Published online Feb 7, 2005. doi: 10.3748/wjg.v11.i5.705
Revised: February 17, 2004
Accepted: March 2, 2004
Published online: February 7, 2005
AIM: To explore the expression of Tiam1 gene in colorectal carcinoma and its correlation with tumor metastasis.
METHODS: Expressions of Tiam1 gene in 8 colorectal carcinoma cell lines were detected by reverse transcriptase-polymerase chain reaction. In vitro invasiveness was determined by means of Matrigel invasion assay. The correlation of Tiam1 expression with the invasive ability was also analyzed.
RESULTS: Tiam1 gene was highly expressed in LoVo and SW620, which were established from metastatic colorectal carcinomas in comparison with LS174T, SW480, HCT116, LST, HRT-18 and Hee8693, which were established from primary colorectal carcinomas. In vitro cell invasivion demonstrated that LoVo and SW620 had a higher invasive ability than LS174T, SW480, HCT116, LST, HRT-18 and Hee8693. The expression of Tiam1 gene was highly related to the metastatic potential of colorectal carcinoma cells.
CONCLUSION: Tiam1 gene may play an important role in invasion and metastasis of colorectal carcinoma and is a metastasis-related gene.
- Citation: Liu L, Wu DH, Ding YQ. Tiam1 gene expression and its significance in colorectal carcinoma. World J Gastroenterol 2005; 11(5): 705-707
- URL: https://www.wjgnet.com/1007-9327/full/v11/i5/705.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i5.705
Colorectal carcinoma is one of the most common malignancies. The majority of colorectal carcinomas arise from a series of somatic genetic changes that involve activation of oncogenes and inactivation of tumor suppressor genes. The delineation of molecular genetic and biological changes that accompany the pathogenesis of colorectal carcinoma will hopefully improve the outcome of patients in the future. Unfortunately, the overwhelming majority of patients with colorectal carcinoma would die of metastatic disease, that is to say, metastasis is the major cause of mortality in the human population[1]. Unlike the molecular events described for the pathogenesis of primary colorectal carcinomas, genes responsible for metastasis in these tumors have not been well characterized. Exploring metastasis-related genes is significantly important in the prevention of tumor metastasis and prolongation of the life expectancy of patients.
A gene designated Tiam1 was originally identified as the invasion- and metastasis-inducing gene by proviral tagging in combination with in vitro selection for invasiveness[2]. Transfection of truncated Tiam1 cDNAs into noninvasive cells can make these cells invasive. Tiam1 protein contains a Dbl homologous (DH) domain and two pleckstrin homologous (PH) domains. DH domain is present in guanine nucleotide exchange factors (GEFs) that activate the Rho-like GTPases[3]. PH domain, present in many signaling proteins, has been reported to be involved in protein-protein and protein-phospholipid interactions and might play a role in translocation of these proteins to membranes[4]. Tiam1 is one of the GEFs and can specifically activate Rac in vitro as well as in vivo[5]. Tiam1 has been implicated to directly bind to a plethora of different cytoplasmic and membrane-associated proteins, which couple Tima1-Rac activity to specific signaling pathways[6,7]. Interestingly, proteins such as Src[8],Myc[9],CD44[10] and nm23[11], claimed to interact with Tiam1 are well known players in the field of cancer. Tiam1 activation has also been shown to be stimulated by certain serum-derived growth activators such as S1P[12] and LPA[13] during cell spreading and motility.
Recent evidence suggests that Tiam1 could influence Rac GTPase signaling specificity in addition to promoting their activation[14]. That is why Tiam1 has a different effect on different cancers. For example, activation of Tiam1 in breast cancer and T-lymphoma cell lines, produces specific structural changes in plasma membrane-cytoskeleton reorganization leading to membrane ruffling, lamellipodia, filopodia, and stresses fiber formation[15,16]. These changes are prerequisite for invasion and metastasis. Conversely, in renal cell carcinoma cell lines, Tiam1 potentiates homotypic cell-cell adhesion and inhibits invasion[17]. In epithelial MDCK cells, Tiam1-Rac1 signaling plays an invasion-suppressor role in Ras-transformed MDCK cells[18]. A Tiam1 knock-out mouse is relatively resistant to chemical induction of skin tumors but paradoxically more prone to malignant histologic progression of those tumors[19].
