Published online Mar 14, 2006. doi: 10.3748/wjg.v12.i10.1529
Revised: September 12, 2005
Accepted: November 18, 2005
Published online: March 14, 2006
AIM: To analyse the Chibby expression and its function in colon carcinoma cell lines and colorectal carcinoma (CRC).
METHODS: Chibby expression levels were investigated by quantitative RT-PCR in a panel of seven different colon carcinoma cell lines. By sequencing, we analysed mutational status of Chibby. To test whether Chibby exhibited effects on β-catenin signalling in colon carcinoma cells, we transfected SW480 cells with Chibby expression plasmid and, subsequently, analysed activity of β-catenin and tested for alterations in cellular phenotype. In addition, we examined Chibby mRNA levels in samples of colorectal carcinomas and adjacent normal tissues by using quantitative RT-PCR and hybridised gene chips with samples from CRC and normal tissues.
RESULTS: Chibby mRNA expression was strongly down-regulated in colon carcinoma cell lines in comparison to normal colon epithelial cells and no mutation in any of the examined colon carcinoma cell lines was found. Further, we could show that Chibby inhibited β-catenin activity in TOPflash assays when over-expressed in SW480 cells. Proliferation and invasion assays with Chibby transfected SW480 cells did not reveal profound differences compared to control cells. In contrast to these in vitro data, quantitative RT-PCR analyses of Chibby mRNA levels in CRC tumor samples did not show significant differences to specimens in adjacent non-cancerous tissue. Consistent with these findings, gene chips analysing tissue samples of tumors and corresponding normal tissue did not show altered Chibby expression
CONCLUSION: Altered Chibby expression might be observed in vitro in different colon carcinoma cell lines. However, this finding could not be confirmed in vitro in CRC tumors, indicating that Chibby is not likely to promote CRC tumor development or progression. As Chibby is an important inhibitor of ß-catenin signalling, our data implicate that the usability of colon carcinoma cell lines for in vitro studies analysing the Wnt/β-catenin pathway in colorectal carcinoma needs extensive verification.
- Citation: Schuierer MM, Graf E, Takemaru KI, Dietmaier W, Bosserhoff AK. Reduced expression of β-catenin inhibitor Chibby in colon carcinoma cell lines. World J Gastroenterol 2006; 12(10): 1529-1535
- URL: https://www.wjgnet.com/1007-9327/full/v12/i10/1529.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i10.1529
The Wnt/Wingless pathway has been shown to exhibit important functions in embryonic and cancer development. In the absence of Wnt signalling, β-catenin is phosphorylated by glycogen synthase kinase-3 (GSK-3) and targeted for ubiquitin-mediated proteasomal degradation[1]. Upon Wnt binding to its cellular receptor Frizzled, the degradation complex is destabilised, and, as a result, unphosphorylated β-catenin accumulates and translocates into the nucleus. There, it functions as a co-factor of lymphoid enhancer factor/T-cell factor (LEF/TCF) transcription factors leading to upregulation of a variety of genes. It has been shown that mutations in different genes of the Wnt pathway are involved in development of various cancers. One of those tumor entities is colorectal carcinoma where approximately 90% of the tumors have been shown to harbour activating mutations within the canonical Wnt signalling pathway[2-4].
The most potent transcriptional activation domain of β-catenin is located within the C-terminal region of β-catenin and has been used as bait in a yeast screening to identify interacting proteins. In this screening, Chibby, a novel protein that is conserved from Drosophila to human, was isolated [5]. It is a nuclear protein of 126 amino acids, and as it inhibited Wnt signalling by preventing the interaction between β-catenin and LEF/TCF factors, it was suggested as a potential tumor suppressor gene. Chibby is encoded by the human C22orf2 gene located at chromosome 22q12-q13. This chromosomal region is known to be mutated frequently in colorectal cancer[6], and therefore, Chibby might be an interesting candidate as a tumor suppressor in CRC. Gad et al[7] performed loss of heterozygosity (LOH) analyses of tumors of 36 patients involving the C22orf2 region in CRC and were not able to detect differences between normal and CRC tumor tissues, but did not analysed effects of Chibby on cellular functions.
In this study, we aimed to analyse whether Chibby is mutated in CRC, to evaluate Chibby mRNA expression levels in colon carcinoma cell lines and tumor samples, and to investigate whether Chibby over-expression in colon carcinoma cell lines has an effect on cell behaviour in functional assays.
Isolation and cultivation of colon epithelial cells (CEC) were performed as previously described[8,9].
The colon carcinoma cell lines SW48 (ATCC CCL-231), SW480 (ATCC CCL-228), CaCo-2 (ATCC HTB-37), LoVo (ATCC CCL-229), HCT116 (ATCC CCL-247), and HT29 (ATCC HTB-38) were used for in vitro experiments. The cell line HT29M3 was chosen as a highly differentiated colon carcinoma cell line[10-13].
For tissue culture, the cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with penicillin (400 U/mL), streptomycin (50 µg/mL), L-glutamine (300 µg/mL), and 100 mL/L fetal calf serum (FCS) (Sigma, Deisenhofen, Germany) and split 1∶5 every three days.
For demethylation assays, the cells were treated with 5-azacytidine at a final concentration of 10 µmol/L for 48 h (Sigma)[8]. HT29 M3 cells were used as control cells in the demethylation experiments.
For RT-PCR, total cellular RNA was isolated from cultured cells or laser microdissected tissue samples using the RNeasy kit (Qiagen, Hilden, Germany). The integrity of the RNA was controlled on 10 g/L agarose/formaldehyde gel, and subsequently cDNAs were generated by reverse transcriptase reactions. The reverse transcription (RT) reaction was performed in 20 µL reaction volume containing 2 µg of total cellular RNA, 4 µL of 5x first strand buffer (Gibco), 2 µL of 0.1 mol/L DTT, 1 µL of dN6 primer (10 mmol/L), 1 µL of dNTPs (10 mmol/L) and DEPC water. The reaction mix was incubated for 10 min at 70 °C. Then 1 µL of Superscript II reverse transcriptase (Gibco) was added and RNAs were reverse transcribed for 1 h at 37 °C. Subsequently, reverse transcriptase was inactivated at 70 °C for 10 min and RNA was degraded by digestion with 1 µL of RNase A (10 mg/mL) at 37 °C for 30 min. cDNAs were controlled by PCR amplification of ß-actin.
The entire coding region of Chibby was amplified by RT-PCR from cDNA using16, ATGCCTTTCTTTGGGAATTACGTTC (forward) and chibby496, TCATTTTCTCTTCCGGCTGATC (reverse), which resulted in a 380-bp fragment. The PCR reaction was performed in 50 µL reaction volume containing 5 µL of 10x Taq buffer, 1 µL of cDNA, 0.5 µL of each primer (0.2 µmol/L), 0.5 µL of dNTPs (10 mmol/L), 0.5 µL of Taq polymerase (5 U/µL), and 41 µL of water. The amplification reactions were performed by 35 repetitive cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 64 °C, extension for 30 s at 72 °C, and a final extension step at 72 °C for 5 min. The PCR products were resolved on 15 g/L agarose gels. For sequencing, the products were purified by PEG precipitation to remove unincorporated primers and dNTPs. Both strands were sequenced for each PCR product from at least two independent PCR reactions. Sequences were compared with the gene data bank by means of BLAST search (National Center of Biotechnology Information).
Quantitative real-time PCR was performed on a LightCycler (Roche, Mannheim, Germany). Two microliters of cDNA template, 1.6 µL of 25 mmol/L MgCl2, 0.2 µmol/L forward and reverse primers {chibby125, TTTGGGAATACGTTCAGTCCG (forward) and chibby395, TCAGCCGCAAGAGATTGTTC (reverse)} and 2 µL of SybrGreen LightCycler mix in 20 µL volume were PCR-amplified through 40 cycles, each cycle consisting of initial denaturation at 95 °C for 30 s, 20 °C/s temperature transition rate up to 95 °C for 15 s, 64 °C for 3 s, 72 °C for 5s, 85 °C acquisition mode single. The PCR product was evaluated by melting curve analysis following the manufacturer’s instructions and by checking the PCR products on 18 g/L agarose gels. All quantitative PCR experiments were repeated three times.
A total of 2×105 cells were seeded onto six-well plates one day before transfection. Transfections with pCMX PL2 or pCMX PL2 flag Chibby plasmid (0.5 µg per well) were performed using the LipofectAMINE plus method (Life Technologies) according to the manufacturer’s instructions. Cells were harvested 48 h after transfection and the amount of Chibby expression was determined by RT-PCR and Western blot analysis.
For protein isolation, 2×106 cells were washed in 1x PBS and lysed in 200 µL RIPA buffer (Roche). The protein concentration was determined using the BCA protein assay reagent (Pierce, USA). Equal amounts of cellular protein (40 µg total protein) were denatured at 94 °C for 10 min after addition of Rotiload buffer (Roth, Karlsruhe, Germany) and subsequently, separated on NuPage SDS gels (Invitrogen, Groningen, The Netherlands). After transferring the proteins onto PVDF membranes (BioRad, Richmond, USA), the membranes were blocked in 50 g/L dry milk in TBS-T and incubated overnight with primary monoclonal anti flag antibody (1∶4 500 dilution, Sigma) at 4 °C. A 1∶4 000 dilution of anti IgG AP (Sigma) was used as secondary antibody. Staining was performed using 5-bromo-4-chloro-3-indolyl phosphate/ nitroblue tetrazolium tablets (Sigma). All Western blot experiments were repeated at least three times.
A total of 2×104 SW480 cells were seeded per chamber of four-well-Falcon chamber slides (Becton Dickinson, Le Pont de Plaix, France) and cultivated overnight before transfection with either control vector or Chibby flag pCMX PL2. After 48 h of transfection, cells were washed with PBS, fixed with 40 g/L paraformaldehyde, permeabelised with 1 ml/L Triton X-100, washed again, and blocked for 30 min in 10 g/L BSA in PBS. Anti flag antibody (dilution 1∶1 000) was used as primary antibody, and anti mouse FITC-conjugated antibody (dilution 1∶40) was used as secondary antibody. Incubations took place at room temperature for 1 h each. Staining of nuclei occurred with DAPI.
For transient transfections, 2×105 cells per well were seeded into six-well plates and transfected with 0.5 µg of TOPflash or FOPflash reporter gene plasmids using the LipofectAMINE plus method (Gibco) according to the manufacturer’s instructions. For cotransfection, 0.125 µg, 0.25 µg, 0.5 µg, and 1 µg of Chibby expression plasmid per well were used, respectively. At 48 h post-transfection, the cells were lysed and the luciferase activity in the lysate was measured. To normalise transfection efficiency, 0.2 µg of a pRL-TK plasmid (Promega, Mannheim, Germany) was cotransfected and renilla luciferase activity was measured by a luminometric assay (Promega). All transfection experiments were repeated at least three times.
Proliferation was measured as previously described[14]. Proliferation of Chibby transfected SW480 cells was compared to non or mock transfected SW480 cells in an XTT assay. Results were expressed as mean ± SD (range). All experiments were repeated three times.
Invasion assays were performed as previouly described[14]. Briefly, the assays were performed in Boyden chambers containing polycarbonate filters with an 8-µm pore size. In brief, filters were coated with a commercially available reconstituted basement membrane (Matrigel; Becton Dickinson, Heidelberg, Germany), and the lower compartment was filled with fibroblast-conditioned medium as chemoattractant. SW480 cells and Chibby transfected SW480 cells were harvested by trypsinization, resuspended in DMEM without FCS at a density of 2×105 cells/mL, and placed in the upper compartment of the chambers. After incubation for 4 h at 37 °C, the filters were removed. The cells adhering to the lower surface were fixed, stained, and counted. Fifteen random fields were counted at 200-fold magnification. Each sample was assayed in triplicate, and all experiments were repeated three times.
It is well known that the Wnt/β-catenin signalling pathway is constitutively active in a large number of colorectal cancers. The most commonly mutated proteins within the pathway are β-catenin and APC. Intriguingly, the activation of the cascade cannot in all cases of CRC be explained by mutations in these proteins. It remains unclear what other proteins of the pathway have to be altered in addition to the aforementioned mutations to lead to activation of the Wnt/β-catenin signalling pathway. Hence, we were interested in analysing other potential endogenous inhibitory factors that might accomplish regulatory functions within the Wnt/β-catenin signalling pathway and thus influence signal transduction. Due to these reasons, we analysed the role of the β-catenin-associated antagonist Chibby in colorectal carcinoma cell lines and compared our in vitro findings of Chibby expression with the in vivo situation by analysing colorectal tumor samples and the corresponding normal tissue.
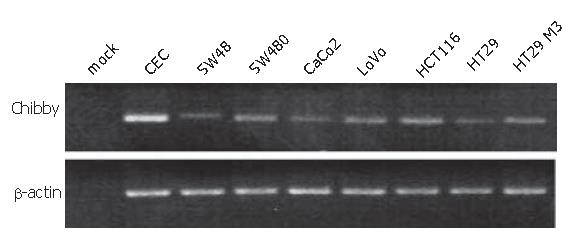
We analysed a panel of seven colon carcinoma cell lines, such as SW48, SW480, CaCo2, LoVo, HCT116, HT29, and HT29 M3, that are frequently used as model systems to study effects in colorectal carcinoma. We evaluated the panel of colon carcinoma cell lines for alterations in the Chibby mRNA expression. The complete Chibby coding region was amplified by RT-PCR. All cell lines examined expressed Chibby mRNA at the expected length, indicating that no deletions or mutational insertions had occurred (Figure 1). As a control the Chibby coding sequence was amplified using CEC cDNA as template. The same cDNAs were used to amplify β-actin as a control for the integrity of the cDNA.
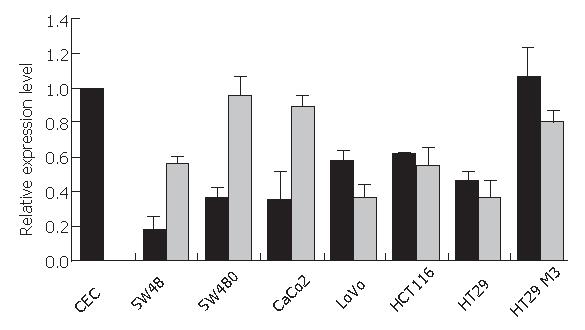
The cell lines were evaluated for levels of Chibby mRNA expression using quantitative RT-PCR in comparison to normal colon epithelial cells (CEC). Chibby mRNA was found to be down-regulated in all cell lines in comparison to CEC and HT29 M3 cells which are considered to be a highly differentiated colon carcinoma cells and express Chibby in amounts similar to those of CEC (Figure 2, black bars). Three of the analysed cell lines, SW48, SW480, and CaCo2 cells, showed residual expressions of Chibby mRNA of 18%, 37%, and 35%, respectively, in comparison to CEC or HT29 M3 (100% and 106%, respectively). LoVo, HCT116, and HT29 cells were also shown to have reduced Chibby mRNA levels, but down-regulation was not as explicit as in the other examined cell lines (57%, 62%, and 46%).
To investigate the reason for down-regulation of Chibby expression in colon carcinoma cell lines, we amplified and analysed the exons of the chibby gene. All PCR products were purified and subsequently sequenced using two different primers. The analysis of sequenced PCR products did not reveal any sequence variation within the coding region of the chibby gene locus (data not shown).
From these results, we concluded that Chibby does not harbour mutations within the coding sequence in all seven analysed colon carcinoma cell lines.
As this reduction in mRNA expression in colon carcinoma cell lines could not be explained by loss or mutation of the chibby gene, we hypothesised that promoter hypermethylation could have silenced gene expression. To test this hypothesis, all cell lines were exposed to the demethylating agent 5-azacytidine. Treated and untreated cells were tested for Chibby mRNA expression which was quantified by quantitative RT-PCR. HT29 M3 cells were used as controls in these experiments. HT29 M3 cells are highly differentiated cells that were used as an additional control as CEC could not be used in demethylation experiments as controls due to culturing issues.
Our results showed that 5-azacytidine treatment did not lead to induction of Chibby expression in LoVo, HCT116, HT29, or HT29 M3 cells, but to a moderately increased expression in SW48, SW480, and CaCo2 cells (Figure 2, grey bars). Therefore, we conclude that promoter hypermethylation might play a minor role for altered Chibby expression in some of the analysed cell lines, but we do not consider this silencing mechanism to be a general event in down-regulation of Chibby mRNA expression in the colon carcinoma cell lines.
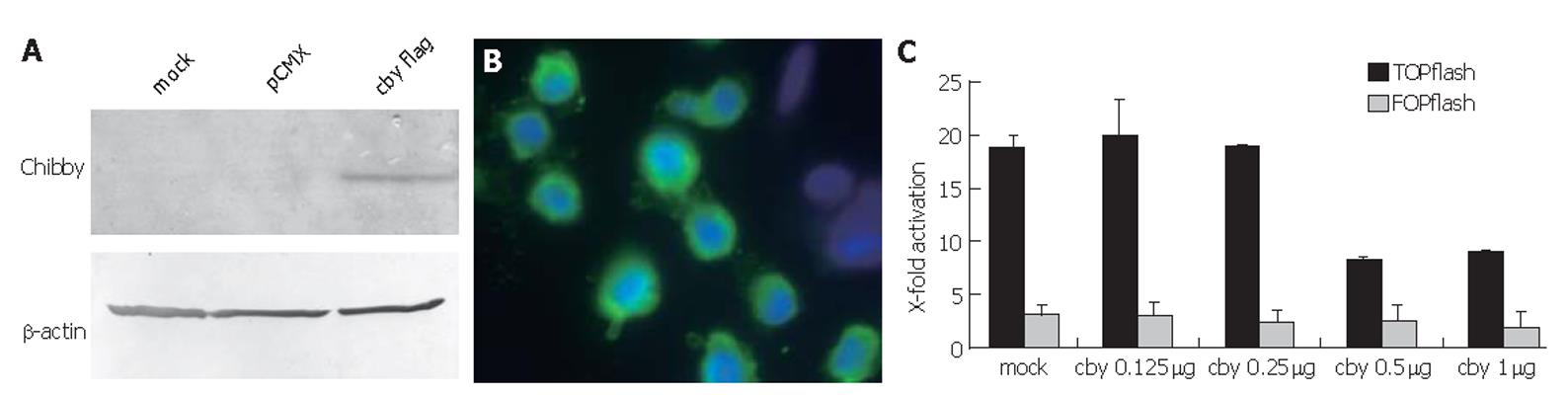
As colon carcinoma cell lines display reduced levels of Chibby expression in comparison to normal CEC, we were interested in whether over-expression of Chibby in these cells influences functional processes. To test for the potential effects, we over-expressed Chibby in the colon carcinoma cell line SW480 by transfecting a flag-tagged Chibby expression construct. Successful over-expression was observed by quantitative RT-PCR (data not shown) and Western blotting (Figure 3A). Exogenous Chibby protein was not detected following mock or control transfection with pCMX PL2 flag plasmid which did not alter Chibby levels. Strong over-expression of Chibby was observed after transfection of SW480 cells with Chibby flag expression plasmid. Immunofluorescence staining revealed correct expression patterns of Chibby flag-tagged protein in the nucleus of SW480 cells (Figure 3B), also displaying high transfection efficiency.
As Chibby as endogenous antagonist of β-catenin is expected to exert negative functions on the Wnt/β-catenin signalling pathway, we performed LEF/TCF luciferase reporter gene assays using the TOPflash/FOPflash system. Analysing the activity of β-catenin in control and Chibby over-expressing cells, we observed reduced activation levels in the TOPflash system in accordance to the amount of transfected Chibby expression plasmid (Figure 3C). Transfection with 0.125 µg or 0.25 µg of Chibby plasmid per well did not affect the activity of TOPflash reporter, whereas activity dropped to 44% and 48%, respectively, when 0.5 µg and 1 µg expression plasmids were applied. FOPflash reporter construct activity was not affected by Chibby expression (Figure 3C). These results showed that transfection of SW480 cells with Chibby expression plasmid reveals functional Chibby protein.
To assay whether altered Chibby levels had an impact on proliferation and invasion potential of SW480 cells, we performed functional experiments utilizing mock and Chibby transfected cells. Boyden chamber assays were performed to examine the effect of Chibby on the invasiveness of SW480 cells. In this assay, the tumor cells have to invade through matrigel mimicking the basal lamina. Comparing mock treated cells, control transfected cells, and Chibby over-expressing cells, we did not observe any differences with regard to their invasiveness.
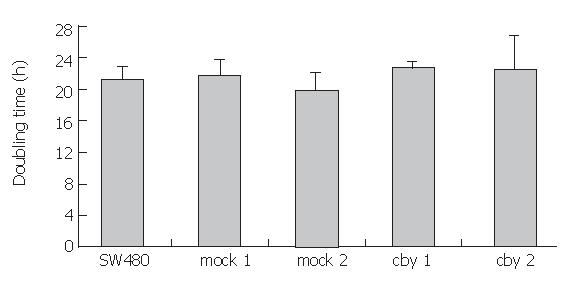
As the Wnt/β-catenin signalling pathway is known to influence cell proliferation, we next assayed for alterations of proliferation and performed in vitro proliferation assays with Chibby transfected SW480 cells and mock or control transfected cells. Modifying the Chibby protein level had no significant effect on cell proliferation (Figure 4). Doubling times were as follows: SW480: 21.2 h, mock 1: 21.7 h, mock 2: 19.7 h, cby 1: 22.6 h, and cby 2: 22.5 h, respectively. Thus, we did not consider proliferation to be affected by Chibby expression levels in SW480 cells.
As our experiments in colon carcinoma cell lines had revealed sound down-regulation of Chibby expression, the absence of effects on cell functions of over-expressed functional Chibby protein in one of these cell lines (SW480) was rather unexpected. We next performed experiments with samples from patients with diagnosed colorectal carcinoma to assess the question whether Chibby expression was altered not only in colon carcinoma cell lines, but also in coloreactal tumors and might therefore contribute to or support development and progression of colorectal tumors.
We analysed Chibby transcript expression levels in tumor samples and corresponding normal colonical tissue from the same patients by hybridisation of an oligonucleotide-based array (Affymetrix HG-U133A) with 43 samples of colorectal cancer and 34 matched normal mucosa (unpublished data). The ratio of expression levels between tumors and normal tissues was 1.11, displaying no altered Chibby expression in CRC tumors in comparison to adjacent normal colon tissue. In addition, when tumor samples with microsatellite stability (MSS; n = 24) and microsatellite instability (MSI; n = 19) were separately assessed, again no difference in Chibby expression was observed.
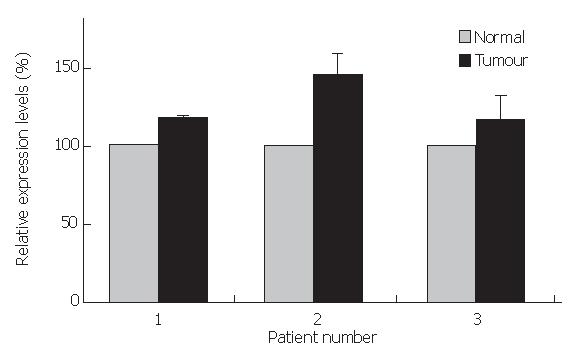
To further validate this result, using quantitative RT-PCR, we tested whether expression levels of Chibby mRNA differed between normal colorectal tissue and adjacent cancerous tissue. We manually microdissected frozen material and analysed the extracted mRNAs. Figure 5 shows the results of 3 examined matched samples, displaying no profound alterations between tumor and adjacent normal tissue. Relative Chibby levels ranged from 100% in normal tissue to 117.4% (patient 1), 146.1% (patient 2), and 116.4% (patient 3), respectively.
The cellular antagonist and β-catenin interacting factor Chibby, a small nuclear protein, has been shown to be an endogenous inhibitor of the canonical Wnt signalling pathway. It binds to the LEF/TCF interaction domain at the C-terminus of β-catenin and thereby eliminates interaction of β-catenin with LEF/TCF transcription factors[5]. This leads to a block of transcriptional activation within the signalling cascade. It has been shown for colorectal carcinoma that more than 90% of all tumors harbour activating mutations within proteins of the canonical Wnt/β-catenin signalling cascade[2] and most mutations affect either the β-catenin or the APC protein[14,15]. Interestingly, tumor development cannot in all cases be solely explained by these mutations, but there need to be further and additional disrupting cellular events. Hence, we were analysing other factors within the Wnt/β-catenin pathway and this study showed our results on the cellular antagonist of β-catenin, Chibby. Due to its function, Chibby has been suggested as a potential tumor suppressor[5]. The aim of our study was to analyse whether Chibby expression is altered in colon carcinoma cell lines and tumors, and if so, whether restoration of Chibby amounts in colon carcinoma cell lines affects cell functions and behaviour.
Examining six colon carcinoma cell lines and one highly differentiated cell line (HT29 M3) that was used as additional control with regard to the mutational status, we could not detect any alterations within Chibby coding sequence. Using quantitative RT-PCR methods, we detected down-regulation of Chibby mRNA expression levels in all six cell lines in comparison to normal colon cells (CEC) or HT29 M3. We demonstrated in another study that gene silencing via promoter hypermethylation might be responsible for down-regulated transcript expression if the coding region does not harbour any mutations[8]. We, therefore, speculated that promoter hypermethylation might be a mechanism for Chibby down-regulation in the examined cell lines, but treating cells with the demethylating agent 5-azacytidine did not lead to sound induction of Chibby mRNA expression in all cell lines. SW48, SW480, and CaCo2 cells showed the lowest default expression levels among all the cell lines tested and displayed a moderate increase in Chibby mRNA expression upon 5-azacytidine treatment. But we do not reckon epigenetic alterations as a common mechanism for Chibby down-regulation.
To assess the functional implication of reduction of chibby expression, we used the cell line SW480 to re-express Chibby via transfection of a Chibby expression plasmid. Tests for correct expression of Chibby were followed by functional assays for proliferation and invasion. We were not able to detect altered invasion potential of Chibby over-expressing cells in comparison to control cells or mock transfected cells. Also, proliferation rate of control and Chibby transfected cells was not different (Figure 4) by means of doubling times. These findings were unexpected as Chibby seemed to be a promising candidate as tumor suppressor in colorectal carcinoma. We speculate that the experimental over-expression of Chibby in SW480 cells does not lead to changes in cell behaviour as mutations in other molecules further downstream within the cascade could overcome Chibby effects. This hypothesis is based on the knowledge that the SW480 cell line is known to harbour several mutations, not only in the Wnt/β-catenin signalling pathway but also in a number of other proto-oncogenes[16,17].
In the very contrast to our findings concerning Chibby expression in a panel of colon carcinoma cell lines, the in situ and in vivo analyses of colorectal tumors did not support the in vitro conclusions. Neither quantitative RT-PCRs, nor hybridisation of an oligonucleotide-based array gave an indication of dysregulation of Chibby in colorectal tumors in comparison to normal colorectal tissue. Our results are supported by a recent study by Gad et al[17] who have reported neither somatic mutations in samples of colorectal tumors from 36 patients, nor have they been able to correlate Chibby expression levels to tumorigenicity. Gad et al[17] have reported very similar findings to our data that also display no differences in expression levels between normal colon tissue and tumor material.
Taking these data together, we conclude that in contrast to colon carcinoma cell lines where Chibby expression was down-regulated in comparison to normal colonical epithelial cells, in vivo Chibby expression is not altered in colorectal tumors and therefore neither a cause nor an indicator for tumor development in colorectal carcinoma.
Our data point out that the use of colon carcinoma cell lines to analyse events within the Wnt/β-catenin signalling pathway has to be considered very carefully. The status of protein expression in these cell lines, at least with regard to Chibby, varies compared to that of tumors and may end up in different phenotypical behaviour of the cells. Thus, direct conclusions on in vivo situations could be misleading. Therefore, these differences have to be taken into account when performing in vitro studies on cellular signalling pathways.
We are indebted to Sibylla Lodermeier and Susanne Wallner for excellent technical assistance, Dr. Gerhard Rogler for help with the isolation and cultivation of CECs, and Randall T. Moon for providing material and helpful discussions.
S- Editor Wang J L- Editor Kumar M E- Editor Bi L
| 1. | Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735-24738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 552] [Cited by in F6Publishing: 583] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 2. | Laurent-Puig P, Blons H, Cugnenc PH. Sequence of molecular genetic events in colorectal tumorigenesis. Eur J Cancer Prev. 1999;8 Suppl 1:S39-S47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1-24. [PubMed] [Cited in This Article: ] |
| 4. | Behrens J, Lustig B. The Wnt connection to tumorigenesis. Int J Dev Biol. 2004;48:477-487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 119] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 5. | Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905-909. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 221] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 6. | Zhou CZ, Qiu GQ, Zhang F, He L, Peng ZH. Loss of heterozygosity on chromosome 1 in sporadic colorectal carcinoma. World J Gastroenterol. 2004;10:1431-1435. [PubMed] [Cited in This Article: ] |
| 7. | Gad S, Teboul D, Lièvre A, Goasguen N, Berger A, Beaune P, Laurent-Puig P. Is the gene encoding Chibby implicated as a tumour suppressor in colorectal cancer ? BMC Cancer. 2004;4:31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Behrmann I, Wallner S, Komyod W, Heinrich PC, Schuierer M, Buettner R, Bosserhoff AK. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol. 2003;163:683-690. [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 9. | Kiessling S, Muller-Newen G, Leeb SN, Hausmann M, Rath HC, Strater J, Spottl T, Schlottmann K, Grossmann J, Montero-Julian FA. Functional expression of the interleukin-11 receptor alpha-chain and evidence of antiapoptotic effects in human colonic epithelial cells. J Biol Chem. 2004;279:10304-10315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Dahiya R, Lesuffleur T, Kwak KS, Byrd JC, Barbat A, Zweibaum A, Kim YS. Expression and characterization of mucins associated with the resistance to methotrexate of human colonic adenocarcinoma cell line HT29. Cancer Res. 1992;52:4655-4662. [PubMed] [Cited in This Article: ] |
| 11. | Kitamura H, Cho M, Lee BH, Gum JR, Siddiki BB, Ho SB, Toribara NW, Lesuffleur T, Zweibaum A, Kitamura Y. Alteration in mucin gene expression and biological properties of HT29 colon cancer cell subpopulations. Eur J Cancer. 1996;32A:1788-1796. [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Lesuffleur T, Kornowski A, Augeron C, Dussaulx E, Barbat A, Laboisse C, Zweibaum A. Increased growth adaptability to 5-fluorouracil and methotrexate of HT-29 sub-populations selected for their commitment to differentiation. Int J Cancer. 1991;49:731-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Lesuffleur T, Porchet N, Aubert JP, Swallow D, Gum JR, Kim YS, Real FX, Zweibaum A. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J Cell Sci. 1993;106:771-783. [PubMed] [Cited in This Article: ] |
| 14. | Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK. Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res. 2004;64:5186-5192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 153] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Higgins KA, Perez JR, Coleman TA, Dorshkind K, McComas WA, Sarmiento UM, Rosen CA, Narayanan R. Antisense inhibition of the p65 subunit of NF-kappa B blocks tumorigenicity and causes tumor regression. Proc Natl Acad Sci U S A. 1993;90:9901-9905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 194] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Rubinfeld B, Albert I, Porfiri E, Munemitsu S, Polakis P. Loss of beta-catenin regulation by the APC tumor suppressor protein correlates with loss of structure due to common somatic mutations of the gene. Cancer Res. 1997;57:4624-4630. [PubMed] [Cited in This Article: ] |
| 17. | Trainer DL, Kline T, McCabe FL, Faucette LF, Feild J, Chaikin M, Anzano M, Rieman D, Hoffstein S, Li DJ. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 1988;41:287-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 132] [Article Influence: 3.7] [Reference Citation Analysis (0)] |