Published online Sep 28, 2013. doi: 10.3748/wjg.v19.i36.6044
Revised: April 4, 2013
Accepted: May 7, 2013
Published online: September 28, 2013
AIM: To evaluate the in vitro effect of amoxicillin and clarithromycin on the cag pathogenicity island (cag PAI).
METHODS: One hundred and forty-nine clinical isolates of Helicobacter pylori (H. pylori) cultured from gastric biopsies from 206 Colombian patients with dyspeptic symptoms from a high-risk area for gastric cancer were included as study material. Antimicrobial susceptibility was determined by the agar dilution method. Resistant isolates at baseline and in amoxicillin and clarithromycin serial dilutions were subjected to genotyping (cagA, vacA alleles s and m), Glu-Pro-Ile-Tyr-Ala (EPIYA) polymerase chain reaction and random amplified polymorphic DNA (RAPD). Images of the RAPD amplicons were analyzed by Gel-Pro Analyzer 4.5 program. Cluster analyses was done using SPSS 15.0 statistical package, where each of the fingerprint bands were denoted as variables. Dendrograms were designed by following Ward’s clustering method and the estimation of distances between each pair of H. pylori isolates was calculated with the squared Euclidean distance.
RESULTS: Resistance rates were 4% for amoxicillin and 2.7% for clarithromycin with 2% double resistances. Genotyping evidenced a high prevalence of the genotype cagA-positive/vacA s1m1. The 3’ region of cagA gene was successfully amplified in 92.3% (12/13) of the baseline resistant isolates and in 60% (36/60) of the resistant isolates growing in antibiotic dilutions. Upon observing the distribution of the number of EPIYA repetitions in each dilution with respect to baseline isolates, it was found that in 61.5% (8/13) of the baseline isolates, a change in the number of EPIYA repetitions lowered antibiotic pressure. The gain and loss of EPIYA motifs resulted in a diversity of H. pylori subclones after bacterial adjustment to changing conditions product of antibiotic pressure. RAPD PCR evidenced the close clonal relationship between baseline isolates and isolates growing in antibiotic dilutions.
CONCLUSION: Antibiotic pressure does not induce loss of the cag pathogenicity island, but it can lead - in most cases - to genetic rearrangements within the 3’ region cagA of the founding bacteria that can affect the level of tyrosine phosphorylation impacting on its cellular effects and lead to divergence of cagA-positive subclones.
Core tip: This study evaluated the in vitro effect of amoxicillin and clarithromycin on the cag pathogenicity island (cag PAI). It was found that the effect of antibiotic pressure does not induce loss of cag PAI, but it can lead - in most cases - to genetic rearrangements (loss or gain of Glu-Pro-Ile-Tyr-Ala motif) within the 3’ region of cagA gene of the founding bacteria that can affect the level of tyrosine phosphorylation impacting on its cellular effects and lead to divergence of cagA-positive subclones, which as a set could alter the pathogenic process of Helicobacter pylori in cases with treatment failure.
-
Citation: Bustamante-Rengifo JA, Matta AJ, Pazos A, Bravo LE.
In vitro effect of amoxicillin and clarithromycin on the 3’ region ofcagA gene inHelicobacter pylori isolates. World J Gastroenterol 2013; 19(36): 6044-6054 - URL: https://www.wjgnet.com/1007-9327/full/v19/i36/6044.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i36.6044
Helicobacter pylori (H. pylori) is a Gram-negative bacteria in spiral form and microaerophilic that infect the gastric mucosa of approximately 50% of the world’s population[1-4]. It is usually acquired during childhood[5] and when not treated it persists for decades causing clinically asymptomatic chronic gastritis[6-8], although histologically apparent. In a subset of infected individuals, the presence of the bacteria is associated to peptic ulcer, adenocarcinoma, and gastric lymphoma[3,6]. Clinical results of the infection are related to the host’s immunological defense mechanisms, environmental factors like cigarette smoking and excessive salt intake, diet low in antioxidants, phylogeographic origin[9], and the bacteria’s virulence capacity[8]. H. pylori uses many modalities to colonize the gastric epithelium and some of these adaptation strategies contribute to the progression of the disease[10], among them gene products encoded by cagA and vacA.
The cagA gene is a marker of the cag pathogenicity island, present in over 50% of the H. pylori strains[7], encodes for the CagA protein one of the main determinants of pathogenicity associated to infection by H. pylori[2,11,12]; followed by H. pylori adhesion to the gastric epithelium, CagA is translocated within the cytoplasm of the epithelial cell via the type IV secretion system, where kinases from the Src and Ab1 family phosphorylate it into specific tyrosine residues inside the Glu-Pro-Ile-Tyr-Ala (EPIYA) repetition motifs[1,2]. These EPIYA motifs can be repeated within the protein’s variable region[1], and are defined as EPIYA-A, -B, -C, and -D according to the amino acids surrounding them[2]. Species of nearby CagA proteins almost always contain EPIYA-A and EPIYA-B sites, followed by one to three repetitions of EPIYA-C in the H. pylori Western-type isolates (ABC, ABCC, and ABCCC) or one EPIYA-D site in East Asian-type isolates (ABD). CagA variability with respect to the EPIYA motifs can play an important role in H. pylori pathogenesis[2]. CagA-positive clinical isolates with increased number of EPIYA motifs obtained from Eastern populations have been associated to higher severity of active chronic gastritis and atrophy. Likewise, an increased number of EPIYA motifs within the Western-type CagA protein has been related to increased levels of phosphorylation, high secretion of interleukin-8, cell elongation, and formation of “hummingbird” phenotype[1,11]. Hence, determination of the number of EPIYA motifs within the cagA variable region in clinical H. pylori isolates is more important than the mere detection of the cagA[1] and can be useful in predicting the bacteria’s pathogenic activity[11].
The vacA gene is present in all H. pylori strains, although it is only expressed in 50%-65% of them[5,12], it encodes a vacuolating toxin of 88-kDa (VacA) that affects the gastric epithelial cells and is important in the pathogenesis of peptic ulcers and gastric adenocarcinoma[7]. Two regions of marked sequence diversity are distinguishable within the vacA gene. The s region (encodes the signal peptide) is present as s1 or s2 allele, while the m region (mid region) can be m1 or m2. The combination of the allele mosaic from the s and m region determines the production of vacuolating cytotoxin and is associated to the bacteria’s pathogenicity. The strains harbored vacA s1m1 have been strongly associated to increased virulence and greater epithelial gastric damage and ulceration than s2m2 strains[7,12]. Thus, cagA-positive and vacA s1m1 genotypes are associated to high risk of gastric cancer[7].
H. pylori has extraordinary allelic diversity and genetic variability[7,13], generated through an elevated rate of point mutations, intragenomic and intergenomic recombination[13], phenomena facilitated by the presence of reactive oxygen species (ROS) and reactive nitrogen species (RNS) produced from inflammatory lesions induced by the H. pylori infection. This complex environment propitiates a second-order selection that implies variation in mutator genes, creating a non-linear diversification system[14]. The fact that virulence factors in H. pylori are linked to disease implies that they are a fixed characteristic, but this is not the case and genotype variations can occur through genetic rearrangements[14] that eliminates particular immune-stimulating genetic regions (cag PAI) or causes variations in potential immune-stimulating molecules (number of EPIYA repetitions in CagA)[3,13], probably reflecting the local selection of H. pylori particular phenotypes[14] as an escape recourse to the immune response of the host induced by environmental pressures.
This variety of strategies and gene products permit H. pylori to persistently colonize its host and cause disease. Within this context, its eradication can contribute to the treatment and prevention of gastric pathologies associated to the infection. Substantial improvements have been accomplished in the efficacy of treatment regimes; nevertheless, all still present faults to completely eradicate the infection[15]. This study evaluated the in vitro effect of the antibiotics used in standard triple therapy over specific virulence factors like cag PAI, in H. pylori isolates from Colombian patients from a high risk region for gastric cancer, to understand the course of the infection in unsuccessful treatments.
During 2009, 206 volunteer subjects were recruited with symptoms of dyspepsia, 44.2% (91/206) men ranging in age from 18-68 years, in a population from Túquerres in the high Andes mountains of Colombia, characterized by a high prevalence of H. pylori and preneoplastic lesions[8,16]. Exclusion criteria included prior gastrectomy, chronic disease, intake of H2-receptor antagonists, proton pump inhibitors, or antibiotic intake during the last four weeks prior to endoscopy. During upper gastrointestinal tract endoscopy, biopsies were obtained of gastric antral and body and embedded in paraffin for histopathological evaluation. Additional biopsies, two antral (greater and lesser curvature) and one gastric body (greater curvature) were taken for H. pylori culture and immediately frozen in thioglycolate with glycerol. The samples were delivered in liquid nitrogen to the Department of Pathology at Universidad del Valle (Cali, Colombia) for analysis. This research was approved by the Ethics Committee at Universidad del Valle. All the participants provided informed consent.
Histopathological diagnosis for each participant was independently evaluated by expert pathologists in antral and body gastric sections stained with hematoxylin and eosin, according to the Sydney classification system[17]. The categories were non-atrophic gastritis (NAG), multifocal atrophic gastritis without intestinal metaplasia (MAG), intestinal metaplasia (IM), and dysplasia. Cases with discordant diagnosis were again revised and consensus was reached.
Fragments of antral and body gastric mucosa were homogenized under sterile conditions in 200 µL of saline solution 0.89% using a homogenizer (Kimble-Kontes, Vinelan, NJ, United States). The homogenized was placed on Columbia agar plates (Oxoid, Basingstoke, Hampshire, England) with defibrinated sheep blood at 7% plus selective supplement for H. pylori (Dent) containing Vancomycin (10 mg/L), Sodium cefsulodin (5 mg/L), Trimethoprim lactate (5 mg/L), and Amphotericin B (5 mg/L) (Oxoid, Basingstoke, Hampshire, England). The agar plates were incubated under microaerophilic conditions (6% O2, 6% CO2, 88% N2, using CampyPak Plus envelope, BBL, Nashville, TN, United States) at 37 °C from four to eight days until observing small gray translucent colonies. The typical colonies were sub-cultured and later identified as H. pylori through their morphology, Gram-stain, and biochemical analyses for oxidase, catalase, and urease activity.
Antimicrobial resistance was evaluated by agar dilution method according to guidelines from the Clinical and Laboratory Standards Institute[18] using Mueller-Hinton (MHA) agar (Merck KGaA, Darmstadt, Germany) supplemented with defibrinated sheep blood at 7%. Double serial clarithromycin and amoxicillin dilutions (Genfar laboratories, Bogotá, Cundinamarca, Colombia) of 0.25, 0.5, 1.0, 2.0, and 4.0 μg/mL were added to MHA plates. Bacterial isolates sub-cultured in Columbia agar for no more than 72 h were re-suspended in saline solution and turbidity was adjusted at a concentration equivalent to a McFarland 2 standard (containing 1 × 107 or 1 × 108 CFU/mL), 1 µL of the adjusted inoculum was placed directly onto each MHA agar plate with dilutions of the antibiotic. The minimum inhibitory concentration (MIC) of clarithromycin and amoxicillin was determined after 72 h of incubating the isolates under microaerophilic conditions at 37 °C, and it was recorded as the lowest concentration of the antibiotic that inhibits visible growth of the colonies[12]. An isolate was considered resistant to clarithromycin or amoxicillin when its MIC was ≥ 1.0 μg/mL. The H. pylori reference strain ATTC® 43504 was used as quality control strain to monitor MIC precision when using the agar dilution method, considering MIC cutoff points < 0.015 µg/mL as sensitive and > 0.12 µg/mL as resistant to clarithromycin and amoxicillin.
Bacterial DNA was extracted from pure H. pylori cultures that showed resistance to clarithromycin and/or amoxicillin in each of the serial dilutions and from the isolates themselves in baseline (without antibiotic pressure). In all cases, single colonies were taken with swabs and washed in a 1.5 mL tube with 200 µL of sterile saline solution (0.89%). Tube contents were homogenized in vortex for 10 s and centrifuged in a 320R universal micro-centrifuge (Hettich Inc, Tuttlingen, Germany) at 13000 rpm for 2 min at 4 °C. Thereafter, the supernatant was discarded and the bacterial pellet was re-suspended in 300 µL of Lysis buffer (3 μL of Tris HCl 1 mol/L, pH 8.0 (Promega, Madison, WI, United States), 3 μL of EDTA 0.5 mol/L (Calbiochem, Gibbstown, NJ, United States), 15 μL of SDS 10% (Calbiochem, Gibbstown, NJ, United States), 3 μL of proteinase K (10 mg/mL) (Invitrogen, Carlsbad, CA, United States), and 276 μL of distilled water) and incubated in dry well (Labnet, Edison, NJ, United States) at 56 °C for 18 h. Then, the proteinase K was inactivated by heating at 70 °C for 10 min and 120 µL of NaCl 5 mol/L (Calbiochem, Gibbstown, NJ, United States) was added, homogenized by vortex and centrifuged for 5 min at 13000 rpm; the supernatant was transferred to another tube in which two volumes of absolute alcohol were added (Mallinckrodt, St.Louis, Mo, United States), again centrifuged at 13000 rpm for 20 min, then discarding the supernatant and adding 200 µL of ethanol at 70%. Once more, it was centrifuged for 5 min at 10000 rpm and then the supernatant was discarded; each tube was dried by inversion at room temperature. Finally, 50 µL of TE buffer were added (Tris-HCl 10 mmol/L, pH 8.0 (Promega, Madison, WI, United States), EDTA 1 mmol/L, pH 8.0 (Calbiochem, Gibbstown, NJ, United States) and the DNA was stored at -20 °C. The yield and purity of the DNA was determined by optical density at 260/280 nm in Gene Quant II® spectrophotometer (Pharmacia Biotech, Piscataway, NJ, United States) according to manufacturer’s instructions.
To genotype the virulence of H. pylori isolates resistant in baseline to clarithromycin and amoxicillin, separate PCR reactions were carried out in a thermocycler (Swift MiniProTM, Esco Technologies, Hatboro, PA, United States) for vacA-s and vacA-m, cagA, cag empty-site. Four sets of primers were initially used in this study. To amplify the s region of the vacA gene, VA1F and VA1R primers were used, which amplify a fragment of 259 bp (allele s1) and 286 bp (allele s2). To detect the m region of the vacA gene, HPMGF and HPMGR primers were employed, which resulted in the amplification of a fragment of 401 bp (allele m1) and 476 bp (allele m2). The cagA gene was amplified to validate the ability of the EPIYA polymerase chain reaction (PCR) to detect the presence of the cag PAI, using cagAF and cagAR primers that amplify a fragment of 183 bp from the 5’ end of the cagA gene[19]. The negative isolates for the cagA gene were in all cases confirmed by cag empty-site PCR employing ES-F and Rnew-1R primers[7], upon detecting a fragment of 106 bp that indicated the loss of the cag PAI. Each experiment included a positive and negative reaction control.
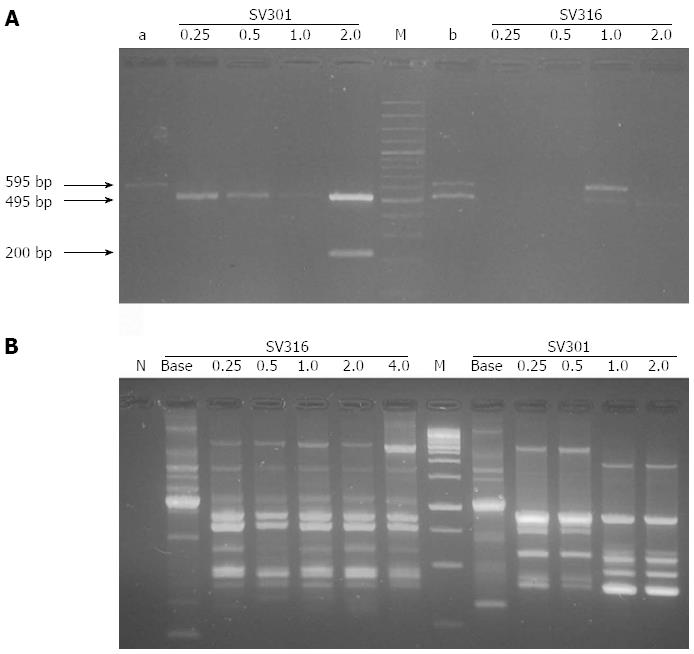
To detect the presence of the cag PAI and characterize the number of EPIYA motifs present in the 3’cagA variable region in resistant isolates at each of the antibiotic dilutions (with antibiotic pressure) and in same isolates in baseline (without antibiotic pressure), a PCR reaction was carried out using cagA2530S and cagA3000AS primers[11]. The size of the amplicons expected varied in the range of 370 bp (2 EPIYA motifs), 470 bp (3 EPIYA motifs), 570 bp (4 EPIYA motifs), 670 bp (5 EPIYA motifs) ± 25 bp and were approximately 100 bp equidistant, indicating the presence of multiple repeated sequences. In cases where a clear band was not evidenced or the presence of amplified, a second PCR was carried out under the same conditions, employing 1 µL of the initial amplified, along with the respective controls to detect possible cross contamination. During the development of the trials, H. pylori strain 26695 (ATCC 700392) were used (number of access AE000511.1) as size control of the nucleotide sequence encoding for three EPIYA motifs-ABC (470 bp ± 25 bp), and H. pylori strain ATCC 43504 that presents an EPIYA-ABCCC motif (670 ± 25 bp); thus, predicting the number of repetitions of the EPIYA motifs was possible by direct comparison of the sizes corresponding to PCR amplified.
The genomic differences between H. pylori isolates found before and after antibiotic pressure were evaluated by using random primers: 1254 (5’-CCGCAGCCAA-3’) and 1281 (5’-AACGCGCAAC-3’)[20]. These oligonucleotides were amplified in a final 12.5 µL reaction volume composed of 2.5 µL of PCR buffer (10 mmol/L of Tris-HCl pH 8.0 and 50 mmol/L of KCl-Promega, Madison, WI, United States), 3 mmol/L MgCl2, 1 U of Go taq polymerase (Promega, Madison, WI, United States), 250 μmol/L of dNTPs (Promega, Madison, WI, United States), 25 pmol of each primer, and 1 µL of bacterial DNA. The amplification was carried out in a thermocycler (Swift MiniProTM, Esco Technologies, Hatboro, PA, United States), prior denaturalization for 5 min at 94 °C, followed by 45 cycles of: 94 °C for 1 min, 36 °C for 1.30 min, 72 °C for 2 min, followed by a final incubation at 72 °C during 10 min. Band size was estimated by using a 100 bp DNA Ladder (Fermentas International Inc., Vilnius, Lithuania). Each isolate was tested with the primers described under the same conditions at least twice and only evident and reproducible bands were evaluated.
All the amplicons obtained in each of the previously described PCR reactions were run on agarose gel (SeaKim, FMC Bioloabs) at 2%, stained with ethidium bromide (Invitrogen, Carlsbad, CA, United States) at 0.5 µg/mL, in an electrophoresis chamber (Fotodyne Inc., Hartland, WI, United States) at 75 V for 40 min provided by a EC-105 Compact Power Supply (Thermo Fisher Scientific Inc, Asheville, NC, United States).
Initially, univariate and bivariate analyses were conducted; the categorical variables were described as proportions. To determine the statistical significance of the differences in the proportions of cag PAI detection via cagA PCR and EPIYA PCR among baseline resistant isolates and in amoxicillin and clarithromycin dilutions, the McNemar test was employed for data related to the SPSS 15.0 statistical package (SPSS Inc., Chicago, IL, United States). Images of the amplicons obtained via RAPD PCR were analyzed through the Gel-Pro Analyzer 4.5 program for Windows (Media Cibernetics, Inc, Rockville, MD, United States). Molecular weights of the well-defined, single-pattern bands were analyzed with this program and a matrix of binary data was constructed based on the presence (1) and absence (0) of the bands observed (polymorphic) among the isolates. Ratios among isolates were established through cluster analysis carried out with the SPSS 15.0 statistical package (SPSS Inc., Chicago, IL, United States), where each of the fingerprint bands were denoted as variables. The dendrograms were designed by following Ward’s clustering method and the estimation of distances between each pair of H. pylori isolates was calculated with the squared Euclidean distance. In the clusters, fingerprints with distances less than or equal to five were considered related and distances above five were unrelated. The cluster analysis and the association to antimicrobial susceptibility were evaluated by using the χ2 exact. The statistical significance was accepted with a P value ≤ 0.05.
