, A.; Chène, C.; Fountoulakis, M.; Garotta, G.; Winkler, F.K.; Ealick, S.E. Observation of An Unexpected Third Receptor Molecule in the Crystal Structure of Human Interferon-Gamma Receptor Complex. Structure 2000, 8, 927–936. [Google Scholar] [CrossRef] [Green Version]
- Boyanova, M.; Tsanev, R.; Ivanov, I. A modified kynurenine bioassay for quantitative determination of human interferon-γ. Anal. Biochem. 2002, 308, 178–181. [Google Scholar] [CrossRef]
- Petrov, S.; Nacheva, G.; Ivanov, I. Purification and refolding of recombinant human interferon-gamma in urea–ammonium chloride solution. Protein Expr. Purif. 2010, 73, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, P.C.; Castell, J.V.; Andus, T. Interleukin-6 and the acute phase response. Biochem. J. 1990, 265, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Taga, T.; Kishimoto, T. Interleukin-6 in Biology and Medicine. Adv. Immunol. 1993, 54, 1–78. [Google Scholar] [PubMed]
- Kishimoto, T. IL-6: From its discovery to clinical applications. Int. Immunol. 2010, 22, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. et Biophys. Acta (BBA)—Bioenerg. 2011, 1813, 878–888. [Google Scholar] [CrossRef] [Green Version]
- Petkov, P.; Rangelov, M.; Ilieva, N.; Todorova, N.; Lilkova, E.; Litov, L. Computational study of IL-6 inhibition by low-molecular-weight heparin. 2021; in preparation. [Google Scholar]
- Savino, R.; Lahm, A.; Salvati, A.; Ciapponi, L.; Sporeno, E.; Altamura, S.; Paonessa, G.; Toniatti, C.; Ciliberto, G. Generation of interleukin-6 receptor antagonists by molecular-modeling guided mutagenesis of residues important for gp130 activation. EMBO J. 1994, 13, 1357–1367. [Google Scholar] [CrossRef]
- Mazák, K.; Beecher, C.N.; Kraszni, M.; Larive, C.K. The interaction of enoxaparin and fondaparinux with calcium. Carbohydr. Res. 2014, 384, 13–19. [Google Scholar] [CrossRef]
- Lin, F.-C.; Young, H.A. Interferon-Gamma in Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2012; pp. 966–972. [Google Scholar]
- Su, X.; Yu, Y.; Zhong, Y.; Giannopoulou, E.G.; Hu, X.; Liu, H.; Cross, J.; Rätsch, G.; Rice, C.M.; Ivashkiv, L.B. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015, 16, 838–849. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E.; et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016, 167, 397–404. [Google Scholar] [CrossRef] [Green Version]
- A Hunter, C.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef]
- Ulhaq, Z.S.; Soraya, G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect. 2020, 50, 382–383. [Google Scholar] [CrossRef]
- Pandolfi, F.; Franza, L.; Carusi, V.; Altamura, S.; Andriollo, G.; Nucera, E. Interleukin-6 in rheumatoid arthritis. Int. J. Mol. Sci. 2020, 21, 5238. [Google Scholar] [CrossRef]
- Van Rhee, F.; Fayad, L.; Voorhees, P.; Furman, C.; Lonial, S.; Borghaei, H.; Sokol, L.; Crawford, J.; Cornfeld, M.; Qi, M.; et al. Siltuximab, a novel anti-interleukin-6 monoclonal antibody, for Castleman’s disease. J. Clin. Oncol. 2010, 28, 3701–3708. [Google Scholar] [CrossRef]
- Deisseroth, A.; Ko, C.-W.; Nie, L.; Zirkelbach, J.F.; Zhao, L.; Bullock, J.; Mehrotra, N.; Del Valle, P.; Saber, H.; Sheth, C.; et al. FDA Approval: Siltuximab for the Treatment of Patients with Multicentric Castleman Disease. Clin. Cancer Res. 2015, 21, 950–954. [Google Scholar] [CrossRef] [Green Version]
- Mihara, M.; Kasutani, K.; Okazaki, M.; Nakamura, A.; Kawai, S.; Sugimoto, M.; Matsumoto, Y.; Ohsugi, Y. Tocilizumab inhibits signal transduction mediated by both mIL-6R and sIL-6R, but not by the receptors of other members of IL-6 cytokine family. Int. Immunopharmacol. 2005, 5, 1731–1740. [Google Scholar] [CrossRef]
- Garbers, C.; Heink, S.; Korn, T.; Rose-John, S. Interleukin-6: Designing specific therapeutics for a complex cytokine. Nat. Rev. Drug Discov. 2018, 17, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Vigant, F.; Santos, N.; Lee, B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Genet. 2015, 13, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-L.; Lei, H.-Y.; Lin, Y.-S.; Yeh, T.-M.; Chen, S.-H.; Liu, H.-S. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antivir. Res. 2002, 56, 93–96. [Google Scholar] [CrossRef]
