Current Taxonomical Situation of Streptococcus suis
Abstract
:1. Introduction
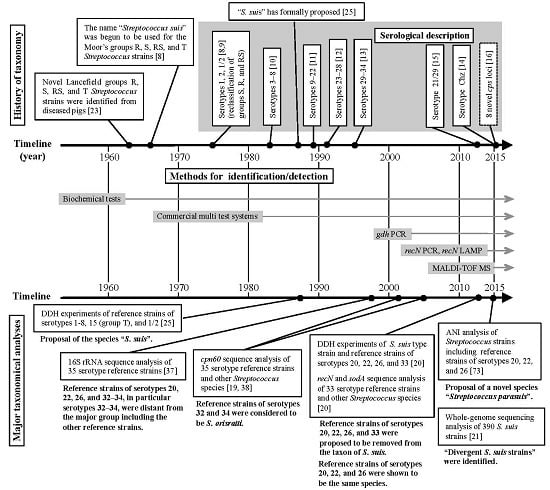
2. Taxonomic and Serological Classification Histories of S. suis
3. Several Taxonomic Analyses Revealed that Six S. suis Serotype Reference Strains are not S. suis
3.1. Taxonomic Standards for Species Delineation and Taxonomic Approaches for Phylogenetic Relationships in Bacteria
3.2. Taxonomic Studies Using S. suis Serotype Reference Strains
4. Methods for Identifying and Detecting S. suis
4.1. Routine Methods for Identifying and Detecting S. suis
4.2. Novel Methods for the Precise Identification and Detection of S. suis
5. Current Topics on the Classification of S. suis-Like Strains
5.1. Whole-Genome Sequencing-Based Taxonomic Analyses in Bacteria
5.2. Streptococcus parasuis and Divergent S. suis Strains
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CPS | capsular polysaccharide |
| ST | Sequence type |
| DNA | deoxyribonucleic acid |
| DDH | DNA–DNA hybridization |
| 16S rRNA | 16 Svedberg units ribosomal-ribonucleic acid |
| API | analytical profile index |
| PCR | polymerase chain reaction |
| MALDI-TOF MS | Matrix-assisted laser desorption ionization time-of-flight mass spectrum |
| LAMP | loop-mediated isothermal amplification |
| ANI | average nucleotide identity |
| UT | unserotypable |
References
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcu suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen. Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M. Streptococcosis. In Diseases of Swine, 10th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Eds.; Wiley-Blackwell: Ames, TA, USA, 2012; pp. 841–855. [Google Scholar]
- Wertheim, H.F.; Nghia, H.D.; Taylor, W.; Schultsz, C. Streptococcus suis: An emerging human pathogen. Clin. Infect. Dis. 2009, 48, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.A.; Haesebrouck, F.; de Herdt, P.; Dom, P.; Ducatelle, R.; Desmidt, M.; Messier, S.; Higgins, R. Streptococcus suis infections in birds. Avian Pathol. 1994, 23, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Risco, D.; Fernández-Llario, P.; Cuesta, J.M.; García-Jiménez, W.L.; Gonçalves, P.; Martínez, R.; García, A.; Rosales, R.; Gómez, L.; de Mendoza, J.H. Fatal case of Streptococcus suis infection in a young wild boar (Sus. scrofa) from southwestern Spain. J. Zoo Wildl. Med. 2015, 46, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.D. Streptococcal infection in young pigs. I. An immunochemical study of the causative agent (PM streptococcus). J. Hyg. 1966, 64, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Windsor, R.S.; Elliott, S.D. Streptococcal infection in young pigs. IV. An outbreak of streptococcal meningitis in weaned pigs. J. Hyg. 1975, 75, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Perch, B.; Pedersen, K.B.; Henrichsen, J. Serology of capsulated streptococci pathogenic for pigs: Six new serotypes of Streptococcus suis. J. Clin. Microbiol. 1983, 17, 993–996. [Google Scholar] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Mittal, K.R.; Henrichsen, J. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 1989, 27, 2633–2636. [Google Scholar] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 1991, 29, 2590–2594. [Google Scholar] [PubMed]
- Higgins, R.; Gottschalk, M.; Boudreau, M.; Lebrun, A.; Henrichsen, J. Description of six new capsular types (29–34) of Streptococcus suis. J. Vet. Diagn. Investig. 1995, 7, 405–406. [Google Scholar] [CrossRef]
