Published online Jun 7, 2009. doi: 10.3748/wjg.15.2595
Revised: April 20, 2009
Accepted: April 27, 2009
Published online: June 7, 2009
AIM: To investigate the promoter methylation status and mRNA expression of DKK-3 and WIF-1 gene in hepatocellular carcinoma (HCC).
METHODS: DKK-3 and WIF-1 acted as Wnt-antagonists and tumor suppressors, but hypermethylation of the gene promoter and low mRNA expression activated Wnt signaling aberrantly and induced the development of HCC. Methylation status of the DKK-3 and WIF-1 gene promoter was investigated using methylation specific polymerase chain reaction (PCR) in tumor and adjacent non-cancerous tissues from 33 HCC patients and 20 normal liver tissues served as control. The expression of DKK-3 and WIF-1 mRNA was also determined by real-time quantitative reverse transcriptase PCR. The relationship between methylation, mRNA expression, and clinical data, as well as methylation and mRNA expression of the two genes were analyzed.
RESULTS: The methylation of DKK-3 and WIF-1 genes in HCC increased significantly compared with adjacent non-cancerous tissues and normal control tissues (χ2 =7.79, P < 0.05; χ2 = 4.89, P < 0.05), and no significant difference in methylation between adjacent non-cancerous tissues and normal control tissues was observed. In HCC tissues, significant differences in the DKK-3 promoter methylation were observed in age and cirrhosis, and significant differences of the WIF-1 promoter methylation were observed in HBsAg and cirrhosis. The average expression of DKK-3 mRNA in HCC and adjacent non-cancerous tissues was increased significantly compared with normal control tissues. The average expression of WIF-1 mRNA showed no significant difference among the three tissues. The mRNA expression of DKK-3 gene in HCC was decreased as the pathological grade increased.
CONCLUSION: The aberrant promoter methylation and decreased expression of DKK-3 and WIF-1 may be an important mechanism in HCC, and may be a far-reaching significance in early diagnosis and therapy of HCC.
-
Citation: Ding Z, Qian YB, Zhu LX, Xiong QR. Promoter methylation and mRNA expression of
DKK-3 andWIF-1 in hepatocellular carcinoma. World J Gastroenterol 2009; 15(21): 2595-2601 - URL: https://www.wjgnet.com/1007-9327/full/v15/i21/2595.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.2595
Hepatocellular carcinoma (HCC) is one of the most fatal human malignancies and the third most frequent cause of tumor-related death[1]. However, the molecular mechanisms of hepatocarcinogenesis remain unclear. At present, α-fetoprotein (AFP) is used as a tumor marker in the screening of HCC, but it is only sensitive to advanced liver cancer or as a marker of the recurrence after hepatectomy. Therefore, identifying a tumor marker for HCC is important for early diagnosis and therapy.
The Wnt/β-catenin protein signal transduction pathway participates in early embryonic development. DKK-3 and WIF-1 are two key genes in the Wnt signal transduction pathway, and the subnormal function of DKK-3 and WIF-1 can contribute to the activation of the Wnt pathway and result in carcinogenesis through dysregulation of cell proliferation and differentiation. CpG islands are short fragments of DNA that contain a higher CG frequency than other regions. The hypermethylation of CpG islands is associated with transcriptional repression of these genes. Aberrant promoter hypermethylation of genes occurs frequently during the pathogenesis of human cancers, and has been found to be a primary mechanism in the down-regulation of these genes. Methylation-specific PCR (MSP) and real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) can detect these epigenetic changes and can be used for cancer detection.
In the present study, MSP and real time quantitative RT-PCR were used to investigate the methylation status and the expression of DKK-3, and WIF-1 in patients with HCC, to explore the potential carcinogenesis of HCC and to investigate its early diagnostic and therapeutic potential.
Thirty-three samples of HCC and adjacent non-cancerous tissues (> 2 cm away from tumor) were obtained from the Department of Surgery, First Affiliated Hospital of Anhui Medical University between July 2006 and July 2007. These samples were documented through a pathology laboratory database. The age of the 33 patients (27 male, 6 female) ranged between 18 and 67 years, with a median age of 51 years. Ten patients were Edmonson stage I, 18 patients were stage II, and five patients were stage III. The diameter of the tumor was smaller than 3 cm in five patients, and ≥ 5 cm in 28 patients. Portal vein tumor thrombus was found in three patients. As a control, 20 samples of normal liver tissues were collected from the resection of hemangiomas between July 2006 and July 2007. The samples were snap-frozen in liquid nitrogen and stored at -80°C until the extraction of DNA and RNA.
Genomic DNA and total RNA were extracted from HCC tissues, adjacent non-cancerous and normal control tissues using the Qiagen kit (Qiagen). The concentration of DNA and RNA were determined with a spectrophotometer and their integrity was assessed by gel electrophoresis.
