Published online Nov 28, 2011. doi: 10.3748/wjg.v17.i44.4917
Revised: August 6, 2011
Accepted: August 13, 2011
Published online: November 28, 2011
AIM: To detect the effects of plasma DNA methylation of Wnt antagonists/inhibitors on recurrence of esophageal squamous cell carcinoma (ESCC).
METHODS: We used methylation-specific polymerase chain reaction to detect hypermethylation of the promoter of four Wnt antagonists/inhibitors (SFRP-1, WIF-1, DKK-3 and RUNX3) using DNA from the plasma of ESCC patients (n = 81) and analyzed the association between promoter hypermethylation of Wnt pathway modulator genes and the two-year recurrence of ESCC.
RESULTS: Hypermethylation of SFRP-1, DKK-3 and RUNX-3 was significantly associated with an increased risk of ESCC recurrence (P = 0.001, 0.003 and 0.001 for SFRP-1, DKK-3 and RUNX3, respectively). Patients carrying two to three methylated genes had a significantly elevated risk of recurrence compared with those not carrying methylated genes (odds ratio = 15.69, 95% confidential interval: 2.97-83). The area under the receiver operating characteristic curve (AUC) was 77.1 for ESCC recurrence prediction (sensitivity = 66.67 and specificity = 83.3). When combining methylated genes and the clinical stage, the AUC was 83.69, with a sensitivity of 76.19 and a specificity of 83.3.
CONCLUSION: The status of promoter hypermethylation of Wnt antagonists/inhibitors in plasma may serve as a non-invasive prognostic biomarker for ESCC.
- Citation: Liu JB, Qiang FL, Dong J, Cai J, Zhou SH, Shi MX, Chen KP, Hu ZB. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol 2011; 17(44): 4917-4921
- URL: https://www.wjgnet.com/1007-9327/full/v17/i44/4917.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i44.4917
Esophageal cancer, predominantly esophageal squamous cell carcinoma (ESCC), is one of the most common malignancies in the world, with 482 300 incident cases and 406 800 deaths estimated in 2008[1]. Most patients with ESCC are diagnosed at an advanced stage. Although therapeutic advances have been designed to improve treatment outcome, the prognosis of patients with ESCC is still poor, with long-term survival rates between 5% and 20%[2]. The poor prognosis is partially due to the high rate of recurrence, which occurs in approximately half of patients after curative surgical resection. Therefore, development of accurate prognostic biomarkers for ESCC is imperative and crucial for improving ESCC prognosis and for guiding treatment.
DNA methylation is one of the most common epigenetic modifications[3]. One of the major changes caused by DNA methylation is transcriptional silencing of tumor suppressor genes resulting from hypermethylation of CpG islands in promoter regions[4]. This change occurs frequently during tumor pathogenesis and progression and has become widely recognized as a mechanism of gene inactivation in cancer[5-7]. The methylation profile may also help predict the response to a chemo-/radio-therapeutic agent and thus the prognosis of cancer[8,9]. In addition, detection of promoter CpG methylation in body fluid DNA is feasible and nearly non-invasive[10].
Wnt signaling operates across cell boundaries via secretion by cells of one tissue type, which results in activation of surface receptors on neighboring cells and tissues, leading to activation of transcription factors that regulate cell proliferation, survival, and differentiation[11]. Dysregulation of these processes in cancer results in aberrant activation of the Wnt pathway[11,12]. Several antagonists of Wnt signaling have been identified, including the secreted frizzled-related protein-1 (SFRP-1) and Wnt inhibitory factor-1 (WIF-1), which bind directly to Wnt proteins, and Dickkopf-3 (DKK-3), which binds to the LDL-receptor-related protein5 (LRP5)/LRP6 component of the Wnt receptor complex[13]. In addition, another Wnt inhibitor, runt-related transcription factor-3 (RUNX3), reportedly forms a ternary complex with β-catenin/transcription factor-4 (TCF4) to attenuate Wnt signaling, which regulates cell proliferation, apoptosis, and invasion[14,15]. Given the important roles of Wnt antagonists/inhibitors in cancer progression and prognosis, we evaluated the association between methylation of promoter CpG islands of the four tumor suppressor genes, SFRP-1, WIF-1, DKK-3 and RUNX3, in the Wnt signaling pathway and ESCC recurrence using plasma DNA from 81 Chinese patients with ESCC.
This study was approved by the institutional reviewer board of Nantong Cancer Hospital, and written informed consent was obtained from each patient or from the patient’s representative. Briefly, patients with histopathologically diagnosed incident ESCC were recruited from Nantong Cancer Hospital from June to December 2008. Exclusion criteria included self-reported previous cancer history, metastasis from other organs, and surgical section, radiotherapy, or chemotherapy before blood collection. Patients were followed up at the 24th month after recruitment by personal or family contacts. All patients were genetically unrelated, ethnic Han Chinese, and each patient donated 5 mL of venous blood at their first admission to the hospital. As a result, 81 ESCC patients who had complete clinical information, adequate DNA samples, and successful follow-up were included.
DNA was extracted from 200 μL plasma from each patient using an SG Spin Column Clinical Sample Genomic DNA MiniPreps kit (Shanghai ShineGene Molecular Biotech, Inc., Shanghai, China). The plasma DNA was modified with sodium bisulfite using the EZ DNA methylation™ kit (Zymo Research, Orange, CA, United States). The methylation status of CpG islands in the promoter region of SFRP-1, WIF-1, DKK-3 and RUNX-3 was determined by methylation-specific polymerase chain reaction (PCR)[16]. In brief, the first universal primer set covered no CpG sites in either the forward or the reverse primer but amplified a DNA fragment of the promoter region containing several sites. Then, a second round of nested methylation-specific PCR or unmethylation-specific PCR was performed using the universal PCR products as templates. Primer sequences are shown in Table 1.
| Gene | Primer sequence (5'–3') | |
| WIF-1 | ||
| U | F | GGGTGTTTTATTGGGTGTATTGT |
| R | AAAAAAACTAACACAAACAAAATACAAAC | |
| M | F | CGTTTTATTGGGCGTATCGT |
| R | ACTAACGCGAACGAAATACGA | |
| RUNX-3 | ||
| U | F | TTATGAGGGGTGGTTGTATGTGGG |
| R | AAAACAACCAACACAAACACCTCC | |
| M | F | TTACGAGGGGCGGTCGTACGCGGG |
| R | AAAACGACCGACGCGAACGCCTCC | |
| SFRP-1 | ||
| U | F | GAGTTAGTGTTGTGTGTTTGTTGTTTTGT |
| R | CCCAACATTACCCAACTCCACAACCA | |
| M | F | GTGTCGCGCGTTCGTCGTTTCGC |
| R | AACGTTACCCGACTCCGCGACCG | |
| DKK-3 | ||
| U | F | TTAGGGGTGGGTGGTGGGGT |
| R | CTACATCTCCACTCTACACCCA | |
| M | F | GGGGCGGGCGGCGGGGC |
| R | ACATCTCCGCTCTACGCCCG |
Differences in demographic and clinical characteristics were evaluated with χ2 tests (or the Fisher’s exact test). The association between methylation and ESCC recurrence was estimated by computing odds ratios (ORs) and 95% confidence intervals (CIs) from logistic regression analyses for adjusted ORs and crude ORs, with or without adjustments for age, gender, smoking, drinking, clinical stage, surgical operation and chemo-/radio-therapy status. Receiver operating characteristic (ROC) curve analysis was conducted by using the “pROC” package in R. All the statistical analyses were performed with R software (version 2.11.1; The R Foundation for Statistical Computing).
The clinicopathologic features of the patients are summarized in Table 2. A total of 81 ESCC patients were included in our current study. The mean age was 63 years (range, 46-80 years), and 61 patients were male (75.3%). Sixty-five (80.2%) patients underwent chemo-/radio-therapy, and 31 (38.3%) patients underwent surgical operation after blood collection. Among the 81 patients, there were 21 recurrences (26%) at the 24th month after recruitment. As expected, advanced stage was a risk factor for ESCC recurrence (P < 0.001). However, there were no significant differences in the recurrence rates among the subgroups of age, gender, smoking status, drinking status, with or without surgical operation, and/or chemo-/radio-therapy.