- Pan, Z.; Ma, J.; Dong, W.; Song, W.; Wang, K.; Lu, C.; Yao, H. Novel variant serotype of Streptococcus suis isolated from piglets with meningitis. Appl. Environ. Microbiol. 2015, 81, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, H.; Gottschalk, M.; Bai, X.; Lan, R.; Ji, S.; Liu, H.; Xu, J. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS ONE 2013, 8, e72070. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ji, S.; Liu, Z.; Lan, R.; Huang, Y.; Bai, X.; Gottschalk, M.; Xu, J. Eight novel capsular polysaccharide synthesis gene loci identified in nontypeable Streptococcus suis isolates. Appl. Environ. Microbiol. 2015, 81, 4111–4119. [Google Scholar] [CrossRef] [PubMed]
- King, S.J.; Leigh, J.A.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: Identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 2002, 40, 3671–3680. [Google Scholar] [CrossRef] [PubMed]
- PubMLST. Available online: http://pubmlst.org/ssuis/ (accessed on 29 April 2016).
- Hill, J.E.; Gottschalk, M.; Brousseau, R.; Harel, J.; Hemmingsen, S.M.; Goh, S.H. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 2005, 107, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Tien le, H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Baig, A.; Weinert, L.A.; Peters, S.E.; Howell, K.J.; Chaudhuri, R.R.; Wang, J.; Holden, M.T.; Parkhill, J.; Langford, P.R.; Rycroft, A.N.; et al. Whole genome investigation of a divergent clade of the pathogen Streptococcus suis. Front. Microbiol. 2015, 6, 1191. [Google Scholar] [CrossRef] [PubMed]
- Lancefield, R.C. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 1933, 57, 571–595. [Google Scholar] [CrossRef] [PubMed]
- De Moor, C.E. Septicaemic infections in pigs, caused by haemolytic streptococci of new Lancefield groups designated R, S and T. Antonie. van Leeuwenhoek 1963, 29, 272–280. [Google Scholar] [CrossRef]
- Skerman, V.B.D.; McGowan, V.; Sneath, P.H.A. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Kilpper-Balz, R.; Schleifer, K.H. Streptococcus suis sp. nov. nom. rev. Int. J. Syst. Bacteriol. 1987, 37, 160–162. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrand, E.; et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; Van de Peer, Y.; Vandamme, P.; Thompson, F.L.; et al. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R.; Mangwani, N.; Chakraborty, J.; Kumari, S. Understanding molecular identification and polyphasic taxonomic approaches for genetic relatedness and phylogenetic relationships of microorganisms. J. Microbiol. Methods 2014, 103, 80–100. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Glazunova, O.O.; Raoult, D.; Roux, V. Partial recN gene sequencing: A new tool for identification and phylogeny within the genus Streptococcus. Int. J. Syst. Evol. Microbiol. 2010, 60, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Glazunova, O.O.; Raoult, D.; Roux, V. Partial sequence comparison of the rpoB, sodA, groEL, and gyrB genes within the genus Streptococcus. Int. J. Syst. Evol. Microbiol. 2009, 59, 2317–2322. [Google Scholar] [CrossRef] [PubMed]
- Chatellier, S.; Harel, J.; Zhang, Y.; Gottschalk, M.; Higgins, R.; Devriese, L.A.; Brousseau, R. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int. J. Syst. Bacteriol. 1998, 48, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, R.; Hill, J.E.; Préfontaine, G.; Goh, S.H.; Harel, J.; Hemmingsen, S.M. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 2001, 67, 4828–4833. [Google Scholar] [CrossRef] [PubMed]
- Higgins, R.; Gottschalk, M. An update on Streptococcus suis identification. J. Vet. Diagn. Investig. 1990, 2, 249–252. [Google Scholar] [CrossRef]