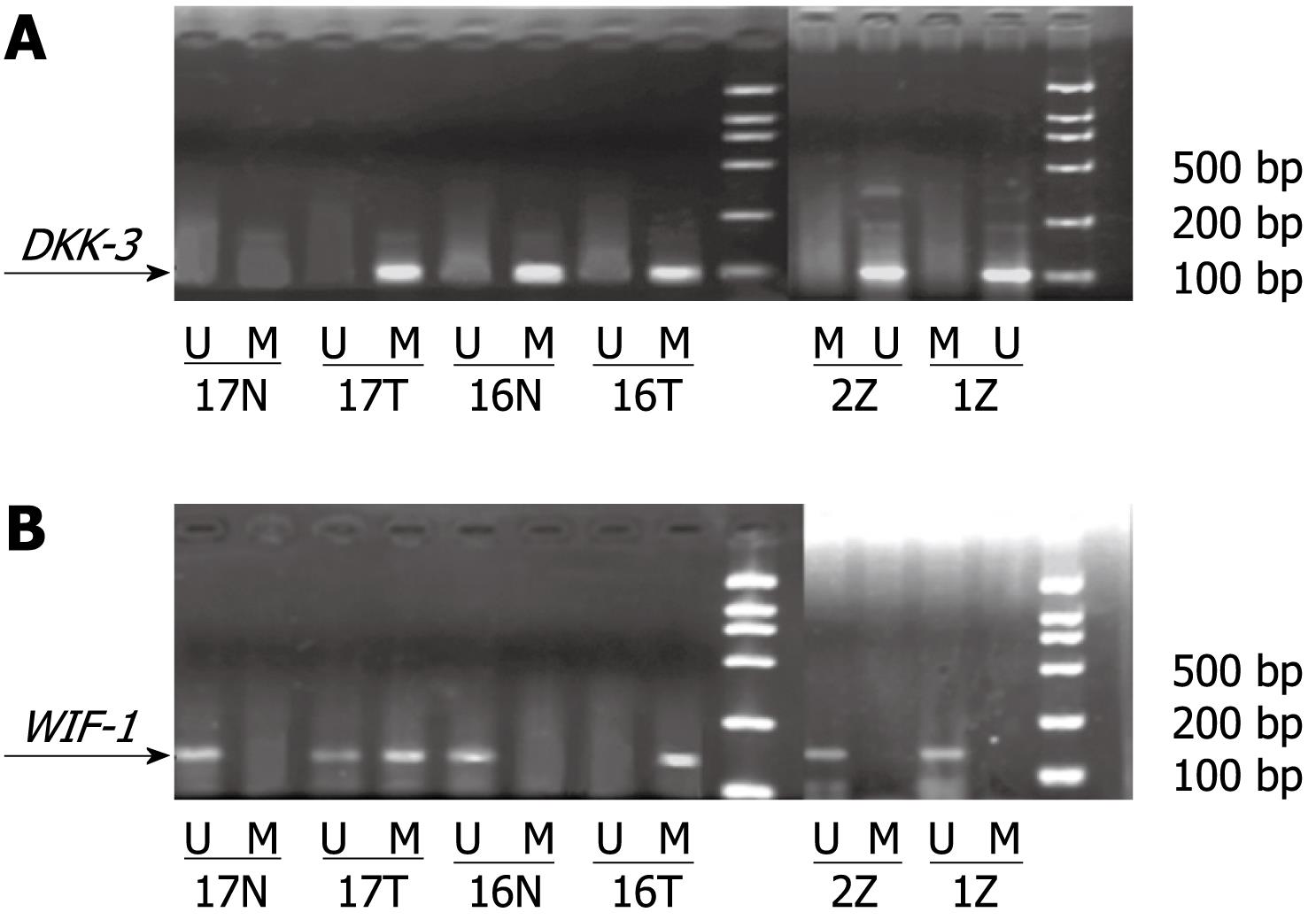
We investigated whether the promoter regions of the two genes were methylated. Genomic DNA was modified with sodium bisulfite using the CpGenome DNA Modification kit (Intergen) according to the specifications of the manufacturer. MSP was performed in a total volume of 25 &mgr;L, containing 2 &mgr;L modified template DNA, AmpliTaq Gold (Roche 5 U/&mgr;L) 0.2 &mgr;L, 10 × buffer 2.5 &mgr;L, dNTP (10 &mgr;mol/L) 1 &mgr;L, each primer (10 &mgr;mol/L) 0.5 &mgr;L, Mg2+ (25 &mgr;mol/L) 2 &mgr;L, and RNase free water 16.3 &mgr;L. MSP reactions were subjected to an initial incubation at 95°C for 10 min, followed by 35 cycles of 95°C for 45 s, and annealing at the 52-57°C for 45 s and 72°C for 45 s. Final extension was completed by incubation at 72°C for 5 min. Primer sequences of MSP and unmethylation-specific PCR (USP) are shown in Table 1[2]. MSP products were separated on 2% agarose gels and visualized after ethidium bromide staining. The product bands of MSP and USP were calculated using a TANON GIS gel image analysis system, and the relative methylation level was determined by MSP ratio = MSP band density/(MSP band density + USP band density).
| Gene | Primer purpose | Primer sequence (5'--3') | Annealing temperature (°C) | Product size (bp) |
| DKK-3 | MSP-M | F: GGGGCGGGCGGCGGGGC | 58 | 120 |
| R: ACATCTCCGCTCTACGCCCG | ||||
| MSP-U | F: TTAGGGGTGGGTGGTGGGGT | 56 | 126 | |
| R: CTACATCTCCACTCTACACCCA | ||||
| Real time RT-PCR | F: GTAAGTTTCCCCTCTGGCTTG | 60 | 90 | |
| R: AAGCACCAGACTGTGAAGCCT | ||||
| Probe: FAM+AGGTGTTGTGCATTTGTTCAGCTCCC+TAMRA | ||||
| WIF-1 | MSP-M | F: CGTTTTATTGGGCGTATCGT | 57 | 145 |
| R: ACTAACGCGAACGAAATACGA | ||||
| MSP-U | F: GGGTGTTTTATTGGGTGTATTGT | 52 | 154 | |
| R: AAAAAAACTAACACAAACAAAATACAAAC | ||||
| Real time RT-PCR | F: TCCAAACACCTCAAAATGCTATC | 60 | 119 | |
| R: GAACCCATCAGGACACTCGC | ||||
| Probe: FAM+ACAAGCTGAGTGCCCAGGCGG+TAMRA | ||||
| GAPDH | Real time RT-PCR | F: CCACTCCTCCACCTTTGAC | 60 | 102 |
| R: ACCCTGTTGCTGTAGCCA | ||||
| Probe: FAM+TTGCCCTCAACGACCACTTTGTC+TAMRA |
Total RNA (1 &mgr;g) was reverse-transcribed with RevertAid First Strand cDNA Synthesis Kit (Fermentas) in a final volume of 20 &mgr;L. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The cDNAs of interest were quantified by real-time quantitative PCR using the Rotor-Gene 3000. Samples were assayed in a 25 &mgr;L reaction mixture containing 1 &mgr;L of cDNA, 12.5 &mgr;L of 2 × QuantiTect probe PCR Master Mix (Qiagen), 1 &mgr;L of each specific primer, 0.5 &mgr;L of fluorogenic probe and 9 &mgr;L of RNase free water. The sequence of the primer and probe is shown in Table 1. Real-time quantitative RT-PCR reactions were subjected to initial incubation at 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, and 60°C for 60 s.
Statistical analyses were performed using SPSS software version 13.0. χ2 test, t test, non-parameter Spearman test and Fisher’s exact test were used to compare the relationship of methylation and clinical data. One-year disease-free survival was analyzed through the log rank test. The level for a statistically significant difference was set at P < 0.05 for all the tests.
Methylation status of DKK-3 and WIF-1 in the three tissues: The methylation status of DKK-3 and WIF-1 in the three kinds of tissues is shown in Table 2 and Figure 1. In HCC and adjacent non-cancerous tissues, the methylation rate of DKK-3 was 37.61% ± 4.26% and 15.96% ± 3.91%, respectively. The methylation status of WIF-1 was 29.70% ± 3.24% in HCC and 18.34% ± 4.02% in adjacent non-cancerous tissues. The methylation rate of DKK-3 and WIF-1 in HCC was higher than that in adjacent non-cancerous tissues (P < 0.05). The methylation of DKK-3 and WIF-1 was not observed in normal control tissues. There was no linear correlation between methylation of DKK-3 and WIF-1 (r = 0.296, P > 0.05).
| Methylation level | DKK-3 | WIF-1 |
| HCC tissues | 37.61 ± 4.26 | 29.70 ± 3.24 |
| Adjacent non-cancerous tissues | 15.96 ± 3.91 | 18.34 ± 4.02 |
| P < 0.05 | P < 0.05 |
Relationship of methylation and clinical data: The relationship between DKK-3 and WIF-1 methylation and clinical data such as sex, age, Child-Pugh score, AFP, HBsAg, tumor number, cirrhosis, pseudo-capsule and pathology class is shown in Tables 3 and 4. The methylation rate of the DKK-3 gene was higher in older (≥ 60) than younger (< 60) patients, higher in non-cirrhosis than that in cirrhosis patients (P < 0.05), and higher in HBsAg (-) than in HBsAg (+) patients. There was no relationship and no associations observed between the methylation status of DKK-3 and WIF-1 in adjacent non-cancerous tissues and clinical data (P < 0.05).
| Clinical data | Methylation in HCC | Methylation in adjacent non-cancerous tissues | ||||
| Negative | Positive | P | Negative | Positive | P | |
| Sex | ||||||
| Male | 12 | 15 | 1.000 | 21 | 6 | 1.000 |
| Female | 3 | 3 | 5 | 1 | ||
| Age (yr) | ||||||
| < 60 | 13 | 7 | 0.011 | 17 | 3 | 0.393 |
| ≥ 60 | 2 | 11 | 9 | 4 | ||
| Child | ||||||
| A | 12 | 17 | 0.308 | 23 | 6 | 1.000 |
| B | 3 | 1 | 3 | 1 | ||
| AFP (ng/mL) | ||||||
| < 20 | 7 | 10 | 0.308 | 15 | 3 | 0.674 |
| ≥ 20 | 8 | 8 | 11 | 4 | ||
| Number of tumor | ||||||
| Single | 11 | 13 | 1.000 | 20 | 4 | 0.358 |
| Multiple | 4 | 5 | 6 | 3 | ||
| Tumor diameter (cm) | ||||||
| < 3 | 2 | 3 | 1.000 | 3 | 2 | 1.000 |
| ≥ 3 | 13 | 15 | 23 | 5 | ||
| HBsAg | ||||||
| (+) | 15 | 14 | 0.108 | 23 | 6 | 1.000 |
| (-) | 0 | 4 | 3 | 1 | ||
| Hepatocirrhosis | ||||||
| (+) | 15 | 10 | 0.004 | 20 | 5 | 1.000 |
| (-) | 0 | 8 | 6 | 2 | ||
| False capsule | ||||||
| (+) | 14 | 14 | 0.346 | 23 | 5 | 0.282 |
| (-) | 1 | 4 | 3 | 2 | ||
| Tumor thrombus | ||||||
| (+) | 1 | 2 | 1.000 | 3 | 0 | 1.000 |
| (-) | 14 | 16 | 23 | 7 | ||
| Edmonson stage | ||||||
| I/II | 13 | 15 | 1.000 | 22 | 6 | 1.000 |
| III | 2 | 3 | 4 | 1 | ||
| Clinical data | Methylation in HCC | Methylation in adjacent non-cancerous tissues | ||||
| Negative | Positive | P | Negative | Positive | P | |
| Sex | ||||||
| Male | 17 | 10 | 0.659 | 23 | 4 | 1.000 |
| Female | 3 | 3 | 5 | 1 | ||
| Age (yr) | ||||||
| < 60 | 7 | 5 | 1.000 | 17 | 3 | 1.000 |
| ≥ 60 | 13 | 8 | 11 | 2 | ||
| Child | ||||||
| A | 17 | 12 | 1.000 | 24 | 5 | 1.000 |
| B | 3 | 1 | 4 | 0 | ||
| AFP (ng/mL) | ||||||
| < 20 | 11 | 8 | 1.000 | 16 | 3 | 1.000 |
| ≥ 20 | 9 | 5 | 12 | 2 | ||
| Number of tumor | ||||||
| Single | 14 | 10 | 1.000 | 21 | 3 | 0.597 |
| Multiple | 6 | 3 | 7 | 2 | ||
| Tumor diameter (cm) | ||||||
| < 3 | 2 | 3 | 1.000 | 5 | 0 | 0.569 |
| ≥ 3 | 18 | 10 | 23 | 5 | ||
| HBsAg | ||||||
| (+) | 20 | 9 | 0.017 | 24 | 5 | 1.000 |
| (-) | 0 | 4 | 4 | 0 | ||
| Hepatocirrhosis | ||||||
| (+) | 18 | 7 | 0.035 | 21 | 4 | 1.000 |
| (-) | 2 | 6 | 7 | 1 | ||
| False capsule | ||||||
| (+) | 18 | 10 | 0.360 | 24 | 4 | 1.000 |
| (-) | 2 | 3 | 4 | 1 | ||
| Tumor thrombus | ||||||
| (+) | 1 | 2 | 0.547 | 2 | 1 | 0.400 |
| (-) | 19 | 11 | 26 | 4 | ||
| Edmonson stage | ||||||
| I/II | 16 | 12 | 0.625 | 23 | 5 | 0.569 |
| III | 4 | 1 | 5 | 0 | ||
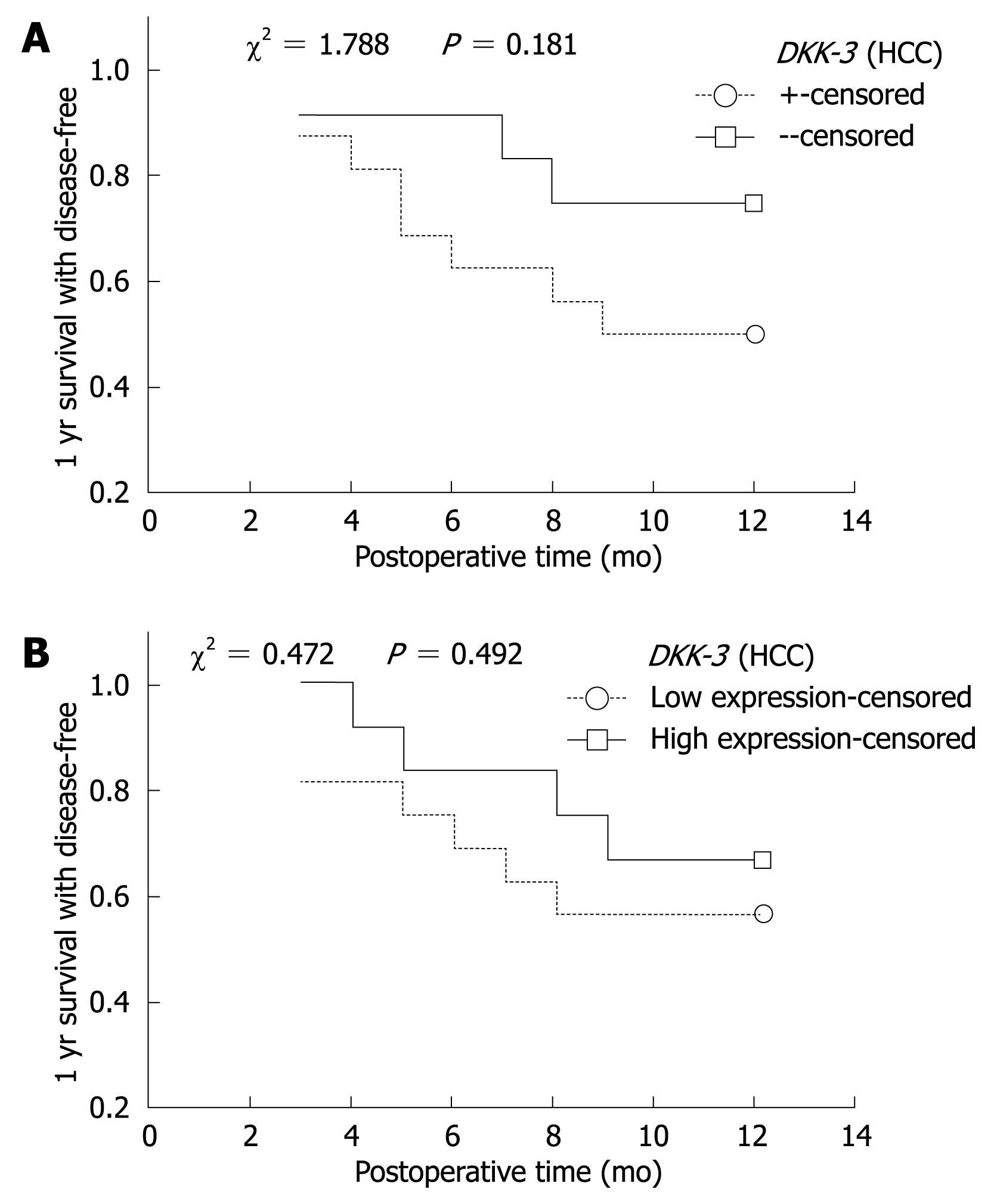
Relationship of methylation and 1-year disease-free survival: Complete follow-up data was obtained from 27 (27/33) cases of HCC, and the follow-up time was > 1 year. Among the 27 cases, 11 cases showed disease recurrence or died. Kaplan-Meier method and Log-rank test were used to calculate the survival rate and the significance of difference. One-year disease-free survival decreased as time passed, but there was no significant difference in the DKK-3 or WIF-1 between HCC and adjacent non-cancerous tissues (Figure 2).
mRNA expression of DKK-3 and WIF-1: The DKK-3 mRNA expression in HCC, adjacent non-cancerous tissues and normal control tissues was 0.773 ± 0.319, 0.833 ± 0.316 and 1.012 ± 0.125, respectively. The expression of DKK-3 mRNA in HCC and adjacent non-cancerous tissues was significantly lower than that in normal control tissues (P < 0.05). There was no significant difference between HCC and adjacent non-cancerous tissues. WIF-1 mRNA expression in the three tissues was 0.853 ± 0.510, 0.820 ± 0.316 and 0.995 ± 0.148, without significant difference among them. Pearson’s product-moment correlation analysis showed that there was no correlation between DKK-3 and WIF-1 in HCC tissues (r = 0.472, P = 0.127).
Relationship between DKK-3 and WIF-1 mRNA expression and clinical data: The relationship between DKK-3 and WIF-1 mRNA expression and clinical data are shown in Tables 5 and 6. DKK-3 mRNA expression in HCC was decreased as the pathological grade increased (P < 0.05), but no significant correlation was found with other clinical data. The expression of DKK-3 mRNA in adjacent non-cancerous tissues, and the expression of WIF-1 mRNA in HCC and adjacent non-cancerous tissues showed no significant correlation with clinical data (P > 0.05). Pearson’s product-moment correlation analysis showed that there was no correlation between DKK-3 and WIF-1 in HCC, adjacent non-cancerous tissues and normal control tissues
| Clinical data | Case | The mRNA expression of DKK-3 in HCC | t | P | The mRNA expression of DKK-3 in adjacent non-cancerous tissues | t | P |
| Sex | |||||||
| Male | 27 | 0.776 ± 0.346 | 0.153 | 0.880 | 0.779 ± 0.319 | 0.888 | 0.404 |
| Female | 6 | 0.761 ± 0.168 | 0.913 ± 0.338 | ||||
| Age (yr) | |||||||
| < 60 | 20 | 0.756 ± 0.356 | 0.334 | 0.741 | 0.819 ± 0.341 | 0.195 | 0.847 |
| ≥ 60 | 13 | 0.795 ± 0.285 | 0.840 ± 0.274 | ||||
| Child | |||||||
| A | 29 | 0.761 ± 0.331 | 0.789 | 0.466 | 0.829 ± 0.329 | 0.171 | 0.869 |
| B | 4 | 0.863 ± 0.225 | 0.813 ± 0.155 | ||||
| AFP (ng/mL) | |||||||
| < 20 | 17 | 0.716 ± 0.287 | 1.069 | 0.294 | 0.840 ± 0.354 | 0.124 | 0.902 |
| ≥ 20 | 16 | 0.835 ± 0.384 | 0.826 ± 0.281 | ||||
| Number of tumor | |||||||
| Single | 24 | 0.740 ± 0.315 | 0.971 | 0.348 | 0.823 ± 0.298 | 0.262 | 0.797 |
| Multiple | 9 | 0.860 ± 0.329 | 0.860 ± 0.377 | ||||
| Tumor diameter (cm) | |||||||
| < 3 | 4 | 0.908 ± 0.300 | 0.943 | 0.399 | 0.792 ± 0.352 | 0.290 | 0.780 |
| ≥ 3 | 29 | 0.755 ± 0.322 | 0.841 ± 0.315 | ||||
| HBsAg | |||||||
| (+) | 29 | 0.660 ± 0.233 | 0.984 | 0.372 | 0.817 ± 0.304 | 0.639 | 0.563 |
| (-) | 4 | 0.789 ± 0.329 | 0.955 ± 0.418 | ||||
| Hepatocirrhosis | |||||||
| (+) | 25 | 0.780 ± 0.319 | 0.046 | 0.964 | 0.761 ± 0.268 | 2.157 | 0.057 |
| (-) | 8 | 0.774 ± 0.365 | 1.060 ± 0.361 | ||||
| False capsule | |||||||
| (+) | 28 | 0.757 ± 0.326 | 1.750 | 0.132 | 0.819 ± 0.307 | 0.790 | 0.466 |
| (-) | 5 | 1.014 ± 0.299 | 0.966 ± 0.395 | ||||
| Tumor thrombus | |||||||
| (+) | 3 | 0.770 ± 0.330 | 0.738 | 0.519 | 0.811 ± 0.307 | 1.088 | 0.379 |
| (-) | 30 | 0.890 ± 0.260 | 1.057 ± 0.379 | ||||
| Edmonson stage | |||||||
| I/II | 28 | 0.807 ± 0.335 | 3.380 | 0.020 | 0.820 ± 0.317 | 0.564 | 0.596 |
| III | 5 | 0.586 ± 0.350 | 0.910 ± 0.333 |
| Clinical data | Case | The mRNA expression of WIF-1 in HCC | t | P | The mRNA expression of WIF-1 in adjacent non-cancerous tissues | t | P |
| Sex | |||||||
| Male | 27 | 0.882 ± 0.542 | 0.942 | 0.365 | 0.828 ± 0.320 | 0.286 | 0.783 |
| Female | 6 | 0.720 ± 0.336 | 0.787 ± 0.318 | ||||
| Age (yr) | |||||||
| < 60 | 12 | 0.767 ± 0.397 | 1.092 | 0.289 | 0.793 ± 0.247 | 0.551 | 0.589 |
| ≥ 60 | 21 | 0.985 ± 0.643 | 0.862 ± 0.407 | ||||
| Child | |||||||
| A | 29 | 0.854 ± 0.543 | 0.067 | 0.948 | 0.793 ± 0.309 | 1.243 | 0.286 |
| B | 4 | 0.845 ± 0.171 | 1.015 ± 0.337 | ||||
| AFP (ng/mL) | |||||||
| < 20 | 19 | 0.873 ± 0.516 | 0.231 | 0.819 | 0.828 ± 0.377 | 0.148 | 0.883 |
| ≥ 20 | 14 | 0.831 ± 0.519 | 0.812 ± 0.247 | ||||
| Number of tumor | |||||||
| Single | 24 | 0.927 ± 0.532 | 1.292 | 0.212 | 0.801 ± 0.288 | 0.496 | 0.630 |
| Multiple | 9 | 0.704 ± 0.408 | 0.872 ± 0.393 | ||||
| Tumor diameter (cm) | |||||||
| < 3 | 5 | 1.002 ± 0.750 | 0.507 | 0.636 | 0.796 ± 0.415 | 0.147 | 0.889 |
| ≥ 3 | 28 | 0.826 ± 0.469 | 0.825 ± 0.304 | ||||
| HBsAg | |||||||
| (+) | 29 | 0.875 ± 0.527 | 0.155 | 0.886 | 0.838 ± 0.312 | 0.754 | 0.497 |
| (-) | 4 | 0.820 ± 0.681 | 0.695 ± 0.360 | ||||
| Hepatocirrhosis | |||||||
| (+) | 25 | 0.840 ± 0.523 | 0.260 | 0.797 | 0.810 ± 0.322 | 0.331 | 0.746 |
| (-) | 8 | 0.893 ± 0.486 | 0.853 ± 0.314 | ||||
| False capsule | |||||||
| (+) | 28 | 0.860 ± 0.490 | 0.152 | 0.886 | 0.816 ± 0.296 | 0.144 | 0.892 |
| (-) | 5 | 0.812 ± 0.676 | 0.846 ± 0.454 | ||||
| Tumor thrombus | |||||||
| (+) | 3 | 0.815 ± 0.505 | 1.392 | 0.276 | 0.806 ± 0.319 | 0.840 | 0.474 |
| (-) | 30 | 1.223 ± 0.481 | 0.960 ± 0.300 | ||||
| Edmonson stage | |||||||
| I/II | 28 | 0.910 ± 0.517 | 1.005 | 0.083 | 0.821 ± 0.308 | 0.141 | 0.894 |
| III | 5 | 0.532 ± 0.360 | 0.848 ± 0.401 |
(P > 0.05).
Relationship of mRNA expression and 1-year survival: The expression of DKK-3 and WIF-1 gene mRNA in HCC and adjacent non-cancerous tissues was divided into two groups: higher expression group (> average expression) and lower expression group (≤ average expression). Kaplan-Meier curve showed that the disease-free survival was decreased. The survival rate of the lower expression group was lower than that of the higher expression group, but without significant difference (P > 0.05, Figure 2).
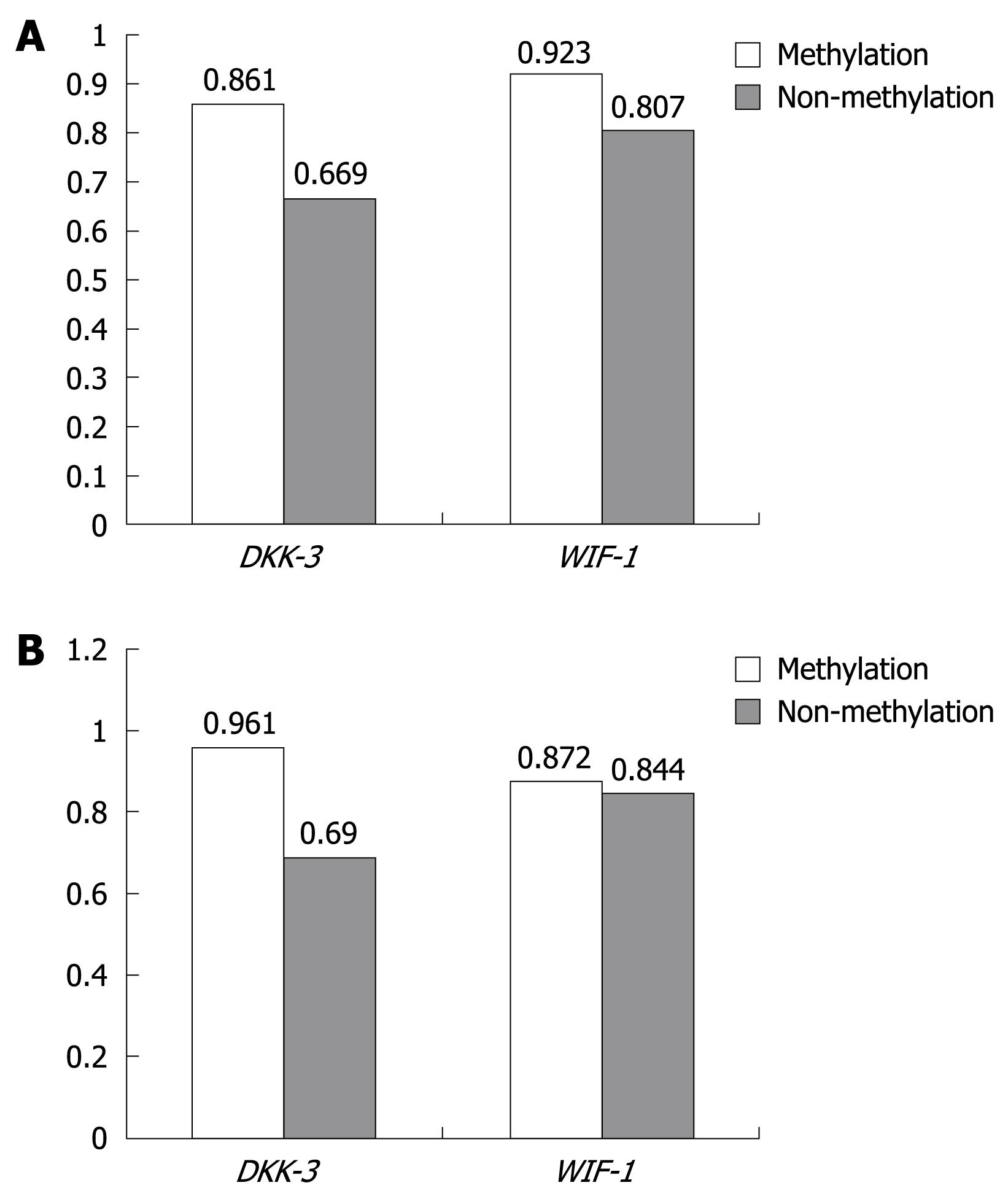
Relationship between methylation and mRNA expression: The relationship between mRNA expression and methylation status in HCC and adjacent non-cancerous tissues is shown in Figure 3, and the relationship of DKK-3 and WIF-1 mRNA expression from HCC and adjacent non-cancerous tissues was not found between methylation and non-methylation (P > 0.05).
The Wnt signal transduction pathway plays a very important role in embryonic development, and abnormalities may lead to developmental defects and cellular malignant transformation[3]. It has been shown that disturbances of the Wnt signal transduction pathway were significantly related to human neoplastic transformation. Overexpression of Wnt genes has been reported in many cancers, including breast cancer, esophagus cancer, colorectal cancer, malignant melanoma, leukemia, prostate cancer, endometrial carcinoma, HCC, thyroid cancer, and pancreatic cancer[45]. Gene silencing related with the hypermethylation of its promoter is an important epigenetic mechanism, and an important mechanism to regulate the expression of genes in the body.
The DKK-3 gene locus maps to 11p15.1 of the human chromosome, which encodes a Wnt complex receptor antagonist. Wnt will combine with Frizzle receptor and activate Wnt signaling if the DKK-3 protein is inactivated. Promoter-hypermethylation and reduced expression of the DKK-3 gene were found in bladder cancer, lung cancer cell lines and tissues[26]. In our experiment, the methylation status of DKK-3 in HCC was significantly higher than that in adjacent non-cancerous tissues and normal control tissues, and the expression of DKK-3 mRNA in HCC and adjacent non-cancerous tissues was significantly lower than that in normal control tissues. These results show that the methylation of the DKK-3 gene promoter plays an important role in the carcinogenesis of HCC. It has been reported that hypermethylation was frequently present in elderly people[78], and HCC was reported to be closely linked to hepatitis B (HBV) infection. The results of previous reports are consistent. Our results show that hypermethylation was more frequent in elderly people (age ≥ 60 years) and non-cirrhotic HCC tissues, which suggests that the silence of the DKK-3 gene resulted from the hypermethylation of its promoter. The expression of DKK-3 mRNA in Edmonson stage III was significantly lower than that in stage I/II, suggesting that the expression of DKK-3 is negatively related to the stage of tumor and cell proliferation. Therefore, the decreased expression of DKK-3 mRNA may affect the invasion and intrahepatic metastasis of HCC. In addition, the expression of DKK-3 was the lowest in late G1 of the cell cycle[9].
The WIF-1 gene locus maps to 12q14.1 of the human chromosome, and encodes a conservative secreted protein that inhibits the activity of Wnt by holding back the regular or non-regular Wnt signal transduction pathway. Promoter-hypermethylation and reduced expression of the WIF-1 gene was found in lung cancer cell lines and tissues, malignant pleural mesothelioma cell lines and tissues, and nasopharyngeal cancer cell lines[10–12]. Our study showed that the methylation status of WIF-1 in HCC was higher than that in adjacent non-cancerous tissues and normal control tissues, and was higher in the HBsAg (-) and non-cirrhosis patients, which implies that the WIF-1 gene silence related to promoter hypermethylation may represent the pathogenesis with HBV infection that results in HCC development. WIF-1 mRNA expression in HCC and non-cancerous tissues was not significantly lower than that of normal control tissues, which may be attributed to gene silence of multiple factors. WIF-1 mRNA expression has no relationship with clinical data, suggesting that HCC development is a complex polygene and multipathway process[13].
In summary, Wnt signaling is a major cell signal transduction pathway, and plays an important role in the proliferation and differentiation of cells. Wnt-antagonists function as tumor suppressors and contribute to the pathogenesis of several human malignancies. In the normal state, DKK-3 and WIF-1 can act as negative regulators of Wnt signaling, however, hypermethylation of the gene promoter and low expression of mRNA will activate Wnt signaling aberrantly, and induce the development of HCC. At the same time, hypermethylation of the DKK-3 gene in the elderly and non-cirrhotic HCC, and hypermethylation of the WIF-1 gene in HbsAg (-) and non-cirrhotic HCC suggests that there may be two independent mechanisms in the carcinogenesis of HCC. One is HBV infection, which induces HCC development, and the other is the aberrant expression of Wnt-antagonist genes, such as DKK-3 and WIF-1, which induces HCC development. A synergistic action of these factors has not been identified, and further study is needed. Our results may provide a reliable way to improve the early diagnosis of HCC and new therapies by blocking this pathway in the treatment of HCC.
The Wnt signal transduction pathway is significantly related to human neoplastic transformation. The Wnt-antagonist genes function as tumor suppressors and contribute to the pathogenesis of several human malignancies. Recently, promoter CpG hypermethylation and gene silencing in DKK-3 and WIF-1 genes have been identified in several human malignancies. The DKK-3 gene locus maps to 11p15.1 of the human chromosome, which encodes a Wnt complex receptor antagonist. Wnt will combine with Frizzle receptor and activate Wnt signaling if the DKK-3 protein is inactivated. The WIF-1 gene locus maps to 12q14.1 of the human chromosome, which encodes a conservatively secreted protein that inhibits the activity of Wnt by holding back regular or non-regular Wnt signal transduction pathway.
Recently, Wnt antagonists have received increasing attention because of their potential role in carcinogenesis. Many studies have reported a relationship between hypermethylation of a gene promoter and low expression of mRNA in several human carcinomas. Promoter hypermethylation and reduced expression of the DKK-3 gene were found in bladder cancer, lung cancer cell lines and tissues.
Few studies have described the correlation between Wnt antagonists and the development of hepatocellular carcinoma (HCC). The results of this study suggest that DKK-3 and WIF-1 may act as negative regulators of Wnt signaling and may be related to the development of HCC. However, this pathogenesis is different from that of hepatitis B virus infection inducing HCC development.
In this study, the authors investigated the methylation status and mRNA expression of DKK-3 and WIF-1 in HCC, adjacent non-cancerous tissues and normal control tissues, and found that the aberrant Wnt antagonists play an important role in carcinogenesis of HCC. This finding may provide a reliable way to improve early diagnosis of HCC and new therapies by blocking this pathway in the treatment of HCC.
MSP: Methylation-specific polymerase chain reaction is a simple, rapid and inexpensive method to determine the methylation status of CpG islands. This approach allows the determination of methylation patterns from very small samples of DNA and can be used in the study of abnormally methylated CpG islands in neoplasia.
Promoter-hypermethylation and the reduced expression of DKK-3 and WIF-1 were shown to be important mechanisms of hepatocarcinogenesis. These results may provide a reliable way to improve early diagnosis of HCC and develop new therapies by blocking this pathway in the treatment of HCC.
| 1. | Durnez A, Verslype C, Nevens F, Fevery J, Aerts R, Pirenne J, Lesaffre E, Libbrecht L, Desmet V, Roskams T. The clinicopathological and prognostic relevance of cytokeratin 7 and 19 expression in hepatocellular carcinoma. A possible progenitor cell origin. Histopathology. 2006;49:138-151. [Cited in This Article: ] |
| 2. | Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109-2116. [Cited in This Article: ] |
| 3. | Suzuki T, Yano H, Nakashima Y, Nakashima O, Kojiro M. Beta-catenin expression in hepatocellular carcinoma: a possible participation of beta-catenin in the dedifferentiation process. J Gastroenterol Hepatol. 2002;17:994-1000. [Cited in This Article: ] |
| 4. | Seidler HB, Utsuyama M, Nagaoka S, Takemura T, Kitagawa M, Hirokawa K. Expression level of Wnt signaling components possibly influences the biological behavior of colorectal cancer in different age groups. Exp Mol Pathol. 2004;76:224-233. [Cited in This Article: ] |
| 5. | Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199-221. [Cited in This Article: ] |
| 6. | Kobayashi K, Ouchida M, Tsuji T, Hanafusa H, Miyazaki M, Namba M, Shimizu N, Shimizu K. Reduced expression of the REIC/Dkk-3 gene by promoter-hypermethylation in human tumor cells. Gene. 2002;282:151-158. [Cited in This Article: ] |
| 7. | Edamoto Y, Hara A, Biernat W, Terracciano L, Cathomas G, Riehle HM, Matsuda M, Fujii H, Scoazec JY, Ohgaki H. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer. 2003;106:334-341. [Cited in This Article: ] |
| 8. | Loeppen S, Koehle C, Buchmann A, Schwarz M. A beta-catenin-dependent pathway regulates expression of cytochrome P450 isoforms in mouse liver tumors. Carcinogenesis. 2005;26:239-248. [Cited in This Article: ] |
| 9. | Tsuji T, Miyazaki M, Sakaguchi M, Inoue Y, Namba M. A REIC gene shows down-regulation in human immortalized cells and human tumor-derived cell lines. Biochem Biophys Res Commun. 2000;268:20-24. [Cited in This Article: ] |
| 10. | Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717-4720. [Cited in This Article: ] |
| 11. | Batra S, Shi Y, Kuchenbecker KM, He B, Reguart N, Mikami I, You L, Xu Z, Lin YC, Clément G. Wnt inhibitory factor-1, a Wnt antagonist, is silenced by promoter hypermethylation in malignant pleural mesothelioma. Biochem Biophys Res Commun. 2006;342:1228-1232. [Cited in This Article: ] |
| 12. | Lin YC, You L, Xu Z, He B, Mikami I, Thung E, Chou J, Kuchenbecker K, Kim J, Raz D. Wnt signaling activation and WIF-1 silencing in nasopharyngeal cancer cell lines. Biochem Biophys Res Commun. 2006;341:635-640. [Cited in This Article: ] |
| 13. | Tannapfel A, Wittekind C. Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis. Virchows Arch. 2002;440:345-352. [Cited in This Article: ] |