Tiam1 has been studied in breast cancer[15], renal carcinoma[17], chondrosarcoma[20], T-lymphoma[16], etc. However, it is not known at present whether there is any relationship between Tiam1 and colorectal carcinoma. Here we showed the expression of Tiam1 in 8 colorectal carcinoma cell lines, and analyzed its correlation with tumor metastasis. Our observations suggest that Tiam1 has a close relationship with metastasis of colorectal carcinoma and may be required for colorectal carcinoma cell invasion and migration.
HRT-18 and Hee8693 cell lines were a kind gift from Cancer Research Institute in Xiangya Medical School of Central South University. LST cell line was a generous gift from Digestive Department in Nanfang Hospital. LoVo, LS174T, HCT116, SW480 and SW620 were obtained from America Type Culture Collection. LoVo, LS174T, LST, HRT-18, HCT116 and Hee8693 cell lines were cultured in RPMI-1640 medium. SW480 and SW620 were cultured in Dulbecco’s modified Eagle’s medium. All mediums supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin. Cultured cells were grown in a 37 °C humidfied incubator with 50 mL/L CO2.
Total RNA was extracted from the cell lines using TRIzol reagent (Invitrogen Corporation). Then DNase I was used to remove genomic DNA from total RNA. Concentration and purity of the RNA samples were examined by a spectrophotometer. Two oligonucleotides, 5’-AATCCCATCACCATCTTCCA-3’ and 5’-CCTGCTTCACCACCTTCTTG-3’, designated GAPDH (a housekeeping gene), were utilized as primers to perform polymerase chain reaction (PCR) in order to check whether the RNA was contaminated with trace genomic DNA.
Reverse transcriptase (RT) was performed using an access RT-PCR system (Promega Corporation) according to the manufacturer’s protocol. Total RNA in the amount of 1 µg was used in each RT reaction to synthesize cDNA. For PCR, we used the forward primer 5’-AAGACGTACTCAGGCCATGTCC-3’ and reverse primer 5’-GACCCAAATGTCGCAGTCAG-3’, which were designed by primer premier 5.0 software, to amplify human Tiam1(NM_003253 from GeneBank). And human GAPDH primers were utilized as an internal control. Thirty cycles of reactions were carried out, each at 94 °C for 45 s, at 58 °C for 45 s, at 72 °C for 45 s. PCR products were electrophoresed in 2% agarose gel and sequenced by Bioasia Biological Corporation. Electrophorectogram was taken by a pickup camera under UV light and analyzed by 1-D advanced software.
This assay was based on the principle of Boyden chamber[21]. The top and bottom of Boyden chamber (corning company) were separated by a polycarbonate filter with 8 μm pore size. The top chamber was prepared by coating the filter with 50 μg of diluted Matrigel and incubated for 30 min. A single cell suspension of 10000 tumor cells in serum-free medium was inoculated in the upper chamber, after 5% FBS was added into the bottom chamber as a chemoattactant. After 24 h incubation, noninvasive cells were removed with a cotton swab. The cells migrated through the membrane and stuck to the lower surface of the membrane were fixed with methanol and stained with hematoxylin. Tumor cell invasiveness was determined by counting all tumor cells in five randomly selected counting fields at ×200.
Relationship between expression of Tiam1 and invasive ability was analyzed by bivariate correlation using SPSS 10.0 Software.
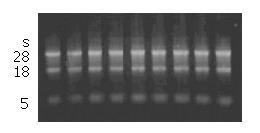
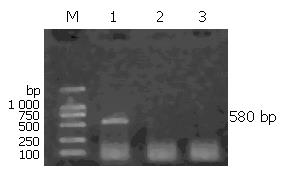
Total RNA extracted from eight colorectal carcinoma cell lines was confirmed to have no degradation by agarose gel electrophoresis (Figure 1). In order to detect whether the RNA was contaminated with gDNA, we used the RNA digested with DNase I and RNA not digested with DNaseIas templates to amplify the housekeeping gene, GAPDH. The results showed that the RNA we extracted was not contaminated with gDNA (Figure 2).
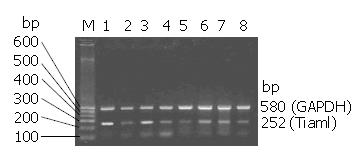
RT-PCR detection of Tiam1expression in eight colorectal carcinoma cell lines was heterogeneous (Figure 3). Analyzed by 1-D Advanced software, relative Tiam1 expression was calculated as the ratio of the densitometry of the Tiam1 band: GAPDH band on RT-PCR. We found that Tiam1 was abundantly expressed in metastastic colorectal carcinoma cell lines, LoVo and SW620, 1.19 and 0.96 respectively. In primary colorectal carcinoma cell lines, SW480 (0.57), LS174T (0.32), LST (0.44), HCT116 (0.60), HRT-18 (0.52) and Hee8693 (0.55) were moderately and lowly expressed. The product of RT-PCR, Tiam1, was confirmed by sequence analysis (date was not shown).
In vitro cell invasive assay was performed based on the principle of the Boyden chamber assay. Matrigel matrix served as a reconstituted basement membrane in vitro. The number of cells migrating through the Matrigel matrix was counted. The result indicated that LoVo (145) and SW620 (130) had a higher invasive ability than SW480 (91), HCT116 (89), LS174T (64), LST (79), HRT-18 (87) and Hee8693 (90).
We used the SPSS 10.0 to analysis the correlation between the expression of Tiam1 and invasive ability. The results showed that the both are highly correlative(r = 0.995, P<0.01).
Tumor invasion is a complex biological process, during which tumor cells detach from the primary tumor and infiltrate its surrounding tissues. This process requires loss of cell contacts between tumor cells, active cell migration, adhesion to the extracellular matrix (ECM) and proteolytic degradation of ECM[22]. At the molecular level, a number of different molecules, including cadherins, integrins, proteases, and growth factors, have been implicated in the regulation of tumor cell invasiveness[23].
It has been found that Tiam1 is capable of activating Rac1 as a ubiquitous guanine nucleotide exchange factor and inducing membrane cytoskeleton-mediated cell shape changes, cell adhesion, and cell motility[24-26]. Rac1 could act on downstream of Tiam1 signaling and regulate the function of several cell adhesion molecules such as laminin receptor, integrin[27], E-cadherin[18], and the hyaluronan receptor, CD44.
In the past, to search new metastasis-associated genes, we prepared cDNA microarray which is highly sensitive and applicable in examining gene expression profile. Then metastatic colorectal carcinoma microarray gene expression data were mined by literature profiling, based on the analysis of literature profiles generated by extracting the frequencies of certain terms from MEDLINE. We found that Tiam1 gene had a potential relation to metastatic colorectal carcinoma.
In this study, we investigated the relation between Tiam1 expression and metastasis in colorectal carcinoma. LoVo cell line was derived from human colonic adenocarcinoma established from the metastatic nodule. SW480 was isolated from a high-grade primary colonic tumor, and SW620 was isolated from a metastatic lymph node from the same patient 1-year later at the time of clinical relapse. LS174T, LST and HCT116 were isolated from primary colonic carcinomas. HRT-18 was isolated from primary rectal cancer and Hee8693 from primary cecal cancer. We found that the expression of Tiam1 in cell lines established from metastatic colorectal carcinomas was much higher than that in cell lines established from primary colorectal carcinomas. We also found that the invasiveness and metastasis in cell lines established from metastatic colorectal carcinomas were much stronger than those in cell lines established from primary colorectal carcinomas. We identified that Tiam1 was highly correlated with invasiveness and metastasis (P<0.01). These data indicate that enhanced expression of Tima1 is associated with increased invasive ability.
In a word, our results suggest that Tiam1 might be a metastasis-related gene in colorectal carcinoma. In the future research, we will study the effect of Tiam1 gene on invasion and metastasis in colorectal carcinoma in vivo and in vitro through stable Tiam1 gene transfection or RNA interfering. The two-dimensional electrophoresis and mass spectrum analysis will be used to study the possible mechanism of Tiam1 gene and its signal transduction.
Edited by Wang XL Proofread by Chen WW
| 1. | Budenholzer B. Screening for colorectal cancer. CMAJ. 2001;164:965-966; author reply 967-968. [PubMed] [Cited in This Article: ] |
| 2. | Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 400] [Cited by in F6Publishing: 407] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 3. | Robbe K, Otto-Bruc A, Chardin P, Antonny B. Dissociation of GDP dissociation inhibitor and membrane translocation are required for efficient activation of Rac by the Dbl homology-pleckstrin homology region of Tiam. J Biol Chem. 2003;278:4756-4762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Crompton AM, Foley LH, Wood A, Roscoe W, Stokoe D, McCormick F, Symons M, Bollag G. Regulation of Tiam1 nucleotide exchange activity by pleckstrin domain binding ligands. J Biol Chem. 2000;275:25751-25759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621-625. [PubMed] [Cited in This Article: ] |
| 6. | Buchsbaum RJ, Connolly BA, Feig LA. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol Cell Biol. 2002;22:4073-4085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 152] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | Buchsbaum RJ, Connolly BA, Feig LA. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J Biol Chem. 2003;278:18833-18841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 99] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 8. | Servitja JM, Marinissen MJ, Sodhi A, Bustelo XR, Gutkind JS. Rac1 function is required for Src-induced transformation. Evidence of a role for Tiam1 and Vav2 in Rac activation by Src. J Biol Chem. 2003;278:34339-34346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 9. | Otsuki Y, Tanaka M, Kamo T, Kitanaka C, Kuchino Y, Sugimura H. Guanine nucleotide exchange factor, Tiam1, directly binds to c-Myc and interferes with c-Myc-mediated apoptosis in rat-1 fibroblasts. J Biol Chem. 2003;278:5132-5140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Bourguignon LY. CD44-mediated oncogenic signaling and cytoskeleton activation during mammary tumor progression. J Mammary Gland Biol Neoplasia. 2001;6:287-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Otsuki Y, Tanaka M, Yoshii S, Kawazoe N, Nakaya K, Sugimura H. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci USA. 2001;98:4385-4390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 12. | Buchanan FG, Elliot CM, Gibbs M, Exton JH. Translocation of the Rac1 guanine nucleotide exchange factor Tiam1 induced by platelet-derived growth factor and lysophosphatidic acid. J Biol Chem. 2000;275:9742-9748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 82] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Van Leeuwen FN, Olivo C, Grivell S, Giepmans BN, Collard JG, Moolenaar WH. Rac activation by lysophosphatidic acid LPA1 receptors through the guanine nucleotide exchange factor Tiam1. J Biol Chem. 2003;278:400-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 144] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 14. | Mertens AE, Roovers RC, Collard JG. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003;546:11-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 154] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 15. | Bourguignon LY, Zhu H, Shao L, Chen YW. Ankyrin-Tiam1 interaction promotes Rac1 signaling and metastatic breast tumor cell invasion and migration. J Cell Biol. 2000;150:177-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 98] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Stam JC, Michiels F, van der Kammen RA, Moolenaar WH, Collard JG. Invasion of T-lymphoma cells: cooperation between Rho family GTPases and lysophospholipid receptor signaling. EMBO J. 1998;17:4066-4074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 178] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Engers R, Springer E, Michiels F, Collard JG, Gabbert HE. Rac affects invasion of human renal cell carcinomas by up-regulating tissue inhibitor of metalloproteinases (TIMP)-1 and TIMP-2 expression. J Biol Chem. 2001;276:41889-41897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 82] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Hordijk PL, ten Klooster JP, van der Kammen RA, Michiels F, Oomen LC, Collard JG. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464-1466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 359] [Cited by in F6Publishing: 367] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 19. | Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867-871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 272] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 20. | Karjalainen HM, Sironen RK, Elo MA, Kaarniranta K, Takigawa M, Helminen HJ, Lammi MJ. Gene expression profiles in chondrosarcoma cells subjected to cyclic stretching and hydrostatic pressure. A cDNA array study. Biorheology. 2003;40:93-100. [PubMed] [Cited in This Article: ] |
| 21. | Itoh F, Yamamoto H, Hinoda Y, Imai K. Enhanced secretion and activation of matrilysin during malignant conversion of human colorectal epithelium and its relationship with invasive potential of colon cancer cells. Cancer. 1996;77:1717-1721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 22. | Engers R, Gabbert HE. Mechanisms of tumor metastasis: cell biological aspects and clinical implications. J Cancer Res Clin Oncol. 2000;126:682-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Takeda A, Stoeltzing O, Ahmad SA, Reinmuth N, Liu W, Parikh A, Fan F, Akagi M, Ellis LM. Role of angiogenesis in the development and growth of liver metastasis. Ann Surg Oncol. 2002;9:610-616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Michiels F, Habets GG, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 451] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 25. | Sander EE, van Delft S, ten Klooster JP, Reid T, van der Kammen RA, Michiels F, Collard JG. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143:1385-1398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 549] [Cited by in F6Publishing: 559] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 26. | Michiels F, Collard JG. Rho-like GTPases: their role in cell adhesion and invasion. Biochem Soc Symp. 1999;65:125-146. [PubMed] [Cited in This Article: ] |
| 27. | Leeuwen FN, Kain HE, Kammen RA, Michiels F, Kranenburg OW, Collard JG. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol. 1997;139:797-807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 309] [Article Influence: 11.4] [Reference Citation Analysis (0)] |