- Hommez, J.; Devriese, L.A.; Henrichsen, J.; Castryck, F. Identification and characterization of Streptococcus suis. Vet. Microbiol. 1986, 11, 349–355. [Google Scholar] [CrossRef]
- Tarradas, C.; Arenas, A.; Maldonado, A.; Luque, I.; Miranda, A.; Perea, A. Identification of Streptococcus suis isolated from swine: Proposal for biochemical parameters. J. Clin. Microbiol. 1994, 32, 578–580. [Google Scholar] [PubMed]
- Tillotson, G.S. An evaluation of the API-20 STREP system. J. Clin. Pathol. 1982, 35, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Freney, J.; Bland, S.; Etienne, J.; Desmonceaux, M.; Boeufgras, J.M.; Fleurette, J. Description and evaluation of the semiautomated 4-hour rapid ID 32 Strep method for identification of streptococci and members of related genera. J. Clin. Microbiol. 1992, 30, 2657–2661. [Google Scholar] [PubMed]
- Okwumabua, O.; O’Connor, M.; Shull, E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 2003, 218, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Lacouture, S.; Bonifait, L.; Roy, D.; Fittipaldi, N.; Grenier, D. Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Québec, Canada. Vet. Microbiol. 2013, 162, 819–825. [Google Scholar] [PubMed]
- Tien le, H.T.; Sugiyama, N.; Duangsonk, K.; Tharavichitkul, P.; Osawa, R. Phenotypic and PCR-based identification of bacterial strains isolated from patients with suspected Streptococcus suis infection in northern Thailand. Jpn. J. Infect. Dis. 2012, 65, 171–174. [Google Scholar] [PubMed]
- Ishida, S.; Tien le, H.T.; Osawa, R.; Tohya, M.; Nomoto, R.; Kawamura, Y.; Takahashi, T.; Kikuchi, N.; Kikuchi, K.; Sekizaki, T. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J. Microbiol. Methods 2014, 107, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Okura, M.; Lachance, C.; Osaki, M.; Sekizaki, T.; Maruyama, F.; Nozawa, T.; Nakagawa, I.; Hamada, S.; Rossignol, C.; Gottschalk, M.; et al. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J. Clin. Microbiol. 2014, 52, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.E.; Veenbergen, V.; van der Velde, J.; Damman, M.; Wisselink, H.J.; Smits, M.A. The cps genes of Streptococcus suis serotypes 1, 2, and 9: Development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 1999, 37, 3146–3152. [Google Scholar] [PubMed]
- Smith, H.E.; van Bruijnsvoort, L.; Buijs, H.; Wisselink, H.J.; Smits, M.A. Rapid PCR test for Streptococcus suis serotype 7. FEMS Microbiol. Lett. 1999, 178, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Wisselink, H.J.; Joosten, J.J.; Smith, H.E. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J. Clin. Microbiol. 2002, 40, 2922–2929. [Google Scholar] [CrossRef] [PubMed]
- Marois, C.; Bougeard, S.; Gottschalk, M.; Kobisch, M. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J. Clin. Microbiol. 2004, 42, 3169–3175. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.M.; Baums, C.G.; Rehm, T.; Wisselink, H.J.; Goethe, R.; Valentin-Weigand, P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 2006, 115, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Fan, W.; Wisselink, H.; Lu, C. The cps locus of Streptococcus suis serotype 16: Development of a serotype-specific PCR assay. Vet. Microbiol. 2011, 153, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Nga, T.V.; Nghia, H.D.; Tu le, T.P.; Diep, T.S.; Mai, N.T.; Chau, T.T.; Sinh, D.X.; Phu, N.H.; Nga, T.T.; Chau, N.V.; et al. Real-time PCR for detection of Streptococcus suis serotype 2 in cerebrospinal fluid of human patients with meningitis. Diagn. Microbiol. Infect. Dis. 2011, 70, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Sun, X.; Lu, C. Development of rapid serotype-specific PCR assays for eight serotypes of Streptococcus suis. J. Clin. Microbiol. 2012, 50, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Akeda, Y.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Fifteen Streptococcus suis serotypes identified by multiplex PCR. J. Med. Microbiol. 2012, 61, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, J.; Ren, H.; Zhu, S.; Zhao, P.; Zhang, F.; Lv, H.; Hu, D.; Hao, L.; Geng, M.; et al. Rapid visual detection of highly pathogenic Streptococcus suis serotype 2 isolates by use of loop-mediated isothermal amplification. J. Clin. Microbiol. 2013, 51, 3250–3256. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Akeda, Y.; Hatrongjit, R.; Detchawna, U.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 2014, 63, 824–830. [Google Scholar] [PubMed]
- Bai, X.; Liu, Z.; Ji, S.; Gottschalk, M.; Zheng, H.; Xu, J. Simultaneous detection of 33 Streptococcus suis serotypes using the luminex xTAG® assay™. J. Microbiol. Methods 2015, 117, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, M.; Huber, I.; Konrad, R.; Busch, U. Application of MALDI-TOF MS for the Identification of Food Borne Bacteria. Open Microbiol. J. 2013, 7, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Cherkaoui, A.; Emonet, S.; Fernández, J.; Schorderet, D.; Schrenzel, J. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Beta-hemolytic streptococci. J. Clin. Microbiol. 2011, 49, 3004–3005. [Google Scholar] [CrossRef] [PubMed]
- Hinse, D.; Vollmer, T.; Erhard, M.; Welker, M.; Moore, E.R.; Kleesiek, K.; Dreier, J. Differentiation of species of the Streptococcus bovis/equinus-complex by MALDI-TOF mass spectrometry in comparison to sodA sequence analyses. Syst. Appl. Microbiol. 2011, 34, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Ikryannikova, L.N.; Filimonova, A.V.; Malakhova, M.V.; Savinova, T.; Filimonova, O.; Ilina, E.N.; Dubovickaya, V.A.; Sidorenko, S.V.; Govorun, V.M. Discrimination between Streptococcus pneumoniae and Streptococcus mitis based on sorting of their MALDI mass spectra. Clin. Microbiol. Infect. 2013, 19, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Werno, A.M.; Christner, M.; Anderson, T.P.; Murdoch, D.R. Differentiation of Streptococcus pneumoniae from non-pneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 2863–2867. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.; Beighton, D.; Klein, J.L. Identification of the ‘Streptococcus anginosus group’ by matrix-assisted laser desorption ionization—Time-of-flight mass spectrometry. J. Med. Microbiol. 2014, 63, 1143–1147. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sancho, M.; Vela, A.I.; García-Seco, T.; Gottschalk, M.; Domínguez, L.; Fernández-Garayzábal, J.F. Assessment of MALDI-TOF MS as alternative tool for Streptococcus suis identification. Front. Public Health 2015, 3, 202. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Tohya, M.; Yamada, R.; Osawa, R.; Nomoto, R.; Kawamura, Y.; Sekizaki, T. Development of loop-mediated isothermal amplification to detect Streptococcus suis and its application to retail pork meat in Japan. Int. J. Food. Microbiol. 2015, 208, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Rainey, F.A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014, 64, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, R.; Maruyama, F.; Ishida, S.; Tohya, M.; Sekizaki, T.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Okura, M.; Takamatsu, D.; Maruyama, F.; Nozawa, T.; Nakagawa, I.; Osaki, M.; Sekizaki, T.; Gottschalk, M.; Kumagai, Y.; Hamada, S. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcu suis: Potential mechanisms for generation of capsular variation. Appl. Environ. Microbiol. 