- Montanuy, I.; Alejo, A.; Alcami, A. Glycosaminoglycans mediate retention of the poxvirus type I interferon binding protein at the cell surface to locally block interferon antiviral responses. FASEB J. 2011, 25, 1960–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-M.; Liao, C.-L.; Lee, Y.-L.; Lin, Y.-L. Highly Sulfated Forms of Heparin Sulfate Are Involved in Japanese Encephalitis Virus Infection. Virology 2001, 286, 206–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skidmore, M.A.; Kajaste-Rudnitski, A.; Wells, N.M.; Guimond, S.E.; Rudd, T.R.; Yates, E.A.; Vicenzi, E. Inhibition of influenza H5N1 invasion by modified heparin derivatives. Med. Chem. Comm. 2015, 6, 640–646. [Google Scholar] [CrossRef]
- Ghezzi, S.; Cooper, L.; Rubio, A.; Pagani, I.; Capobianchi, M.R.; Ippolito, G.; Pelletier, J.; Meneghetti, M.C.Z.; Lima, M.A.; Skidmore, M.; et al. Heparin prevents Zika virus induced-cytopathic effects in human neural progenitor cells. Antivir. Res. 2017, 140, 13–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Guimond, S.E.; Miller, G.; Meneghetti, M.C.Z.; Nader, H.B.; Li, Y.; et al. Heparin inhibits cellular invasion by SARS-CoV-2: Structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar]
- Negri, E.M.; Piloto, B.; Morinaga, L.K.; Jardim, C.; Lamy, S.A.E.-D.; Ferreira, M.A.; D’Amico, E.A.; Deheinzelin, D. Heparin therapy improving hypoxia in COVID-19 patients—A case series. Front. Physiol. 2020, 11, 573044. [Google Scholar] [CrossRef]
- Yan, Y.; Ji, Y.; Su, N.; Mei, X.; Wang, Y.; Du, S.; Zhu, W.; Zhang, C.; Lu, Y.; Xing, X.-H. Non-anticoagulant effects of low molecular weight heparins in inflammatory disorders: A review. Carbohydr. Polym. 2017, 160, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Page, C. Heparin and Related Drugs: Beyond Anticoagulant Activity. ISRN Pharmacol. 2013, 2013, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Weitz, J.I. Low-molecular-weight heparins. N. Engl. J. Med. 1997, 337, 688–698. [Google Scholar] [CrossRef]
- Lever, R.; Page, C.P. Non-anticoagulant Effects of Heparin: An Overview. In Heparin—A Century of Progress. Handbook of Experimental Pharmacology; Lever, R., Mulloy, B., Page, C.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 207. [Google Scholar]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Allegra, A.; Di Gioacchino, M.; Tonacci, A.; Musolino, C.; Gangemi, S. Immunopathology of SARS-CoV-2 infection: Immune cells and mediators, prognostic factors, and immune-therapeutic implications. Int. J. Mol. Sci. 2020, 21, 4782. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Al-Horani, R.; Kar, S.; Aliter, K. Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials. Int. J. Mol. Sci. 2020, 21, 5224. [Google Scholar] [CrossRef]
- Dixon, B.; Smith, R.J.; Artigas, A.; Laffey, J.; McNicholas, B.; Schmidt, E.; Nunes, Q.; Skidmore, M.; de Lima, M.A.; Moran, J.L.; et al. Can Nebulised Heparin Reduce Time to Extubation in SARS CoV 2 (CHARTER)—Study Protocol. Available online: https://www.medrxiv.org/content/10.1101/2020.04.28.20082552v2 (accessed on 25 August 2021).
- Tang, N.; Bai, H.; Chen, X.; Gong, J.; Li, D.; Sun, Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020, 18, 1094–1099. [Google Scholar] [CrossRef]
- Perna, A.F.; Capolongo, G.; Trepiccione, F.; Simeoni, M.; Zacchia, M.; Ingrosso, D. COVID-19, Low-Molecular-Weight Heparin, and Hemodialysis. Kidney Blood Press. Res. 2020, 45, 357–362. [Google Scholar] [CrossRef]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Wang, C.; Wang, H.; Yang, C.; Cai, F.; Zeng, F.; Cheng, F.; Liu, Y.; Zhou, T.; Deng, B.; et al. The Potential of Low Molecular Weight Heparin to Mitigate Cytokine Storm in Severe COVID-19 Patients: A retrospective clinical study. Available online: https://www.medrxiv.org/content/10.1101/2020.03.28.20046144v3 (accessed on 25 August 2021).
- Tan, M.; Liu, Y.; Zhou, R.; Deng, X.; Li, F.; Liang, K.; Shi, Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology 2020, 160, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, E.; Hess, B.; Van Der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Perlin, A.; Mackie, D.; Dietrich, C. Evidence for a (1→4)-linked 4-O-(α-L-idopyranosyluronic acid 2-sulfate)-(2-deoxy-2-sulfoamino-D-glucopyranosyl 6-sulfate) sequence in heparin: Long-range H-H coupling in 4-deoxy-hex-4-enopyranosides. Carbohydr. Res. 1971, 18, 185–194. [Google Scholar] [CrossRef]
- Gupta, R.; Ponnusamy, M.P. Analysis of sulfates on low molecular weight heparin using mass spectrometry: Structural characterization of enoxaparin. Expert Rev. Proteom. 2018, 15, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sheng, A.; Liu, X.; Shi, F.; Jin, L.; Xie, S.; Zhang, F.; Linhardt, R.J.; Chi, L. Comprehensive Identification and Quantitation of Basic Building Blocks for Low-Molecular Weight Heparin. Anal. Chem. 2016, 88, 7738–7744. [Google Scholar] [CrossRef]
- Park, S.-J.; Lee, J.; Qi, Y.; Kern, N.R.; Lee, H.S.; Jo, S.; Joung, I.; Joo, K.; Lee, J.; Im, W. CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 2019, 29, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, I.F.W.; MacKerell, J.A.D. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate–Protein Modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef] [Green Version]
- Case, D.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Petkov, P.; Lilkova, E.; Ilieva, N.; Nacheva, G.; Ivanov, I.; Litov, L. Computational Modelling of the Full Length hIFN-gamma Homodimer in Lecture Notes in Computer Science; Lirkov, I., Margenov, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Volume 10665. [Google Scholar]
- Lilkova, E.; Petkov, P.; Ilieva, N.; Krachmarova, E.; Nacheva, G.; Litov, L. Molecular modeling of the effects of glycosylation on the structure and dynamics of human interferon-gamma. J. Mol. Model. 2019, 25, 127. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Size-independent comparison of protein three-dimensional structures. Proteins: Struct. Funct. Bioinform. 1995, 22, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Somers, W.; Stahl, M.; Seehra, J.S. 1.9 Å crystal structure of interleukin 6: Implications for a novel mode of receptor dimerization and signaling. EMBO J. 1997, 16, 989–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comp. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Boulanger, M.J.; Chow, D.-C.; Brevnova, E.E.; Garcia, K.C. Hexameric Structure and Assembly of the Interleukin-6/IL-6 α-Receptor/gp130 Complex. Science 2003, 300, 2101–2104. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE). Chemical Computing Group ULC; 1010 Sherbooke St. West, Suite #910: Montreal, QC, Canada, 2021. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.M.; Labute, P. 2D Depiction of Protein−Ligand Complexes. J. Chem. Inf. Modeling 2007, 47, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [Green Version]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Litov, L.; Petkov, P.; Rangelov, M.; Ilieva, N.; Lilkova, E.; Todorova, N.; Krachmarova, E.; Malinova, K.; Gospodinov, A.; Hristova, R.; et al. Molecular Mechanism of the Anti-Inflammatory Action of Heparin. Int. J. Mol. Sci. 2021, 22, 10730. https://doi.org/10.3390/ijms221910730
Litov L, Petkov P, Rangelov M, Ilieva N, Lilkova E, Todorova N, Krachmarova E, Malinova K, Gospodinov A, Hristova R, et al. Molecular Mechanism of the Anti-Inflammatory Action of Heparin. International Journal of Molecular Sciences. 2021; 22(19):10730. https://doi.org/10.3390/ijms221910730
Chicago/Turabian StyleLitov, Leandar, Peicho Petkov, Miroslav Rangelov, Nevena Ilieva, Elena Lilkova, Nadezhda Todorova, Elena Krachmarova, Kristina Malinova, Anastas Gospodinov, Rossitsa Hristova, and et al. 2021. "Molecular Mechanism of the Anti-Inflammatory Action of Heparin" International Journal of Molecular Sciences 22, no. 19: 10730. https://doi.org/10.3390/ijms221910730