Global prevalence of infection by H. pylori was 85.4% (176/206) and 72.3% (149/206) by histology and culture, respectively. Isolates positive for H. pylori were characterized by antimicrobial susceptibility.
Among the 176 participants diagnosed through histopathology, 140 (79.5%) presented non-atrophic chronic gastritis, four (2.3%) multifocal atrophic gastritis without intestinal metaplasia, and 32 (18.2%) were diagnosed with multifocal atrophic gastritis with intestinal metaplasia (data not shown).
The antimicrobial susceptibility of the 149 isolates is shown in Table 1; MIC50 and MIC90 values are indicated. A total of 136 (91.3%) of the bacterial isolates were sensitive to clarithromycin and amoxicillin, while six (4%) of the isolates were AmxR, and four (2.7%) were ClaR. Only three (2%) of the isolates were resistant to both antibiotics. Global resistance was at 8.7% (13/149). The histopathological diagnosis of the participants with isolates that showed resistance was as follows: nine (69.2%) presented non-atrophic chronic gastritis, two (15.4%) had multifocal atrophic gastritis without metaplasia and two (15.4%) had multifocal atrophic gastritis with intestinal metaplasia. It is interesting to find that 50% (2/4) participants diagnosed through histopathology with multifocal atrophic gastritis presented AmxR/ClaR double-resistant isolates (SV323 and SV399) (Table 2).
| Antibiotic | MIC (μg/mL) | Resistance (n = 149) | ||
| MIC50 | MIC90 | Range | n (%) | |
| Amoxicillin | 2 | 4 | 0.25-4.0 | 6 (4) |
| Clarithromycin | 2 | 4 | 0.25-4.0 | 4 (2.7) |
| Antibiotic resistance | Isolates | Patients (n = 13) | Genotype | Diagnosis | MIC | EPIYA motifs (molecular weight in pairs of bases) | |||||
| Baseline | 0.25 | 0.5 | 1 | 2 | 4 | ||||||
| Amoxicillin | Single | SV301 | cagA+/vacA s1m1 | NAG | 4 | 495 | 495 | 495 | 495 | 495/200 | - |
| SV310 | cagA+/vacA s1m1 | NAG | 2 | 495 | 495 | 495 | 495 | - | - | ||
| SV333 | cagA+/vacA s1m1 | NAG | 2 | 495 | 495 | 595/495 | 595/495 | - | - | ||
| SV328 | cagA+/vacA s1m1 | IM | 2 | 695/595/495 | 495/300/200 | 495 | NA | - | - | ||
| SV338 | cagA+/vacA s1m1 | NAG | 2 | 595/495 | NA2 | NA | NA | - | - | ||
| Mixed | SV316 | cagA+/vacA s1m1 cagA-/vacA s1m1 | NAG | > 4 | 495 | NA | NA | 595/495 | 495 | NA | |
| Clarithromycin | Single | SV336 | cagA+/vacA s1m1 | NAG | 2 | 595 | 595 | 595 | 595 | - | - |
| SV440 | cagA+/vacA s2m2 | NAG | > 4 | 595 | 595/495 | 595/495 | NA | NA | NA | ||
| Mixed | SV415 | cagA+/vacA s1m1 cagA-/vacA s1m1 | NAG | > 4 | 495 | 495 | 495 | 495 | 495 | 2NA | |
| SV438 | Undefined | IM | 2 | NA2 | NA1 | NA1 | NA | - | - | ||
| SV323 | cagA+/vacA s1m1 | MAG | 2 | 495 | NA | NA | 495/200 | - | - | ||
| 4 | NA2 | NA2 | NA2 | NA2 | |||||||
| Amoxicillin + Clarithromycin | Single | SV399 | cagA+/vacA s1m1 | MAG | 2 | 495 | NA | 495 | 495 | - | - |
| 2 | 495/200 | NA | 495 | - | - | ||||||
| SV433 | cagA+/vacA s1m1 | NAG | > 4 | 495 | NA | 1NA | 495 | 595/495 | 200 | ||
| > 4 | 495 | 495 | 495 | 495 | 495 | ||||||
The genotyping results of H. pylori isolates resistant to amoxicillin and clarithromycin are shown in Table 2. In 23% (3/13) of the participants with isolates resistant to the antibiotics evaluated, colonization was documented with multiple H. pylori strains; in one of the cases (SV438) it was impossible to determine the infecting bacterial genotype. It was observed that in all isolates the cagA-positive genotype predominated. vacA s1m1 alleles were predominant, present in 84.6% (11/13), while s2m2 alleles were only observed in 7.7% (1/13) of the isolates. None of the resistant isolates obtained in this study presented genotypes s1m2 or s2m1.
Successful amplification took place of the 3’ variable region of the cagA encoding for EPIYA phosphorylation motifs of the CagA protein in 92.3% (12/13) of the baseline resistant isolates (without antibiotic pressure) and in 60% (36/60) of the resistant isolates growing at each of the serial amoxicillin and clarithromycin dilutions (with antibiotic pressure) (Table 2). This permitted confirming the presence of the cag PAI previously detected by cagA-specific PCR, as well as characterizing the number of EPIYA motifs present. Amplicon size was in the range of 470-670 bp (± 25 bp), according to that suggested by Panayotopoulou et al[11] except for some cases where strong bands were obtained -reproducible with unexpected molecular weights close to 170 and 270 bp (± 25 bp) (Figure 1A and Table 2). A single PCR product was observed in 49.3% (36/73) of the isolates obtained between baseline resistant samples and antibiotic dilutions and more than one PCR product with sizes corresponding to different numbers of EPIYA repetitions in 16.43% (12/73) of the isolates. All isolates negative for EPIYA PCR (n = 25) were confirmed by cag empty-site PCR, finding only four isolates as true cagA-negative. When comparing amplification proportions of EPIYA PCR vs cagA PCR in detecting cag PAI in isolates growing in the amoxicillin and clarithromycin serial dilutions, significant differences were found (McNemar test P = 0.006 and P = 0.031, respectively), contrasting with the baseline resistant isolates where no significant differences were found between both PCR methods (P > 0.05) (Table 3).
| Method | Detection of the cag PAI | ||
| Dilutions | |||
| Baseline | Amoxicillin | Clarithromycin | |
| (n = 13) | (n = 32) | (n = 28) | |
| cagA PCR | 11 (84.6) | 30 (93.8) | 22 (78.6) |
| EPIYA PCR | 12 (92.3) | 20 (62.5) | 16 (57.1) |
| P value | > 0.05 | 0.006 | 0.031 |
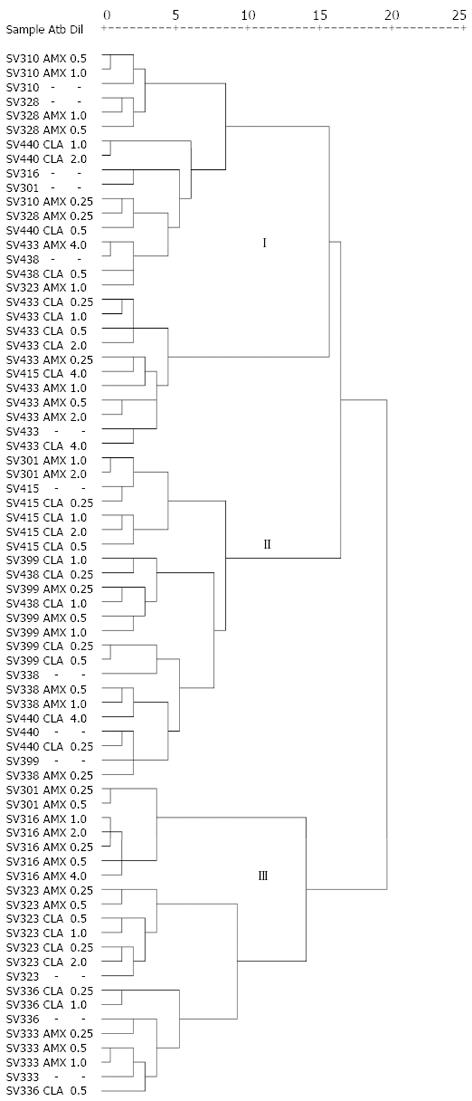
Primers 1281 and 1254 generated a reproducible RAPD fingerprints; they were capable of discriminating 73 different profiles. The number and size of the bands obtained with primer 1281 varied from 3 to 20 and from 76 to 1111 bp, respectively. It was determined if antibiotic pressure was associated to RAPD clusters. Most of the baseline resistant isolates (12/13; 92.3%) and under antibiotic pressure of amoxicillin (18/32; 56.3%) and clarithromycin (18/28; 64.3%) were included in cluster I in comparison to the distribution of these isolates in clusters II and III (P = 0.003).
With primer 1254, well-defined patterns were generated of two to 19 fragments in a range from 49 to 1950 bp (Figure 1B). The dendrogram based on RAPD profiles obtained with this primer includes the antimicrobial susceptibility for each isolate (Figure 2). Cluster analyses for this primer showed three main clusters in a parsimonious solution. In none of the clusters was the segregation of the baseline isolates and under antibiotic pressure significant (P = 0.704).
Upon observing the distribution of the number of EPIYA repetitions in each of the serial dilutions of the antibiotics with respect to the baseline isolates, change at the number of EPIYA repetitions was found in 61.5% (8/13) of the baseline isolates under antibiotic pressure. In six of these isolates (SV301, SV328, SV440, SV323, SV399 y SV433), the change consisted in loss of EPIYA repetitions evidenced by the low molecular weight of the amplified bands. In contrast, two isolates (SV316 and SV333) revealed gain of EPIYA repetitions. The gain and loss of EPIYA motifs resulted in a diversity of H. pylori subclones after bacterial adjustment to changing conditions product of antibiotic pressure. In all cases, the genotyping results and the presence of a single band in the EPIYA PCR for the baseline isolate permitted discarding the presence of more than one infecting cagA-positive isolate, except for isolate SV328 that presented in baseline three CagA species that differed in the number of EPIYA repetitions; after antibiotic pressure, alteration of the number of EPIYA repetitions was evidenced in two of the CagA species present (MIC 0.25 µg/mL) and its subsequent loss with increased concentration of the antimicrobial (MIC 0.5 µg/mL). Also, in three isolates no change was noted in the 3’ variable region of the cagA and in the two remaining cases (SV338 and SV438) change could not be documented due to absence of EPIYA PCR amplification. We used RAPD PCR to evaluate the clonal relationship of the isolates and verify that these strains had the same origin. Specifically, the RAPD fingerprints generated by both primers reflected a close clonal relationship between baseline isolates (without pressure) and isolates growing in antibiotic dilutions (under pressure).
Gastric infection by H. pylori is a prominent risk factor for gastric cancer; the outcome of the infection is determined by the pathogen’s characteristics in combination with environmental and host factors. It has been demonstrated that the precursor lesions that lead to the development of intestinal-type gastric adenocarcinoma have a series of sequential changes in the gastric mucosa[21]. The prolonged process of these precancerous lesions provides an opportunity to prevent the progression to more advanced stages; hence, efforts must be aimed at primary prevention. Anti-H. pylori treatment regimes combine two or more antibiotics with a proton pump inhibitor; however, they are not 100% effective in infection resolution[12]. The purpose of this research was to evaluate the in vitro effect of amoxicillin and clarithromycin on cag PAI and the 3’ variable region of the cagA of H. pylori, to understand the course of the infection in unsuccessful treatments.
We found H. pylori prevalence of 85.4% and 72.3% through histology and culture, respectively. Differences between the results could be due to the unequal distribution of H. pylori in the stomach. These results also coincide with the prevalence of the infection (73.8%) previously reported by Bravo et al[16] using histology within the same community. Differences observed through histology between both studies can be due to participant age and sex. Some 79.5% of the subjects diagnosed via histopathology presented non-atrophic chronic gastritis and the proportion observed of gastric cancer precursor lesions was low; findings unexpected in a population at high risk for gastric cancer. This could be due to sample bias; over half (55.8%) of the subjects recruited were women and the intestinal-type gastric adenocarcinoma is predominant in men[5].
Also, a low resistance rate to clarithromycin was noted in this population (2.7%), which agrees with the resistance rate reported in another Colombian region with similar geographic conditions, Pereira (2.2%)[22], and other regions in Latin America[23,24], but differing from the rates reported in Ecuador (9.5%)[25], Brazil (9.8%), the metropolitan region in Chile (20%)[26], Mexico (25%)[27], and Argentina (27.7%)[12]. A relatively high resistance rate was also noted for amoxicillin (4%), similar to rates documented within the country by other authors in patients with dyspepsia symptoms in Bogotá (3.8%)[28] and (7%)[29], although contrasting with the results obtained by Álvarez et al[22] who found no isolates resistant to amoxicillin in Pereira, agreeing with that described in other parts of the world, where resistance rates to this antibiotic are null or below 2%[12,23,30,31], implying an added difficulty to manage infection by H. pylori in our environment. In general, increased resistance to these two antibiotics could be a risk factor to consider for unsuccessful treatment within this population; thus, inasmuch as treatment is based on tests to antibiotic susceptibility, it may be more efficient in achieving high eradication rates. Additionally, studies conducted by our group have found that cases where treatment was not successful was associated to increased prevalence of less virulent strains[15,32]; it is not clearly known if this phenomenon is because of the effect of positive selection of certain genotypes (cagA-negative/vacA s2m2) in cases of multiple infection or because of loss of specific virulence factors (cag PAI) induced by antibiotic pressure, and this is the question we seek to respond.
The combination of different cagA genotypes and vacA s and m alleles illustrate the genomic variability of H. pylori[12]. This study found high prevalence of the cagA gene and vacA s1m1 alleles in baseline amoxicillin and clarithromycin resistant isolates; these observations agree with previous findings reported by Sicinschi et al[7] who genotyped gastric biopsies embedded in paraffin of two Colombian populations with contrast in the risk of gastric cancer, finding that the most virulent cagA-positive/vacA s1m1 strains are more prevalent in the high-risk area. In 23% of these resistant isolates, genotyping of cagA and vacA permitted our documenting infection by multiple strains of H. pylori. Mixed infection with two or more H. pylori strains colonizing the same patient possibly represent a stable association during long-term colonization.
Also, although no theoretical bases exist to predict an association between antibiotic resistance and virulence markers, several studies have been carried out to find an association. Most studies have shown no significant correlation[33]; however, studies conducted in northern Wales[34], central Italy[35] and western Argentina[12,36] found a clear association between virulent genotypes and antibiotic resistance. The correlation between virulence factors and antimicrobial resistance varies in different countries and although we did not evaluate this possible relationship, we found that the cagA-positive/vacA s1m1 genotype predominated in resistant isolates; nevertheless, this could be a product of the high prevalence of circulating virulent strains in this region[2,7,16] and not a reflection of a possible association between virulence factors and resistance.
Over 98% of the cagA-positive isolates harbored a CagA protein with three (495 bp) or four (595 bp) EPIYA motifs located quite probably on the ABC or ABCC combination, respectively. Although in none of the cases were PCR products sequenced, with this methodology we can certainly predict the presence of these combinations, as demonstrated by that reported by Panayotopoulou et al[11]. Nonetheless, in amplicons with unexpected sizes of 170 and 270 bp (+25 bp) and over four EPIYA repetitions (695 bp), sequencing is indispensable to establish the exact type of EPIYA motif combinations; although in the last case, predicting the correct number of EPIYA motifs was possible, being one of the limitations in our study. In no case do we expect to find East Asian-type strains with EPIYA-D motif circulating in the population, although the primers used were also designed for its detection, these predictions are supported by a prior study performed by our group with 41 individuals from the same location, which evaluated through sequencing the variations in the cagA carboxyl-terminal end, which varied considerably in size, number, and structure of EPIYA motifs (ABC, ABCC, ABCBC, ABBCC, or ABCCC) and no H. pylori strain contained an EPIYA-D motif[2].
A total of 25 cases were EPIYA PCR negative and were then confirmed by cagA-specific PCR and cag empty-site PCR, finding only four of them as true cagA-negative, which evidences: (1) the low sensitivity of EPIYA PCR compared to cagA PCR, which contrasts with the observations by Panayotopoulou et al[11] who suggest that the lack of amplification of the EPIYA PCR can precisely identify the cagA-negative cases. Perhaps, the differences observed in our case can be the effect of the random degradation of the bacterial DNA extracted from some samples -hindering amplification of large-size fragments (> 200 bp); in any case, it is wise to validate the detection of the cagA gene by cagA-specific PCR; and (2) the inefficiency of the DNA extraction method used in those cases where amplification was not obtained[37]. However, EPIYA PCR is a simple strategy to detect the cagA status and for precise prediction of the number and type of EPIYA motifs, involving a single amplification step[11] and permitting rapid screening of multiple isolates. Additionally, the EPIYA PCR facilitated the identification of cagA-positive H. pylori subclones closely related within the same isolate, harbored different number of EPIYA repetitions, indistinguishable in genotyping.
Because of the methodological approach employed in this study, it was possible to document in vitro that the effect of antibiotic pressure does not induce loss of cag PAI, but it can lead - in most cases - to genetic rearrangements within the 3’ region of cagA gene of the founding bacteria that produces a non-directed alteration (loss or gain) of EPIYA motifs. H. pylori has shown a unique genetic variability and these genetic changes could hypothetically be guided by intra-specific homologous recombination[11,14] and favored by their panmictic population structure[3,6], permitting the bacteria to quickly adapt to changing conditions, being able to: (1) eliminate particular immune-stimulating genetic regions (EPIYA motifs) that would result in the production of a non-phosphorylatable form of CagA that does not induce profound morphological changes in the host cell[38] supported by amplicons of unexpected molecular weights (170 and 270 bp) obtained in this study; and (2) induce variations within the 3’ terminal of the cagA encoding sequence that lead to differentiation in subclones with different numbers of EPIYA repetitions that can act as potential reservoirs of genetic elements for their cohabitants, with dominant clones favored by natural selection and additionally serve as escape recourse to the host’s immune response; thus, increasing their possibilities to adapt, evolve, and persist. In cases in which the 3’ variable region of the cagA remained intact following antibiotic pressure, suggests the presence of unknown extrinsic factors that can be influencing the phenomenon.
In most cases, the RAPD fingerprints confirmed the close clonal relationship existing among the isolates growing under antibiotic pressure and their corresponding baseline isolate, gathering them within the same cluster; for this reason, the cluster analysis for primer 1254 did not show differences in the segregation of isolates without pressure (baseline) and with pressure (AmxR and/or ClaR) among the three clusters formed, and although the cluster analysis based on the profile generated by the other primer (1281) did reveal significant differences (P = 0.003) among the three clusters, the differences observed are attributed to gathering most of the isolates within cluster I, without discriminating between isolates without pressure and those under pressure, likewise, always gathering the isolates under antibiotic pressure along with the baseline isolate. Some isolates under specific antibiotic concentration were not grouped along with their other clones at different antibiotic concentrations within the same cluster, probably due to the presence of more than one subclone within that isolate that induced a distortion in the RAPD pattern with this being a technique that generates a profile based on the complete DNA. Even though these observations agree with that expected, no differences should exist between isolates growing in each of the antibiotic dilutions and the baseline isolate because they are the same isolate.
These findings are specially important because of their possible implications for the treatment and evolution of gastro-duodenal diseases caused by H. pylori, suggesting that acquisition or deletion of EPIYA motifs product of the antibiotic effect can affect the phosphorylation level of tyrosine, impacting on its cellular effects[1] and leading to the divergence of cagA-positive H. pylori subclones, which as a set could alter the pathogenic process of H. pylori in cases with treatment failure. Such pool of H. pylori clones may exist in dynamic equilibrium within any potential host[11], cooperating through quorum sensing and recombination, which can lead to downregulation of interaction with the host[14] to induce lesser damage, even when some damage is inevitable, given that in these subclones the divergent CagA species would be normally expressed by inducing several degrees of “hummingbird” phenotype upon the infected epithelial cells[11]. This hypothesis could explain - in part - the observations registered by Mera et al[39] who during a follow up of a cohort for 12 years found that those subjects receiving anti-H. pylori treatment but remaining infected showed decreased histopathological score -0.19 over the average histopathology score at baseline. Additional in vitro studies including a higher number of resistant isolates are necessary to determine the direct effect of clarithromycin and amoxicillin upon the cagA gene and evaluate their effect on the vacA gene, which was not considered in this study, even though it is a gene susceptible to genetic modifications according to that reported by Argent et al[40]. Likewise, new studies are needed aimed at evaluating the in vivo effect of anti-H. pylori treatment upon virulence factors previously described in H. pylori isolates from patients with treatment failure and deciphering the complex molecular mechanisms through which these genomic alterations are produced. Upon proving its effect on cagA, this methodological approach could constitute a useful prognosis tool in clinical practice, when detecting the presence of multiple subclones and determining the number and type of EPIYA motifs, permitting the prediction of pathogenic activity of the bacteria in unsuccessful treatments.
Finally, given the generalized use of antibiotics in our environment to combat diverse infectious agents, it is tempting to speculate that inadequate treatments (not H. pylori-targeted) can, likewise, significantly impact upon H. pylori strains co-existing along with their host.
We are very grateful to Professor Francis Mégraud, INSERM - U853, Université Bordeaux Segalen, France, for comments on the manuscript.
Helicobacter pylori (H. pylori) is a prominent risk factor for gastric cancer, higher risk in individuals infected with cagA-positive strains. Its eradication with antibiotics constitutes a chemoprevention strategy; however, treatments are not completely effective and seem to contribute to increasing the prevalence of less virulent strains.
Virulence-associate genotypes of H. pylori are increasingly recognized as determinants of the clinical outcome of the infection. The fact that virulence factors are linked to disease implies that they are a fixed characteristic, but this is not the case and genotype variations can occur through genetic rearrangements that either eliminates particular immunostimulatory genetic regions (cag pathogenicity island) or causes variations in potential immunostimulatory molecules (number of Glu-Pro-Ile-Tyr-Ala motifs in CagA) probably reflecting the local selection of H. pylori particular phenotypes as an escape resource to the immune response of the host induced by environmental pressures.
Some studies indicate that failure to eradicate H. pylori from the stomach may result in an increase in the prevalence of less virulent genotypes. It is unknown if this phenomenon is due to the positive selection of certain genotypes (cagA-negative/vacA s2m2) in cases of multiple infection or loss of specific virulence factors (cag PAI) induced by antibiotic pressure. This study shows that antibiotic pressure does not induce loss of the cag PAI, but it can lead to genetic rearrangements within of the cagA variable region that can affect the level of tyrosine phosphorylation impacting on its cellular effects and lead to divergence of cagA-positive subclones, which as a set could alter the pathogenic process of H. pylori in cases with treatment failure.
These in vitro findings are specially important because of their possible implications for the treatment and evolution of gastro-duodenal diseases caused by H. pylori, suggesting that acquisition or deletion of EPIYA motifs product of the antibiotic effect can affect the phosphorylation level of tyrosine and leading to the divergence of cagA-positive H. pylori subclones. Further studies are needed aimed at evaluating the effect of anti-H. pylori treatment on the cagA variable region in H. pylori isolates from patients with treatment failure and deciphering the complex molecular mechanisms through which these genomic alterations are produced. Upon proving its effect on cagA, this methodological approach could constitute a useful prognosis tool in clinical practice, when detecting the presence of multiple subclones and determining the number and type of EPIYA motifs, permitting the prediction of pathogenic activity of the bacteria in unsuccessful treatments.
The cag pathogenicity island is a genomic fragment of approximately 40 kb that contains 30 genes that encode a type IV secretion apparatus, which export proteins from bacterial cytoplasm to host epithelial cells. The terminal gene in the island, cagA, is commonly used as a marker for the entire cag locus.
This manuscript deals with the evaluation of the in vitro effect of amoxicillin and clarithromycin on the cag PAI. The authors found that antibiotic pressure does not induce loss of the cag PAI and leads in most cases to genetic rearrangements within the 3’ region of cagA of the bacteria that lead to divergence of cagA-positive subclones. This study is interesting and original. Methods are appropriate, results are clearly presented and conclusions are corroborated by the results.
P- Reviewer Romano M S- Editor Song XX L- Editor A E- Editor Zhang DN
| 1. | Argent RH, Zhang Y, Atherton JC. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J Clin Microbiol. 2005;43:791-795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 2. | Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A, Mera R. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16:369-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Merrell DS, Falkow S. Frontal and stealth attack strategies in microbial pathogenesis. Nature. 2004;430:250-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 4. | Greenberg ER, Anderson GL, Morgan DR, Torres J, Chey WD, Bravo LE, Dominguez RL, Ferreccio C, Herrero R, Lazcano-Ponce EC. 14-day triple, 5-day concomitant, and 10-day sequential therapies for Helicobacter pylori infection in seven Latin American sites: a randomised trial. Lancet. 2011;378:507-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 5. | Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1317] [Cited by in F6Publishing: 1298] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 6. | Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747-765. [PubMed] [Cited in This Article: ] |
| 7. | Sicinschi LA, Correa P, Peek RM, Camargo MC, Delgado A, Piazuelo MB, Romero-Gallo J, Bravo LE, Schneider BG. Helicobacter pylori genotyping and sequencing using paraffin-embedded biopsies from residents of colombian areas with contrasting gastric cancer risks. Helicobacter. 2008;13:135-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Ek C, Whary MT, Ihrig M, Bravo LE, Correa P, Fox JG. Serologic evidence that ascaris and toxoplasma infections impact inflammatory responses to Helicobacter pylori in Colombians. Helicobacter. 2012;17:107-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, Bravo LE, Sicinschi LA, Delgado AG, Mera RM. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60:1189-1195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 10. | Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 11. | Panayotopoulou EG, Sgouras DN, Papadakos K, Kalliaropoulos A, Papatheodoridis G, Mentis AF, Archimandritis AJ. Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J Clin Microbiol. 2007;45:488-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Vega AE, Cortiñas TI, Puig ON, Silva HJ. Molecular characterization and susceptibility testing of Helicobacter pylori strains isolated in western Argentina. Int J Infect Dis. 2010;14 Suppl 3:e85-e92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 264] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 14. | Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119:2475-2487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 367] [Cited by in F6Publishing: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 15. | Correa P, van Doorn LJ, Bravo JC, Ruiz B, Bravo LE, Realpe JL. Unsuccessful treatment results in survival of less virulent genotypes of Helicobacter pylori in Colombian patients. Am J Gastroenterol. 2000;95:564-566. [PubMed] [Cited in This Article: ] |
| 16. | Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839-2842. [PubMed] [Cited in This Article: ] |
| 17. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [PubMed] [Cited in This Article: ] |
| 18. | Dixon MF; CLSI. Performance standards for antimicrobial susceptibility testing: Seventeenth Informational Supplement. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute 2007; . [Cited in This Article: ] |
| 19. | van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa JC, Carneiro F, Quint WG. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271-1276. [PubMed] [Cited in This Article: ] |
| 20. | Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137-5142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 565] [Cited by in F6Publishing: 595] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 21. | Cuello C, Correa P, Haenszel W, Gordillo G, Brown C, Archer M, Tannenbaum S. Gastric cancer in Colombia. I. Cancer risk and suspect environmental agents. J Natl Cancer Inst. 1976;57:1015-1020. [PubMed] [Cited in This Article: ] |
| 22. | Alvarez A, Moncayo JI, Santacruz JJ, Corredor LF, Reinosa E, Martínez JW, Beltrán L. [Antimicrobial susceptibility of Helicobacter pylori strains isolated in Colombia]. Rev Med Chil. 2009;137:1309-1314. [PubMed] [Cited in This Article: ] |
| 23. | González C, García A, Daroch F, Kawaguchi F, Solar H, Rivera N, Vega E. [In vitro antimicrobial susceptibility of Helicobacter pylori strains: isolation of strains resistant to clarithromycin]. Rev Med Chil. 2001;129:643-646. [PubMed] [Cited in This Article: ] |
| 24. | Fariña N, Kasamatsu E, Samudio M, Morán M, Sanabria R, Laspina F. [Antimicrobial susceptibility of H pylori strains obtained from Paraguayan patients]. Rev Med Chil. 2007;135:1009-1014. [PubMed] [Cited in This Article: ] |
| 25. | Debets-Ossenkopp YJ, Reyes G, Mulder J, aan de Stegge BM, Peters JT, Savelkoul PH, Tanca J, Peña AS, Vandenbroucke-Grauls CM. Characteristics of clinical Helicobacter pylori strains from Ecuador. J Antimicrob Chemother. 2003;51:141-145. [PubMed] [Cited in This Article: ] |
| 26. | Vallejos C, Garrido L, Cáceres D, Madrid AM, Defilippi C, Defilippi C, Toledo H. [Prevalence of metronidazole, clarithromycin and tetracycline resistance in Helicobacter pylori isolated from Chilean patients]. Rev Med Chil. 2007;135:287-293. [PubMed] [Cited in This Article: ] |
| 27. | Torres J, Camorlinga-Ponce M, Pérez-Pérez G, Madrazo-De la Garza A, Dehesa M, González-Valencia G, Muñoz O. Increasing multidrug resistance in Helicobacter pylori strains isolated from children and adults in Mexico. J Clin Microbiol. 2001;39:2677-2680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 84] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Trespalacios AA, Otero Regino W, Mercado Reyes M. Helicobacter pylori resistance to metronidazole, clarithromycin and amoxicillin in Colombian patients (in Spanish). Rev Col Gastroenterol. 2010;25:31-38. [Cited in This Article: ] |
| 29. | Yepes CA, Varón AR, Morales ÁR, Ariza B. Antibiotics resistance of Helicobacter pylori at the San Ignacio University Hospital in Bogota (in Spanish). Acta Méd Colomb. 2008;33:11-14. [Cited in This Article: ] |
| 30. | Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Meyer JM, Silliman NP, Wang W, Siepman NY, Sugg JE, Morris D, Zhang J, Bhattacharyya H, King EC, Hopkins RJ. Risk factors for Helicobacter pylori resistance in the United States: the surveillance of H. pylori antimicrobial resistance partnership (SHARP) study, 1993-1999. Ann Intern Med. 2002;136:13-24. [PubMed] [Cited in This Article: ] |
| 32. | Fontham ET, Correa P, Mera R, Bravo LE, Bravo JC, Piazuelo MB, Camargo MC. Duration of exposure, a neglected factor in chemoprevention trials. Cancer Epidemiol Biomarkers Prev. 2005;14:2465-2466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 33. | Godoy AP, Ribeiro ML, Benvengo YH, Vitiello L, Miranda Mde C, Mendonça S, Pedrazzoli J. Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates. BMC Gastroenterol. 2003;3:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Elviss NC, Owen RJ, Breathnach A, Palmer C, Shetty N. Helicobacter pylori antibiotic-resistance patterns and risk factors in adult dyspeptic patients from ethnically diverse populations in central and south London during 2000. J Med Microbiol. 2005;54:567-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Capodicasa S, Marzio L. Analysis of genetic variability, antimicrobial susceptibility and virulence markers in Helicobacter pylori identified in Central Italy. Scand J Gastroenterol. 2006;41:280-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Stege PW, Vega AE. Analysis of resistance to clarithromycin and iceA status in Helicobacter pylori isolates from San Luis, Argentina. Int J Antimicrob Agents. 2006;28:477-478. [PubMed] [Cited in This Article: ] |
| 37. | Bustamante JA, Astudillo M, Pazos AJ, Bravo LE. Evaluation of Two Methods DNA Extraction from Formalin-Fixed, Paraffin-Embedded Tissues on Non-Optimal Conditions (in Spanish). Acta biol Colomb. 2011;16:83-98. [Cited in This Article: ] |
| 38. | Aras RA, Lee Y, Kim SK, Israel D, Peek RM, Blaser MJ. Natural variation in populations of persistently colonizing bacteria affect human host cell phenotype. J Infect Dis. 2003;188:486-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 55] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Mera R, Fontham ET, Bravo LE, Bravo JC, Piazuelo MB, Camargo MC, Correa P. Long term follow up of patients treated for Helicobacter pylori infection. Gut. 2005;54:1536-1540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 40. | Argent RH, Thomas RJ, Aviles-Jimenez F, Letley DP, Limb MC, El-Omar EM, Atherton JC. Toxigenic Helicobacter pylori infection precedes gastric hypochlorhydria in cancer relatives, and H. pylori virulence evolves in these families. Clin Cancer Res. 2008;14:2227-2235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |