INTRODUCTION
Survivin, a structurally unique member of the inhibitors of apoptosis protein (IAP)family, is involved in the control of cell division and inhibition of apoptosis. Survivin inhibits apoptosis via its baculovirus inhibitor of apoptosis repeat (BIR) protein domain by either directly or indirectly interfering with the function of caspases. Survivin is also a chromosomal passenger protein that is required for cell division. Survivin is expressed in embryonic tissues as well as in the majority of human cancers, but not in most normal adult tissues. The cancer-specific expression of survivin, coupled with its importance in inhibiting cell apoptosis and regulating cell division, makes it a useful diagnostic marker and a potential target for cancer treatment[1].
RNA interference (RNAi) is a process of post transcriptional gene silencing (PTGS) in which double-stranded RNA (dsRNA) inhibits gene expression in a sequence-dependent manner via degradation of the corresponding mRNA [2-5]. RNAi is initiated by an event while dsRNAs are recognized by Dicer, a member of the RNase III protein family[6]. The Dicer enzyme cleaves dsRNAs into 21 to 23 nucleotide short siRNAs. These siRNA duplexes are incorporated into a protein complex called the RNA-induced silencing complex, which recognizes and cleaves the cognate mRNA[7,8]. Recently, Elbashir and his colleagues reported that introduction of synthetic 21nt siRNA into mammalian cells could suppress gene expression in a sequence-specific manner[9]. By using dsRNAs of such a small size, the activation of dsRNA-dependent protein kinase and 2’, 5’- oligoadenylate synthetase, which occurs as an interferon response to long dsRNA (i.e. longer than 30nt), leading to a global decrease in protein synthesis, is avoided [10,11]. Thus, siRNA has now become a powerful tool for studies on gene function, cancer and viral disease therapy. In this study, a siRNA against survivin plasmid expression vector was constructed and transfected into PC-2 cells. The changes of survivin expression following RNAi and the role of siRNA in inducing PC-2 cell apoptosis and enhancing its radiosensitivity were investigated, which may lay a foundation for further studies on the functions of survivin and genetic therapy involved in human pancreatic cancer.
MATERIALS AND METHODS
Cell line and culture
Human pancreatic cancer cell line PC-2 was purchased from the Medical Experimental Animal Center of the Fourth Military Medical University. The cells were cultured in RPMI 1640 medium supplemented with 100 mL/L fetal bovine serum (FBS), 1×105 U/L penicillin and 0.1 g/L streptomycin in a humidified incubator containing 50 mL/L CO2 at 37 °C.
Construction of siRNA plasmid expression vector against survivin
Plasmid vector Pgenesil-1 was purchased from Wuhan Genesil Biotechnology Co, Ltd. Aiming at the sequence of survivin[12] (5’-GGA CCA CCG CAT CTC TAC A-3’), two DNA chains were synthesized as following: 5’-GATCC GGA CCA CCG CAT CTC TAC A TTCAAGACG TGT AGA GAT GCG GTG GTC C TTTTTT GAATTC A-3’ and 3’-G CCT GGT GGC GTA GAG ATG T AAGTTCTGC ACA TCT CTA CGC CAC CAG G AAAAAA CTTAAG TTCGA-5’. The structure of the DNA chains is BamHI + sense chain + loop + antisense chain + termination signal + EcoRI + HindIII. Equal amount of the two DNA chains was annealed and phosphorylated, then the double-stranded DNA was ligated with the linearized plasmid vector Pgenesil-1 completely digested with BamHI and HindIII. The ligated plasmid DNA was transfected into competent cells DH5α which were then seeded on solid LB medium containing 0.05 g/L kanamycin and cultured at 37 °C overnight. Three monoclonal colonies were picked out and seeded in 3 mL LB culture fluid containing 0.05 g/L kanamycin and cultured at 37 °C overnight in a rocking bed. The plasmid was extracted and verificated by digesting with restriction endonucleases and sequencing. The new plasmid was named Pgenesil-sur(+). The negative control plasmid Pgenesil-sur(-) was constructed as the same process. The target sequence is 5’-GAC TTC ATA AGG CGC ATG C-3’ and does not have any homology with human beings or mice. The sequences of the two DNA chains are 5’-GATCC GAC TTC ATA AGG CGC ATG C TTCAAGACG GCA TGC GCC TTA TGA AGT C TTTTTT GTCGAC A-3’ and 3’-G CTG AAG TAT TCC GCG TAC G AAGTTCTGC CGT ACG CGG AAT ACT TCA G AAAAAA CAGCTG TTCGA -5’. Their structure is BamHI + sense chain + loop + antisense chain + termination signal + SalI + Hind III.
Extraction of plasmids
The plasmid extraction kit without endotoxin was purchased from Beijing Tianwei Corporation. The plasmids were extracted following the manufacturer’ s instructions. The concentration and purity of the plasmids were detected with an ultraviolet spectrophotometer. The plasmids were stored at -20 °C for following experiments.
Transfection of plasmids
Transfection reagent LipofectamineTM2000 was purchased from Invitrogen Corporation. Transfection was performed following the manufacturer’ s instructions. The ratio of plasmids: LipofectamineTM2000 was 1:3. Twenty-four hours before the transfection, the common complete medium was replaced by the antibiotics- free medium containing serum. Six hours after the transfection, the medium was replaced by the common complete medium again. Twelve hours after the transfection, expression of EGFP in PC-2 cell was observed under an inverse fluorescence microscope.
Expression of survivin mRNA detected by semi-quantitive RT-PCR
PC-2 cells were seeded in 6cm culture capsules and divided into blank control group, negative control group and positive experiment group. Each group contained 3 culture capsules. Only LipofectamineTM2000 was used for the transfection in the blank control group, plasmid Pgenesil-sur(-) was used for the transfection in the negative control group and plasmid Pgenesil-sur(+) was used for the transfection in the positive experiment group. Twenty-four hours after the transfection, 1×106 cells were collected and total RNA was extracted using the Trizol reagent following the manufacturer’ s instructions. The concentration and purity of the total RNA were detected with ultraviolet spectrophotometer. RT-PCR was performed by two-step method. Synthesis of cDNA was performed using the RevertAidTM first chain cDNA synthesis kit following the manufacturer’ s instructions. PCR was performed using the one tube convenient PCR kit purchased from Beijing Tianwei Corporation. Amplification of human GAPDH served as an internal standard. The primers used are 5’-CGA AGT CAA CGG ATT TGG TCG TAT-3’ (forward primer) and 5’-AGC CTT CTC GGT GGT GAA GAC-3’ (reverse primer). The amplification product was 306bp.The primers of survivin are 5’-GCA TGG GTG CCC CGA CGT TG-3’ (forward primer) and 5’-GCT CCG GCC AGA GGC CTC AA -3’ (reverse primer). The amplification product was 447bp. The PCR products were separated in 1.5% agarose gels, visualized by staining with ethidium bromide. Semi-quantitative analysis was performed with the Dolphin 1D software. The expression intensity of survivin was denoted with the ratio of the photodensity of the RT-PCR products of survivin and GAPDH. The inhibition ratio of survivin expression was calculated with the following formula: inhibition ratio of survivin expression = (1-the expression intensity of survivin in the observation group/ the expression intensity of survivin in the blank control group)×100%.
Expression of survivin protein detected by immunohistochemical method
PC-2 cells were seeded in 24-well plates with a small square cover slip in each well. Grouping and transfection were performed the same as above. Each group contained 8 wells. Twenty-four hours after the transfection, the cells were fixed with cold 65% liquor pyroaceticus. The expression of survivin protein was detected by immunohisto- chemical SP method. The anti-survivin antibody was a rabbit anti-human monoclonal antibody purchased from Beijing Zhongshan Corporation. The working concentration was 1:50. A petroline section of breast cancer expressing survivn positively was used as a positive control, PBS was used to replace the anti-survivn antibody to make a negative control. Cells with brown-yellow granulations in endochylema or nuclei were considered as positive cells. According to the coloration, the cells were scored as following: 0, no staining; 1, faint-yellow; 2, brown-yellow; 3, dark-brown. Five fields of vision of each cover slip were observed under high power lens randomly. The cells were calculated and scored. The sum of the scores was divided by the number of cells, the result was considered as the expression intensity of survivin. The inhibition ratio of survivin expression was calculated with the following formula: inhibition ratio of survivin expression = (1-the expression intensity of survivin in the observation group/ the expression intensity of survivin in the blank control group)×100%.
Radiation and apoptosis of PC-2 cells detected by flow cytometry
Cells were seeded in 25 cm2 culture flasks and divided into blank control group, negative control group, positive experiment group, blank control + radiation group, negative control + radiation group, and positive experiment + radiation group. Each group contained 5 culture flasks. Transfection was performed the same as above. Twenty-four hours after the transfection, cells of groups 4, 5, 6 were radiated with 15 Gy of X-ray(6MV) at room temperature using a Varian 2300 C/D linear accelerator. The radiation conditions were ounce-skin distance = 100 cm, radiation dose rate =2 Gy/min. After radiation, the cells were continuously cultured for another 24 h, digested, collected and washed twice with PBS. Then the cells were fixed with cold 75% ethanol at 4 °C overnight. The cells were centrifuged at 1000 r/ min for 5 min, the supernate was poured out, the cells were washed twice with PBS. The cells were resuspended with PBS containing 0.05 g/L RNase and incubated for 30 min at room temperature avoiding light to eliminate the intracellular RNA. The cells were centrifuged at 1000 r/min for 5 min, the supernate was poured out. The cells were resuspended with PBS containing 0.06 g/L propidium iodode again and incubated for 30 min at room temperature avoiding light, then cell apoptosis was detected by flow cytometry. The management was the same in cells of groups 1-3 except for no radiation.
Statistical analysis
Data were expressed as mean ± SD. All statistical analyses were performed using SPSS 10.0 software( one-way ANOVA). P < 0.05 was considered statistically significant.
RESULTS
Verification of plasmids
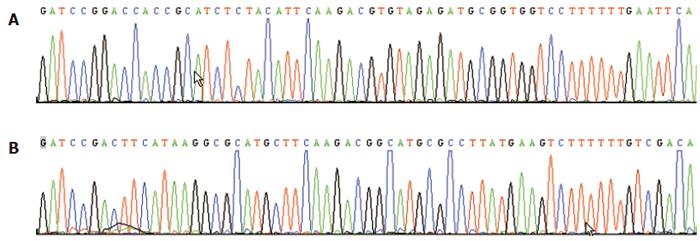
The multiclone sites of plasmid Pgenesil-1 were as following:HindIII-XbaI-SalI -PstI-BamHI-U6 Promotor-EcoRI-SalI-XbaI-DraIII. An EcoRI site for plasmid Pgenesil-sur(+) and a SalI site for plasmid Pgenesil-sur(-) were designed in the inserted fragments between the sites of BamHI and HindIII. If the insertion was correct, a band about 400 bp should be cut off by EcoRI or SalI. The results of digestion with restriction endonucleases and sequencing showed correct plasmids(Figure 1).
Figure 1 Reports of the sequencing of plasmid Pgenesil-sur(+) (A) and plasmid Pgenesil-sur (-) (B).
Results of transfection
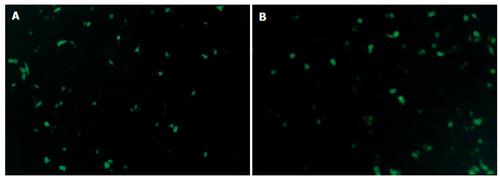
Twelve hours after plasmid Pgenesil-sur(+) or plasmid Pgenesil-sur(-) was transfected into PC-2 cells using LipofectamineTM2000, expression of EGFP could be observed under an inverse fluorescence microscope. Twenty-four hours after the transfection, the expression of EGFP was the strongest. Seventy-two hours after the transfection, the expression of EGFP was attenuated gradually(Figure 2).
Figure 2 Expression of EGFP in PC-2 cells 24 h after trancfection of plasmid Pgenesil-sur(+) (A) and plasmid Pgenesil-sur(-) (B) (original magnification×200).
Significant down regulation of survivin mRNA expression by siRNA
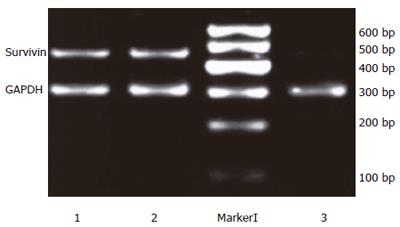
Twenty-four hours after the transfection, the expression intensities of survivin mRNA in blank control, negative control and positive experiment groups were 0.96±0.02, 0.98 ± 0.03 and 0.18 ± 0.03, respectively. The statistical analysis showed that the expression of survivin mRNA in PC-2 cells was down regulated significantly after transfection with plasmid Pgenesil-sur(+) (P < 0.05), the inhibition ratio was 81.25%. Then expression of PC-2 cells transfected with plasmid Pgenesil-sur(-) had no effect on the expression of survivin mRNA (P > 0.05)(Figure 3).
Figure 3 Down-regulation of survivin mRNA expression detected by semi-quantitive RT-PCR.
1: The blank control group; 2: The negative control group; M: Marker I; 3: The positive experiment group.
Significant down regulation of Survivin protein expression by siRNA
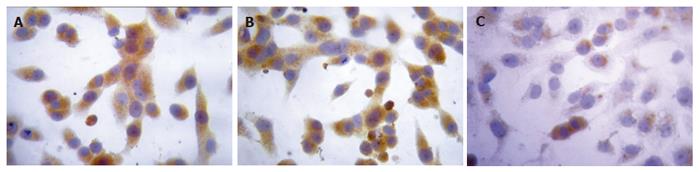
Twenty-four hours after the transfection, the expression intensities of survivin protein in blank control, negative control and positive experiment groups were 2.29 ± 0.23, 2.34 ± 0.15 and 0.59 ± 0.16, respectively. The statistical analysis showed that the expression of survivin protein in PC-2 cells was down- regulated significantly after transfected with plasmid Pgenesil-sur(+) (P < 0.05), the inhibition ratio was 74.24%. The expression of PC-2 cells transfected with plasmid Pgenesil-sur(-) had no effect on the expression of survivin protein (P > 0.05) (Figure 4).
Figure 4 Down-regulation of survivin expression detected by immunohisto- chemical SP method in the blank control group (A), negative control group (B), and positive experiment group (C) (coloration with DAB, original magnification × 400)
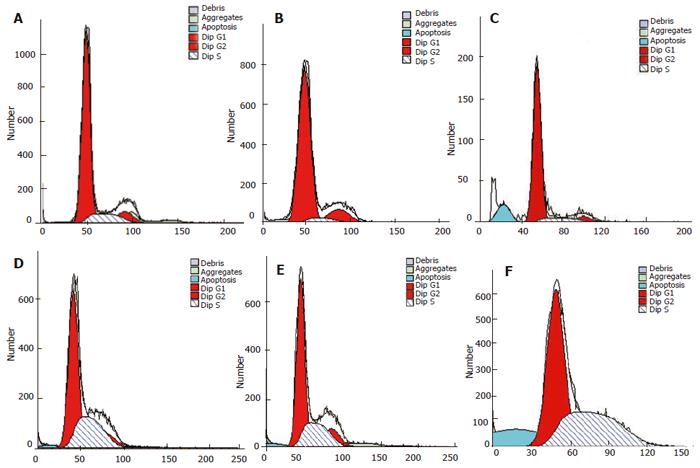
Role of siRNA in inducing PC-2 cell apoptosis and enhancing its radiosensitivity detected by flow cytometry
Forty-eight hours after transfection, no cell apoptosis was detected in blank control and negative control groups, but in (7.03 ± 1.12)% cells in the positive experiment group. The apoptosis rate in blank control + radiation group, negative control + radiation group and positive experiment + radiation group was (1.77 ± 0.30 )%, (1.66 ± 0.46 )% and (14.58 ± 2.72)%, respectively. The statistical analysis showed that transfection with plasmid Pgenesil-sur(+) could induce spontaneous apoptosis of PC-2 cells in a certain degree and the effect was better than that of 15 Gy of X-ray radiation. Inhibiting the expression of survivin could enhance the radiosensitivity of PC-2 cells significantly. siRNA against survivin combined with radiation could induce significant apoptosis of PC-2 cells (Figure 5).
Figure 5 Apoptosis of PC-2 cells detected in 6 groups by flow cytometry (stained with PI)
DISCUSSION
Pancreatic cancer is one of the most malignant tumors with a very poor prognosis. The 5-year survival rate of pancreatic cancer patients receiving surgery and chemotherapy ranges 1%-2%. One of the reasons for this low survival rate is the insensitivity of pancreatic cancer to most oncologic therapies such as chemotherapy, radiotherapy and immunotherapy. Tumor development and progression as well as resistance to most oncologic therapies result mainly from lacking response to apoptotic stimuli[13].
Survivin, a member of the IAP family, is a bifunctional protein that suppresses apoptosis and regulates cell division. Recent studies showed that there is a close relationship between survivin and malignant tumors. The expression of survivin is highly cancer-specific, and is one of the top four transcripts uniformly up-regulated in human cancers, but not in normal tissues[14]. The overexpression of survivin appears to correlate with aggressive tumor behavior and poor prognosis in colorectal cancer[15-17], neuroblastomas[18], melanomas[19], non small cell lung cancers[20], breast carcinomas[21], esophageal squamous cell carcinomas[22] and soft-tissue sarcoma[23]. Additionally, survivin overexpression is also correlated with insensitivity to chemotherapy and radiotherapy in cancers[24,25]. Inhibiting the expression of survivin can induce tumor cell apoptosis, sensitize tumor cells to chemotherapy, radiotherapy and inhibit tumor-angiogenesis[26-29]. Survivin has become an ideal target for the diagnosis and treatment of cancer.
Studies on the relationship between the expression of survivin and pancreatic cancer showed that survivin is also overexpressed in pancreatic cancer, and participates in the development and progress of pancreatic cancer by inhibiting apoptosis. Its overexpression is associated with the insensitivity of pancreatic cancer to chemotherapy and radiotherapy, and is a poor prognosis marker[30-33]. All these studies suggest that survivin is of great potential value in the treatment of pancreatic cancer.
The mechanisms by which survivin inhibits cell apoptosis and cell division are still highly controversial. Survivin may regulate apoptosis by directly or indirectly inhibiting the activity of caspases (caspase-3, -7, -6, -8, -9, -10), or by interacting with cdk4 which releases P21 from its complex with cdk4 making it possible for P21 to complex with caspase-3, the initial step in inactivating caspase-3 in mitochondria. Survivin may possibly function to inhibit the activity of caspases indirectly via binding to and sequestering Smac/DIABLO, thus preventing Smac/DIABLO binding to other IAPs. Survivin might be an additional chromosomal passenger protein just like inner centromere protein (INCENP), TD-60 and Aurora-B, and is important for cytokinesis and chromosome movement during cell division. Overexpression of survivin may overcome the G2/M phase checkpoint to enforce progression of cells through mitosis[1].
RNAi is characterized by high efficiency, high specificity and low toxicity. siRNA has become a powerful tool for studies on gene function, cancer and viral disease therapy. In this study, we designed and constructed the siRNA plasmid expression vector. Its transcript can form a short hairpin RNA(shRNA) with inverted repeat sequence separated by a short loop sequence, then the shRNA is processed into functional siRNA to degradate target mRNA and silences its expression. The siRNA plasmid expression vector constructed in this study contains the EGFP gene that makes it convenient to observe the result of transfection. By aiming at different sequence of the target gene, the effect of RNAi may be greatly different [34,35] suggesting that it is very important to find the proper sequence. The sequence of Kappler consulted in this study has been confirmed effective in multiple tumor cell lines[12,36]. The results of our studies also demonstrated that it was effective both in pancreatic cancer cell line PC-2 and in breast cancer cell line MCF-7 (data not shown), suggesting that this sequence may be of universality in malignant tumors and should be a preferential sequence when study on the RNAi against surviving is carried out.
In this study, we investigated whether the sensitivity to X-irradiation could be affected by inhibiting survivin expression with siRNA. The results showed that the siRNA against survivin plasmid expression vector constructed in this study had highly specificity, and could suppress the expression of survivin in PC-2 cells efficiently suggesting that inhibiting the expression of survivin can induce spontaneous apoptosis of pancreatic cancer cells in a certain degree and enhance its radiosensitivity significantly. siRNA against survivin is of potential value in the treatment of pancreatic cancer.