2013, 79, 2796–2806. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, D.; Wongsawan, K.; Osaki, M.; Nishino, H.; Ishiji, T.; Tharavichitkul, P.; Khantawa, B.; Fongcom, A.; Takai, S.; Sekizaki, T. Streptococcus suis in humans, Thailand. Emerg. Infect. Dis. 2008, 14, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Puangpatra, P.; Sripakdee, S.; Chumla, K.; Boonkerd, N.; Polwichai, P.; Tanimura, S.; Takeuchi, D.; Nakayama, T.; et al. Genotypic profile of Streptococcus suis serotype 2 and clinical features of infection in humans, Thailand. Emerg. Infect. Dis. 2011, 17, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Schultsz, C.; Jansen, E.; Keijzers, W.; Rothkamp, A.; Duim, B.; Wagenaar, J.A.; van der Ende, A. Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in The Netherlands. PLoS ONE 2012, 7, e33854. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Sawanpanyalert, P.; Boonnark, A.; Noithachang, W.; Sriyakum, D.; Simkum, S.; Chokngam, S.; Gottschalk, M.; Akeda, Y.; et al. Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 2011, 378, 960. [Google Scholar] [CrossRef]
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Hamada, S.; Oishi, K.; Akeda, Y. Development of a multiplex PCR assay to detect the major clonal complexes of Streptococcus suis relevant to human infection. J. Med. Microbiol. 2016, 65, 392–396. [Google Scholar] [CrossRef] [PubMed]

| Method | Detecting Serotypes and Descriptions a | Year | Reference |
|---|---|---|---|
| PCR (3 assays) | Assay 1: Serotypes 1 and 14; | 1999 | [49] |
| Assay 2: Serotypes 2 and 1/2; | |||
| Assay 3: Serotype 9 | |||
| PCR | Serotype 7 | 1999 | [50] |
| Multiplex-PCR | Serotypes 1, 2, 1/2, 7, 9, and 14 | 2002 | [51] |
| epf (a virulence-associated marker of S. suis) is also detected | |||
| Multiplex-PCR | Serotypes 2 and 1/2 | 2004 | [52] |
| S. suis-specific sequence of the 16S rRNA gene is also detected | |||
| Multiplex-PCR | Serotypes 1, 2, 1/2, 7, 9, and 14 | 2006 | [53] |
| epf, sly, mrp, arcA (virulence-associated markers of S. suis), and S. suis-specific sequence of gdh are also detected | |||
| PCR | Serotype 16 | 2011 | [54] |
| Real-time PCR | Serotypes 2 and 1/2 | 2011 | [55] |
| PCR (8 assays) | Assay 1: Serotype 3; Assay 2: Serotype 4; | 2012 | [56] |
| Assay 3: Serotype 5; Assay 4: Serotype 8; | |||
| Assay 5: Serotype 10; Assay 6: Serotype 19; | |||
| Assay 7: Serotype 23; Assay 8: Serotype 25; | |||
| Multiplex-PCR (2 reaction sets) | 15 serotypes (serotypes 1−5, 7−10, 14, 16, 19, 23, 25, and 1/2) | 2012 | [57] |
| Reaction 1: Serotypes 1, 2, 1/2, 3, 4, 7, 9, 14, and 16 | |||
| Reaction 2: Serotypes 5, 8, 10, 19, 23, and 25 | |||
| In both reactions, the S. suis-specific sequence of the gdh gene is also detected | |||
| Multiplex-PCR (4 reaction sets) | 33 serotypes (serotypes 1−31, 33, and 1/2) and variant serotype 21/29 | 2013 | [15] |
| Reaction 1: Serotypes 1−10, 14, and 1/2 | |||
| Reaction 2: Serotypes 11−21 | |||
| Reaction 3: Serotypes 22−33 | |||
| Reaction 4: Serotype 21/29 | |||
| In all reactions, the S. suis-specific sequence of the thrA gene is also detected | |||
| LAMP | Serotypes 2 and 1/2 | 2013 | [58] |
| Multiplex-PCR (2-step assay) | 35 serotypes (serotypes 1−34 and 1/2) | 2014 | [48] |
| Step 1: classified into 7 groups (Group I−VII) | |||
| Group I: serotypes 3, 13, and 18 | |||
| Group II: serotypes 1, 2, 1/2, 6, 14, 16, and 27 | |||
| Group III: serotypes 21, 28, 29, and 30 | |||
| Group IV: serotypes 4, 5, 7, 17, 19, and 23 | |||
| Group V: serotypes 8, 15, 20, 22, and 25 | |||
| Group VI: serotypes 9, 10, 11, 12, 24, 26, and 33 | |||
| Group VII: serotypes 31, 32, and 34 | |||
| Step 2: classified into respective serotypes of each group | |||
| In all reactions, universally shared sequences of the 16S rRNA gene are also detected | |||
| Multiplex-PCR (4 reaction sets) | 29 serotypes (serotypes 1−19, 21, 23−25, 27−31, 33, and 1/2) | 2014 | [59] |
| Reaction 1: Serotypes 1, 2, 1/2, 3, 7, 9, 11, 14, and 16 | |||
| Reaction 2: Serotypes 4, 5, 8, 12, 18, 19, 24, and 25 | |||
| Reaction 3: Serotypes 6, 10, 13, 15, 17, 23, and 31 | |||
| Reaction 4: Serotypes 21, 27, 28, 29, and 30 | |||
| In all reactions, the S. suis-specific sequence of the gdh gene is also detected | |||
| PCR | Serotype Chz | 2015 | [14] |
| luminex xTAG® assay™ | 33 serotypes (serotypes 1−31, 33, and 1/2) | 2015 | [60] |
| Strains | Serotype a | Source | Descriptions b | Reference |
|---|---|---|---|---|
| EA1172.91 | 32 | Diseased pig (septicemia) | Serotype reference strains
Considered to be S. orisratti by cpn60 analysis | [19] |
| 92-2742 | 34 | Diseased pig (aborted fetus) | ||
| EA1832.92 | 33 | Diseased lamb (arthritis) | Serotype reference strain
Shown to be a non-S. suis strain by DDH | [20] |
| 86-5192 | 20 | Diseased pig (unknown) | Serotype reference strains
Shown to be non-S. suis strains and the same species by DDH and ANI Reclassified as S. parasuis [73] or considered to be divergent S. suis strains (Classified into Clade 3 on the basis of the whole-genome-based phylogeny [21]) | [20,21,73] |
| 88-1861 | 22 | Diseased calf (unknown) | ||
| 89-4109-1 | 26 | Diseased pig (unknown) | ||
| SUT-286 | 20 | Healthy pigs (saliva) | S. parasuis strains (SUT-286 is the type strain) Shown to be the same species by ANI | [73] |
| SUT-380 | 22 | |||
| SUT-319, 328 | 20/22 | |||
| SUT-7 | 22/26 | |||
| LSS7 | UT | Healthy pigs | Divergent
S. suis strain Classified into Clade 1 by the whole genome-based phylogeny | [21] |
| SS007 | 4 | Diseased pig (systemic-brain infection) | Divergent
S. suis strains cps losus of LSS6 was similar to cps4 locus Classified into Clade 2 by the whole genome-based phylogeny | [21] |
| LSS19 | 4 | Healthy pig | ||
| LSS6 | UT | Healthy pig | ||
| SS1003 | 22 | Diseased pig (respiratory infection) | ||
| LSS17 | UT | Healthy pig | Divergent
S. suis strain Classified into Clade 3 by the whole genome-based phylogeny | [21] |
| SUT-283 | 20 | Healthy pig | recN PCR negative but gdh PCR positive strains | [47] |
| FUT-29 | 20 | Pork | ||
| GUT-182 | 22 | Diseased pig (endocarditis) | ||
| GUT-183−193 (11 strains) | 33 | Diseased calves (endocarditis) | ||
| More than 70 isolates | 33 and UT | Diseased cattle, sheep, and a goat (endocarditis, arthritis, and pneumonia)
Healthy cattle (tonsil and nasal cavity) | recN PCR negative but gdh PCR positive strains Twenty of them analyzed by whole genome sequencing were shown to be the same species with serotype 33 reference strain by ANI | Unpublished |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current Taxonomical Situation of Streptococcus suis. Pathogens 2016, 5, 45. https://doi.org/10.3390/pathogens5030045
Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, Takamatsu D. Current Taxonomical Situation of Streptococcus suis. Pathogens. 2016; 5(3):45. https://doi.org/10.3390/pathogens5030045
Chicago/Turabian StyleOkura, Masatoshi, Makoto Osaki, Ryohei Nomoto, Sakura Arai, Ro Osawa, Tsutomu Sekizaki, and Daisuke Takamatsu. 2016. "Current Taxonomical Situation of Streptococcus suis" Pathogens 5, no. 3: 45. https://doi.org/10.3390/pathogens5030045