| Variables | Recurrence | P value | |
| No | Yes | ||
| Age (yr) | 0.804 | ||
| < 63 | 31 | 10 | |
| ≥ 63 | 29 | 11 | |
| Gender | 0.569 | ||
| Female | 16 | 4 | |
| Male | 44 | 17 | |
| Smoking | 0.614 | ||
| No | 31 | 9 | |
| Yes | 29 | 12 | |
| Drinking | 0.799 | ||
| No | 26 | 8 | |
| Yes | 34 | 13 | |
| Operation | 0.614 | ||
| No | 38 | 12 | |
| Yes | 22 | 9 | |
| Chemo-/radio-therapy | 0.751 | ||
| No | 11 | 5 | |
| Yes | 49 | 16 | |
| Stage | < 0.001 | ||
| I | 25 | 3 | |
| II | 22 | 3 | |
| III | 13 | 15 | |
The percentage of promoter methylation in the genes we analyzed was as follows: 29.6% for SFRP-1, 35.8% for WIF-1, 37.4% for DKK-3, and 35.8% for RUNX-3. Three genes showed significant associations with ESCC recurrence after adjusting for age, gender, smoking, drinking, stage, surgical operation, and chemo-/radio-therapy. As shown in Table 3, patients with methylated SFRP-1 had a 10.8-fold increased risk of recurrence compared with those with unmethylated SFRP-1 (95% CI: 2.54-46, P < 0.001). Similarly, methylated DKK-3 was associated with a 6.07-fold increased risk of recurrence (95% CI: 1.73-21.28, P = 0.003), and methylated RUNX-3 was associated with a 7.81-fold increased recurrence risk (95% CI: 2.30-47, P = 0.001). Methylation of WIF-1, however, was not significantly associated with an increased risk of recurrence (P = 0.107).
| Gene | n | Recurrence | Crude OR (95% CI) | Pvalue | Adjusted OR1 (95% CI) | P value1 | |
| No | Yes | ||||||
| SFRP-1 | |||||||
| Negative | 57 | 49 | 8 | 1.00 | < 0.001 | 1.00 | < 0.001 |
| Positive | 24 | 11 | 13 | 7.24 (2.42-21.68) | 10.8 (2.54-46) | ||
| WIF-1 | |||||||
| Negative | 52 | 42 | 10 | 1.00 | 0.069 | 1.00 | 0.107 |
| Positive | 29 | 18 | 11 | 2.57 (0.93-7.11) | 2.61 (0.8-8.47) | ||
| DKK-3 | |||||||
| Negative | 51 | 44 | 7 | 1.00 | 0.001 | 1.00 | 0.003 |
| Positive | 30 | 16 | 14 | 5.50 (1.88-16.08) | 6.07 (1.73-21.28) | ||
| RUNX-3 | |||||||
| Negative | 52 | 45 | 7 | 1.00 | < 0.001 | 1.00 | 0.001 |
| Positive | 29 | 15 | 14 | 6.00 (2.04-17.65) | 7.81 (2.30-47) | ||
We then performed combined analysis to assess the effect of methylation of the three genes (SFRP-1, DKK-3 and RUNX-3) on ESCC recurrence. As shown in Table 4, compared with patients without methylated genes, those with two to three methylated genes had a significantly increased risk of recurrence (OR = 15.69, 95% CI: 2.97-83). Furthermore, there was a dose-response effect between plasma DNA methylation status and ESCC recurrence (OR = 4.1, 95% CI: 1.73-9.72, P for trend: 0.001).
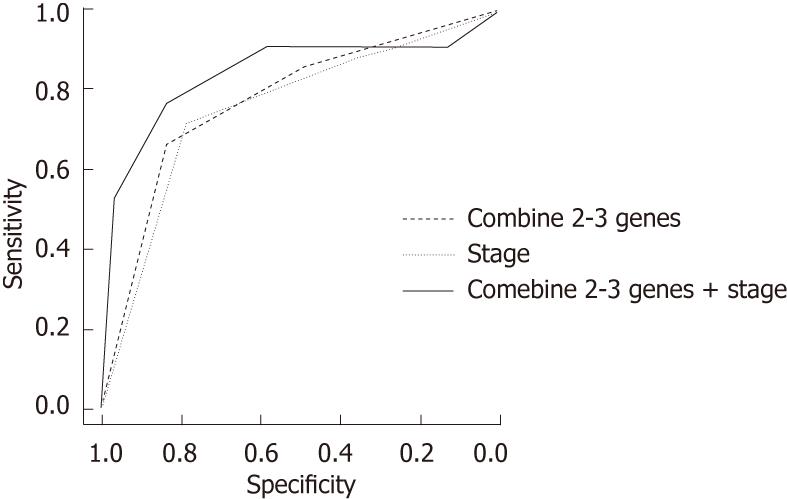
We also conducted a ROC curve analysis and area under curve (AUC) analyses to assess the sensitivity and specificity of the status of methylated genes individually and in combination for predicting ESCC recurrence. For single genes, the AUC were 71.79 (SFRP-1), 70 (DKK-3) and 70.83 (RUNX3). When we considered two to three methylated genes, the AUC increased to 77.1, with a sensitivity of 66.67 and a specificity of 83.3. As a risk factor for ESCC recurrence, the AUC for stage was 75.24 (sensitivity = 71.43, specificity = 78.3). For the combination of methylated genes and clinical stage, the AUC was 83.69 (sensitivity = 76.19, specificity = 83.3) (Figure 1).
Wnt antagonists/inhibitors function as tumor suppressors, and thus hypermethylation of their promoters and subsequent silencing of these genes may be implicated in the pathogenesis or progression of a broad spectrum of human malignancies[17-19]. In our current study, we found that the methylation status of SFRP-1, DKK-3 and RUNX-3 promoters in plasma DNA can individually and jointly predict ESCC recurrence.
Most published studies on the association between hypermethylation of Wnt antagonist/inhibitor gene promoters and cancer development have focused on tumor tissues. Yu et al[17] found that epigenetic silencing of DKK-3 is a common event in gastric cancer and is associated with a poor disease outcome. Urakami et al[18] used a methylation score to analyze the combined effects of hypermethylated Wnt antagonist family genes on bladder cancer detection, and they found that the score was significantly higher in bladder tumors than in bladder mucosa. Hamilton et al[9] assessed the methylation status of nine genes, including RUNX-3, in esophageal cancer patients and found that increased methylation of this gene correlated with poor responsiveness to therapy, suggesting potential clinical application of these biomarkers in guiding prognosis and management. A growing body of evidence suggests that the methylation signature is consistent between DNA derived from tissue and DNA derived from serum/plasma[19,20]. However, serum/plasma DNA has obvious advantages compared with DNA derived from tissue samples, i.e., with respect to the ease to obtain, non-invasiveness, and relative reproducibility. Therefore, serum/plasma DNA may be an excellent source of samples for assessing biomarkers, especially for patients who have not undergone surgery.
Previous studies have shown that methylation of various genes in serum/plasma can be a highly specific biomarker for human cancers[19,21-26]. Herbst et al[24] reported that NEUROG1 is frequently methylated in sera of patients with colorectal cancers independent of the tumor stage and is a suitable non-invasive screening approach for detecting asymptomatic colorectal cancer. Göbel et al[25] found that methylated PITX2 and RASSF1A in plasma are therapy-independent prognostic factors in breast cancer patients. Salazar et al[26] showed that the methylation status of CHFR in serum can influence the outcome of chemotherapy in stage IV non-small-cell lung cancer patients and that unmethylated CHFR predicts increased survival to EGFR TKIs. Urakami et al[19] reported that hypermethylation of Wnt antagonists can serve as an excellent epigenetic biomarker panel for detection, staging and prognosis of renal cell carcinoma using serum DNA. These studies all indicate the available and feasibility of using plasma DNA to establish the methylation status of gene promoters as a biomarker for cancer.
Given that ESCC is one of the most aggressive cancers in the world and that no non-invasive test is available, our preliminary study yielded a promising result. Limitations of our study include the lack of detection of DNA methylation in tissue and the unpredictability of the link between methylation status and gene expression. Nevertheless, the methylation status of Wnt antagonists/inhibitors may serve as a valuable biomarker for predicting ESCC recurrence.
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignancies in the world. The prognosis of patients with ESCC is poor, which is partially due to the high rate of recurrence. Therefore, development of accurate prognostic biomarkers for ESCC is imperative and crucial for improving ESCC prognosis and for guiding treatment.
Wnt antagonist/inhibitor genes function as tumor suppressors, and hypermethylation of their promoters and subsequent silencing of the genes, which can be detected in plasma DNA, may serve as a valuable non-invasive biomarker for predicting ESCC recurrence.
The association between methylation of promoter CpG islands of four tumor suppressor genes (SFRP-1, WIF-1, DKK-3 and RUNX3) in the Wnt signaling pathway and ESCC recurrence was evaluated using plasma DNA from ESCC patients.
This result offers great potential for using plasma DNA to analyze methylation as a non-invasive biomarker for predicting ESCC recurrence.
Epigenetic changes: Heritable changes in the gene structure that do not change the gene sequence. CpG islands: CpG-rich areas located in the promoter regions of many genes. CpG island methylation: The addition of a methyl group to a cytosine residue that is next to guanine.
The study aimed to verify the promoter hypermethylation status of four Wnt [antagonist genes odds ratio (OR) antagonists] (using methylation-specific polymerase chain reaction with OR with the methylation-specific polymerase chain reaction technique using) DNA from plasma of 81 ESCC patients. The article reports interesting results that are important to the field.
Peer reviewer: Dr. Yukie Sato-Kuwabara, Hospital A. C. Camargo, Teaching and Research International Center, Sao Paulo 01508-010, Brazil
S- Editor Sun H L- Editor Cant MR E- Editor Xiong L
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (3)] |
| 2. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 3. | Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632-4642. [DOI] [Cited in This Article: ] [Cited by in Crossref: 797] [Cited by in F6Publishing: 822] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 4. | Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China. 2010;4:378-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042-2054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2489] [Cited by in F6Publishing: 2369] [Article Influence: 112.8] [Reference Citation Analysis (0)] |
| 6. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2210] [Cited by in F6Publishing: 2104] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 7. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1674] [Cited by in F6Publishing: 1498] [Article Influence: 74.9] [Reference Citation Analysis (0)] |
| 8. | Hamilton JP, Sato F, Jin Z, Greenwald BD, Ito T, Mori Y, Paun BC, Kan T, Cheng Y, Wang S. Reprimo methylation is a potential biomarker of Barrett's-Associated esophageal neoplastic progression. Clin Cancer Res. 2006;12:6637-6642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | Hamilton JP, Sato F, Greenwald BD, Suntharalingam M, Krasna MJ, Edelman MJ, Doyle A, Berki AT, Abraham JM, Mori Y. Promoter methylation and response to chemotherapy and radiation in esophageal cancer. Clin Gastroenterol Hepatol. 2006;4:701-708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 64] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511-5517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 174] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 11. | Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394-1404. [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 443] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 12. | Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59-63. [PubMed] [Cited in This Article: ] |
| 13. | Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627-2634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1193] [Cited by in F6Publishing: 1205] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 14. | Subramaniam MM, Chan JY, Yeoh KG, Quek T, Ito K, Salto-Tellez M. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta. 2009;1796:315-331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 192] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 16. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [PubMed] [Cited in This Article: ] |
| 17. | Yu J, Tao Q, Cheng YY, Lee KY, Ng SS, Cheung KF, Tian L, Rha SY, Neumann U, Röcken C. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer. 2009;115:49-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109-2116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 147] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 19. | Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989-6997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 111] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 20. | Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-1665. [PubMed] [Cited in This Article: ] |
| 21. | Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67-70. [PubMed] [Cited in This Article: ] |
| 22. | Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, Lai PB, Lau WY, Hjelm NM, Johnson PJ. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59:71-73. [PubMed] [Cited in This Article: ] |
| 23. | Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892-895. [PubMed] [Cited in This Article: ] |
| 24. | Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110-1118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Göbel G, Auer D, Gaugg I, Schneitter A, Lesche R, Müller-Holzner E, Marth C, Daxenbichler G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res Treat. 2011;130:109-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Salazar F, Molina MA, Sanchez-Ronco M, Moran T, Ramirez JL, Sanchez JM, Stahel R, Garrido P, Cobo M, Isla D. First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer. 2011;72:84-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |