Published online May 28, 2014. doi: 10.3748/wjg.v20.i20.5962
Revised: January 11, 2014
Accepted: April 1, 2014
Published online: May 28, 2014
MicroRNAs are small endogenously expressed RNA molecules which are involved in the process of silencing gene expression through translational regulation. The polycistronic miR-17-92 cluster is the first microRNA cluster shown to play a role in tumorigenesis. It has two other paralogs in the human genome, the miR-106b-25 cluster and the miR-106a-363 cluster. Collectively, the microRNAs encoded by these clusters can be further grouped based on the seed sequences into four families, namely the miR-17, the miR-92, the miR-18 and the miR-19 families. Over-expression of the miR-106b-25 and miR-17-92 clusters has been reported not only during the development of cirrhosis but also subsequently during the development of hepatocellular carcinoma. Members of these clusters have also been shown to affect the replication of hepatitis B and hepatitis C viruses. Various targets of these microRNAs have been identified, and these targets are involved in tumor growth, cell survival and metastasis. In this review, we first describe the regulation of these clusters by c-Myc and E2F1, and how the members of these clusters in turn regulate E2F1 expression forming an auto-regulatory loop. In addition, the roles of the various members of the clusters in affecting relevant target gene expression in the pathogenesis of hepatocellular carcinoma will also be discussed.
Core tip: The polycistronic miR-17-92 cluster has been characterized to play a role in tumorigenesis. Over-expression of the miR-17-92 cluster and its paralog, the miR-106b-25 cluster, has been reported in the development of cirrhosis and hepatocellular carcinoma. Various targets of these microRNAs have been identified, and these targets are involved in tumor growth, cell survival and metastasis. We describe the regulation of these clusters by c-Myc and E2F1, and discuss the roles of the various members of the clusters in affecting relevant target gene expression in the pathogenesis of hepatocellular carcinoma.
-
Citation: Tan W, Li Y, Lim SG, Tan TM.
miR-106b-25/miR-17-92 clusters: Polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol 2014; 20(20): 5962-5972 - URL: https://www.wjgnet.com/1007-9327/full/v20/i20/5962.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i20.5962
Liver cancer is currently the fifth most common cancer in men. It is also the third most common cause of death from cancer with 694000 deaths in 2008[1]. The majority of liver cancers are hepatocellular carcinoma (HCC) and the occurrence is most prevalent in developing nations in Asia and Africa. The incidence of HCC is also sex associated as HCC affects more males than females.
The majority of HCC cases develop from chronic liver disease, which includes cirrhosis caused by long-term infection with hepatitis B (HBV) and C (HCV) viruses, exposure to hepatotoxins, excessive alcohol intake and steatohepatitis[2]. Damage to hepatocytes leads to increase in the generation of reactive oxygen species (ROS) and oxidative stress, promoting necrosis and apoptosis of hepatocytes. The ROS produced activates Kupffer cells to generate more ROS and also secrete cytokines such as transforming growth factor-beta (TGF-β) and platelet-derived growth factor (PDGF), which in turn activate hepatic stellate cells (HSCs) to become proliferative, releasing pro-fibrogenic, proinflammatory, and promitogenic cytokines[3,4]. The increased production of extracellular matrix (ECM) components by these activated stellate cells leads to fibrosis, and chronic fibrosis results in cirrhosis, characterized by the formation of regenerative nodules and the distortion of liver parenchyma and vascular architecture[4].
In recent years, research on post-transcriptional gene silencing has progressed greatly following the discovery of endogenously coded microRNAs (miRNAs). MiRNAs are small, RNA molecules of approximately 20-24 nucleotides in length. The genes encoding the miRNAs can be located in the introns of protein-coding genes, or in both introns and exons of non-coding transcripts[5]. In several instances, miRNAs are clustered together and transcribed as a single polycistronic primary transcript.
Most miRNA genes are transcribed by RNA Polymerase II to generate primary transcripts[6]. The primary transcripts are first processed to smaller approximately 70-nucleotide hairpin looped precursor molecules (pre-miRNAs)[7,8]. The pre-miRNAs are then transported from the nucleus to the cytoplasm[9] and are further cleaved by the enzyme, Dicer[10,11], releasing approximately 22-nucleotide duplexes. The two arms of the duplexes do not have perfect complementation. One arm gives rise to the mature miRNA while the opposing arm is denoted as miRNA*. The duplex is recruited into the RNA-induced silencing complex (RISC) where it unwinds and miRNA* is degraded[12,13]. The mature miRNA that is bound to RISC then serves as a guide to provide the binding specificity to the target RNA. Binding of the miRNA-RISC to the target RNA can result in two possible outcomes. When the miRNA is able to base-pair extensively with the target RNA, cleavage of a single phosphodiester bond occurs in the target across nucleotides 10 and 11 of the miRNA[14,15]. In vertebrates including humans, there is usually only partial complementation between the miRNA and its target RNA, and cleavage of the target does not occur. Instead, translation of the target RNA is repressed[14,16]. It is now well documented that miRNAs are able to regulate the expression of target genes either by blocking mRNA translation, or inducing mRNA decay, or by affecting chromatin structure[17].
In 2001, three groups of researchers simultaneously reported the identification of hundreds of miRNAs in human, fly and worm cells, triggering the upsurge of miRNA studies[18-20]. It soon became evident that miRNAs are involved in multiple cellular and physiological processes and dys-regulation of miRNAs could lead to disease. Current evidence suggests that miRNAs are contributors to oncogenesis. They could serve as either oncogenes or tumor suppressors depending on their target genes and contexts. miR-15 and miR-16 were the first two miRNAs shown to be involved in the development of chronic lymphocytic leukemia (CLL). Both miRNAs were deleted or down-regulated in CLL, consistent with the idea that miRNAs may act as tumor suppressors[21]. Following this initial study, a large number of miRNAs has since been shown to be associated with human cancers.
The polycistronic miR-17-92 cluster is one of the earliest characterized miRNA clusters. The study of this cluster began with the identification of a novel gene designated as chromosome 13 open reading frame 25 (C13orf25) by Ota et al[22] in 2004. This gene resides within the frequently amplified 13q31-q32 region in malignant lymphoma. Based on sequence analysis of the gene, it was unlikely that the C13orf25 gene encoded for any protein product. Instead, the miR-17-92 cluster which is located in the third intron of the C13orf 25 gene was hypothesized to be the actual effectors of the 13q31-q32 amplification and could play a role in tumorigenesis. Subsequent work by various groups showed that this hypothesis is indeed true.
In MYC transgenic mice, lymphoma development was accelerated following the transduction of the miR-17-92 cluster into hematopoietic cells[23]. This observation strongly supported the concept that certain miRNA alterations could predispose a cell to carcinogenesis and abnormal expressions of miRNAs are not just the consequence of cellular transformation. The tumorigenic activity of the miR-17-92 cluster was thus demonstrated and this cluster was termed as “OncomiR-1”.
Subsequently, c-Myc was shown to regulate the expression of the miR-17-92 cluster by binding directly to it and up-regulating the expression of all six miRNAs of the cluster. Furthermore, the transcription factor E2F1 was identified as the target of two members of the miR-17-92 cluster: miR-17-5p and miR-20a. Thus, despite the induction of E2F1 transcription by c-Myc resulting in high levels of E2F1 mRNA, the E2F1 protein level was only modestly increased[24]. Interestingly, c-Myc is in turn positively regulated by E2F1[25].
Following these two studies, there was a subsequent report on the oncogenic role of the miR-17-92 cluster in lung cancers, especially those with small-cell lung cancer histology. An increase in the gene copy number of this miRNA cluster resulted in its over-expression. Again it was confirmed that it was the marked expression of the miR-17-92 cluster and not the putative C13orf25 gene product that promoted cell growth. These findings demonstrated the oncogenic role of the miR-17-92 cluster in the development of lung cancers and confirmed that the real effectors of the 13q31-q32 amplification in cancers are the miRNAs encoded within the miRNA-17-92 cluster[26].
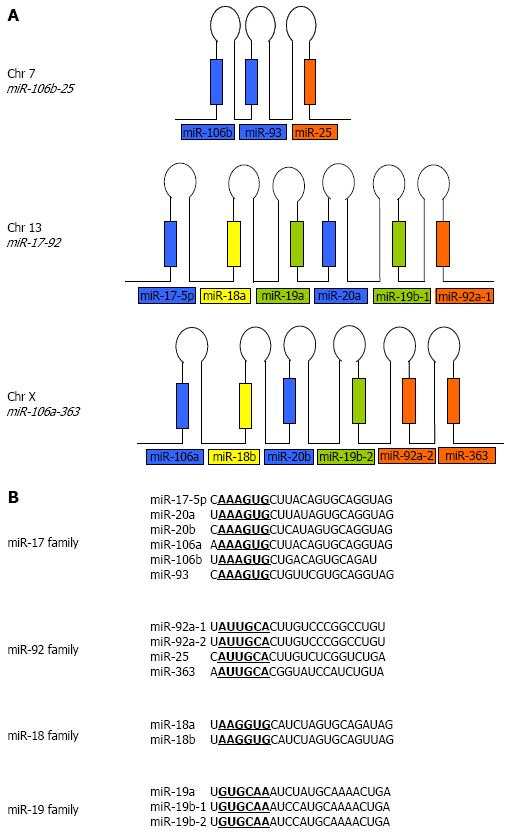
Many of the known miRNAs appear in clusters on single polycistronic transcripts[27-29]. The human miR-17-92 cluster contains six precursor miRNAs within about 1 kb on chromosome 13. It has two paralogs in the human genome: the miR-106b-25 cluster on chromosome 7 and the miR-106a-363 cluster on chromosome X. Reconstruction of the evolutionary history of the miR-17-92 cluster and its two paralogs revealed that the evolution of this cluster arose from tandem duplications of individual miRNAs, followed by duplications of entire clusters and subsequent loss of individual miRNAs[30]. The miR-17-92 cluster encodes for miR-17-5p, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1 while the miR-106a-363 cluster encodes for miR-106a, miR-18b, miR-20b, miR-19b-2, miR92a-2 and miR-363. The miR-106b-25 cluster encodes for only three miRNAs; miR-106b, miR-93, and miR-25 and is embedded within the minichromosome maintenance protein 7 (MCM7) gene (Figure 1A). These miRNAs can be further grouped into four seed families, namely the miR-17, the miR-92, the miR-18 and the miR-19 families (Figure 1B).
To provide insights into the functionality and redundancy of these clusters, studies in mice following the knock-out of the clusters individually or in combination were carried out[31]. Defective embryonic development and postnatal death were observed in mice with homozygous deletion of the miR-17-92 cluster. Deletions of both the miR-17-92 and the miR-106b-25 clusters led to more severe phenotypes including apoptosis in the fetal liver and regions of the central nervous system. In contrast, there was normal development following the deletion of either the widely expressed miR-106b-25 cluster, or the rarely expressed miR-106a-363 cluster, or both clusters. Collectively, these observations indicate that members of the miR-17 and miR-92 families in both the miR-17-92 and the miR-106b-25 clusters may perform specific yet overlapping functions while the miR-18 and miR-19 families which are only present in the miR-17-92 clusters are likely to be functionally important during development.
Interestingly, although most of the studies point to the oncogenic property of the miR-17-92 and miR-106b-25 clusters, particular members of the cluster may also serve as tumor suppressors in certain contexts. One example is miR-17. Over-expression of miR-17 can reduce cell proliferation, affect cell adhesion and migration, and suppress the expression of fibronectin and the fibronectin type-III domain containing 3A (FNDC3A)[32]. Another example is miR-25, a member of the miR-106b-25 cluster which can confer oncogenic or tumor suppressive properties in different types of human cancers. miR-25 is up-regulated and promotes tumorigenesis in stomach, liver and prostate tumor via its inhibition of the target genes Bim, p57 and DR4[33-37]. In contrast, miR-25 is down-regulated in colon cancer where it targets SMAD7 and exerts anti-tumor effects[38]. miR-93, another member of the miR-106b-25 cluster, is be able to adversely affect cell proliferation and colony formation of colon cancer stem cells[39]. All these findings raised the possibility that miRNAs encoded by these clusters can either positively or negatively affect tumorigenesis and depending on the tumor, may exhibit either oncogenic or tumor suppressive activities.
Approximately 80% of HCC cases are associated with chronic HBV or HCV infections. It is well documented that the risk of HCC development is greatly increased in individuals with such infections and anti-viral treatments can effectively reduce the risk[40].
miRNAs have been shown to affect HBV gene expression and inhibit HBV replication. miR-199a-3p, miR-210 and miR-122 can suppress HBV replication[41,42]. In contrast, HBV gene products may also affect miRNAs expressions. The hepatitis B virus X protein (HBx) has been shown to suppress the expression of miR-148 in a murine model of HCC and consequently promote proliferation and metastasis via the upregulation of hematopoietic pre-B cell leukemia transcription factor-interacting protein (HPIP)[43]. Members of the miR-17-92 cluster have also been shown to affect HBV replication. miR-20a and miR-92a-1 can directly bind to HBV genes and inhibition of these miRNAs enhances HBV replication[44].
Although the miR-106-25 cluster which is hosted in intron 13 of the MCM7 gene has not been shown to directly affect HBV or HCV replication, it was interesting to note that a single nucleotide polymorphism (SNP), rs999885 in the promoter region of MCM7 can confer a protective effect against chronic HBV infection. In patients who have achieved clearance of HBV infection, the A to G base change is associated with decreased risk of chronic HBV infection. However, for persistent HBV carriers, this change results in increased risk for HCC with increased expression of the miR-106b-25 cluster in the liver[45]. Another SNP, rs2596452, located within the 5’ flanking region of the MHC class I polypeptide-related sequence A (MICA), is associated with HBV- and HCV-related progression to HCC[46,47]. MICA is usually highly expressed in viral-infected cells and in tumor cells. Its expression is necessary for the activation of cell killing by natural killer cells and CD8 T cells. Over-expression of the miR-106b-25 cluster has been shown to suppress MICA expression and inhibit the cell-killing function[48].
To date, it is also well documented that miR-122 is critical for the replication of HCV. miR-122 protects the HCV genome by binding to 2 sites on the 5’ non-coding region of the HCV RNA and in doing so enhances the propagation of the virus[49]. This makes targeting miR-122 for anti-HCV therapy very attractive and phase 2a trials of miravirsen, a locked nucleic acid modified antisense oligonucleotide complementary to part of miR-122, have resulted in significant reduction in viral RNA with no emergence of resistant viruses[50].
As had been observed for HBV, HCV infection could also alter hepatic miRNA expressions. The expression of miR-27 has been shown to be induced by HCV replication in Huh7.5 cells and in vivo HCV infection models. Consequently, this over-expression results in hepatic steatosis[51]. The roles of the miR-17-92 and the miR-106b-25 clusters have not been examined for HCV infection. However, serum levels of miR-20a and miR-92a have been shown to increase during acute and chronic HCV infections. Significant up-regulation of these miRNAs were also observed in the sera of HCV-infected patients with fibrosis[52].
Liver cirrhosis is the single most significant predisposing factor for HCC. In addition, death from liver cirrhosis itself is estimated at 800000 annually[53]. A complex network of signaling pathways together with contributions from HSCs and inflammatory cells such as Kupffer cells during the course of repetitive and chronic liver damage eventually results in liver cirrhosis. Thus, liver cirrhosis arises from chronic liver disease and is characterized by the development of regenerative nodules which are surrounded by fibrous connective tissues[54].
Differential miRNA expressions have been shown to be not only associated with hepatic viral infections but also with different stages of liver diseases including cirrhosis. Members of the miR-29 family were found to be significantly down-regulated in murine and human fibrotic livers. Down-regulation of miR-29 is dependent on TGF-β and nuclear factor kappa B (NF-κB) in HSCs while over-expression of miR-29 can suppress activation of the HSCs leading to reduced collagen expression[55]. In contrast, the expression of the miR-199 and -200 families increases with progression of liver fibrosis[56]. Interestingly, during rat liver regeneration after 70% partial hepatectomy, selective up-regulation followed by global down-regulation of miRNAs was observed in a negative feedback mechanism[57]. These studies reiterated the possible functional importance of miRNAs during liver regeneration and liver fibrosis.
Members of the miR-17-92 and the miR-106b-25 clusters have been implicated in the progression of liver fibrosis. In fibrotic rat and human livers, the expression of miR-19b was significantly down-regulated. miR-19b has been shown to play an inhibitory role in HSC-mediated fibrogenesis through its effect on the TGF-β signaling pathway. Over-expression of miR-19b inhibited the expression of TGF-β receptor II (TGF-βRII) leading to decreased SMAD3 expression and reduced type-1 collagen production[58]. In contrast, miR-93 and miR-106b were observed to be consistently up-regulated during the development of cirrhosis, although the actual targets have yet to be confirmed[59]. Serum levels of miR-106b have also been shown to be useful for the prediction of liver cirrhosis. Together with miR-181, both miR-106b and miR-181 were shown to serve as biomarkers for liver cirrhosis irrespective of the etiology[60].
Changes in the expression of miRNAs have been observed in all human cancers. Dys-regulation in the expression of the miR-106b-25 and miR-17-92 clusters have been reported for various tumors, including B-cell lymphoma and solid tumors derived from breast, colon, lung, pancreas, prostate and stomach[23,26,61,62]. The first few reports on miRNA expression in human and rodent HCC also showed that members of the miR-17-92 and miR-106b-25 clusters were aberrantly expressed. These include significant up-regulation of miR-18 together with its precursor, pre-miR-18, miR-17, miR-20, miR-93, and miR-106[63,64]. Subsequent studies from various other groups also showed this to be true[34,59,65-71].
The role of c-Myc in liver and HCC has been well studied and documented. c-Myc is a transcription factor which is frequently up-regulated in HCC, in patients with advanced liver fibrosis, and in experimental liver fibrosis in mice[72,73]. The expression of c-Myc is controlled by the different phosphorylated forms of Smad2 and Smad3. During acute liver injury, TGF-β binds to TGF-β type I receptor (TβRI), which then phosphorylates Smad2 and Smad3 at their C-termini. These Smads then complex with Smad4 and translocate to the nucleus to repress the expression of its target genes, including c-Myc. In chronic liver injury, PDGF activates c-Jun N-terminal kinase (JNK), and the activated JNK then phosphorylates Smad2 and Smad3 in their linker regions. These dually phosphorylated Smads can up-regulate the expression of c-Myc[74]. The up-regulation of c-Myc in hepatocytes during chronic fibrosis or as a consequence of gene amplification may promote or repress the expression of many genes including those encoding for miRNAs. Indeed recent studies have characterized Myc-repressed as well as Myc-induced miRNAs in HCC[44,75]. The latter includes miRNAs of the miR-17-92 cluster and the miR-106b-25 cluster.
Given that the miR-106b-25 cluster is embedded in the thirteenth intron of the MCM7 gene, it is not surprising that in HCC, gastric and prostate cancers, the expression of the precursors of the miR-106b-25 cluster has been shown to correspond to that of Mcm7 transcripts[34,76,77]. The transcription of the miR-106b-25 cluster is thus regulated through the expression of the MCM7 gene and both c-Myc and E2F1 have been shown to up-regulate Mcm7 expression[77-80]. The expression of the miR-17-92 cluster, which is located on human chromosome 13, can also be promoted by c-Myc and the E2F family of transcription factors[24,81,82]. In HCC, there is significant up-regulation of members of the miR-17-92 and miR-106b-25 clusters. In addition, in a study by Pineau et al[59], miR-93 and miR-106b were observed to be consistently up-regulated during the development of cirrhosis and subsequently during the development of HCC. The consistent up-regulation of the miR-106b-25 and miR-17-92 clusters from chronic liver fibrosis onwards indicate that these two miRNA clusters are likely to be involved in the initiation of tumorigenesis, as well as at subsequent stages of HCC.
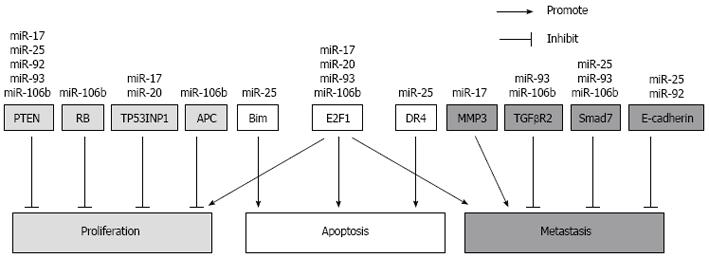
To date, various targets of these two clusters have been identified in HCC and other cancers, and these targets are involved in tumor growth, cell survival and metastasis (Figure 2). It is also noteworthy that the expression of these clusters is likely to be regulated by the expression of the oncogene Myc and that the targets of these clusters include genes which exhibit significant changes during HCC. Such targets include phosphatase tensin homolog (PTEN) and members of the Wnt/β-catenin pathway. Multiple genetic alterations have been documented for HCC and these can be fitted broadly into four distinct groups. These include the inactivation of tumor suppressor genes such as p53, retinoblastoma (RB), PTEN and runt-related transcription factor 3, as well as the activation of oncogenes such as that of Myc, Ras and Raf. The activation of growth factors and their receptors such as transforming growth factor-α, insulin-like growth factor-2 receptor as well as the reactivation of developmental pathways including that of the hedgehog and Wnt/β-catenin pathways are also contributors to HCC[83].
These two miRNA clusters can suppress the protein expression of tumor suppressor PTEN[35,84-86]. Down-regulation of PTEN leads to hyper-proliferative and anti-apoptotic activities via the AKT pathway[87]. PTEN-deficient mice exhibit hepatomegaly, steatohepatitis, fibrosis and eventually develop HCC[88]. PTEN down-regulation was also observed in fibrotic livers of bile duct ligated mice[89]. In contrast, over-expression of PTEN inhibited HSC activation[90]. In HCC tissues, there is usually low or no PTEN expression[86,87]. Together, these data indicate that the up-regulation of these two miRNA clusters can result in hepatic down-regulation of PTEN during fibrosis and in the progression towards tumorigenesis.
The transcription factor E2F1 is a target gene for the miR-17 seed family. Members of this family can interact with the 3’ UTR of the E2F1 transcripts and inhibit its translation in HCC cell lines. In HCC tumors, it is likely that the miR-17 family prevents excessively high E2F1 expression, which may then cause apoptosis[24,34]. Both the miR-17-92 and the miR-106b-25 clusters have been shown to participate in the auto-regulatory feedback loop involving c-Myc and E2F transcription factors. These factors regulate the expression of the clusters while members of the clusters in turn regulate the expression of E2F1. This mode of action has been also observed for these clusters in other tumors including prostate cancer and gastric cancer[76,77]. In addition, the RB protein which regulates the transactivation function of E2F transcription factors, was also found as a target of miR-106b in laryngeal carcinoma[91]. In mice, RB loss significantly increased liver tumor multiplicity and proliferation, and had higher expression of E2F target genes compared to the wild-type controls[92].
Current evidence also showed that members of these clusters can confer resistance to apoptosis. This is achieved by miR-25 targeting components of the mitochondrial-dependent and the death receptor-dependent apoptotic pathways. In HCC, miR-25 has been shown to target the pro-apoptotic Bim[34]. Bim is a member of the BH3-only domain family of proteins which regulates the mitochondrial-dependent apoptosis pathway. It inhibits the anti-apoptotic activities of Bcl-2 or Bcl-xL by its interactions with these molecules on the mitochondria membrane[93]. In addition, miR-25 has also been shown to target the TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 (DR4) in cholangiocarcinoma[37]. The binding of TRAIL to DR4 results in the formation of the death-inducing signaling complex and the activation of caspases in the apoptotic signaling pathway[94].
The Wnt/β-catenin signaling pathway is often activated in HCC and is regulated by the miR-106b-25 cluster. Adenomatous polyposis coli (APC), which is a member of the complex of proteins that targets cytosolic β-catenin for proteasomal degradation, is targeted by miR-106b in HCC[70]. Over-expression of miR-106b decreased APC protein expression, promoted nuclear accumulation of β-catenin and activated the transcription of cyclin D1, resulting in the proliferation and anchorage-independent growth of hepatoma cells. The epithelial cell adhesion molecule E-cadherin, which links β-catenin and α-catenin to actin to stabilize cell adhesion[95], is also targeted by miR-92a and miR-25 in esophageal cancer[96,97]. Loss of E-cadherin in HCC has been shown to correlate with intrahepatic metastasis[98].
During HCC progression, both intra- and extra-hepatic metastasis can occur and efforts have been made to identify miRNAs that may contribute to metastasis in HCC. Some miRNAs are pro-metastatic while others are considered to be anti-metastatic. For the former, various miRNAs including miR-29a, miR-135a and miR-155 have been shown to promote cell migration, cell invasion and metastasis in HCC. miR-29a targets PTEN while miR-135a inhibits metastasis suppressor 1 (MTSS1)[99,100]. Expression of miR-155 has been shown to be predictive of poor survival in HCC patients following liver transplant[101]. In contrast, miR-124 and miR-29b are examples of anti-metastatic miRNAs. miR-124 is able to inhibit HCC invasion and migrating by targeting the 3’-UTR of Rho-associated, coiled-coil containing protein kinase 2 (ROCK2) and enhancer of zeste homolog 2 (Drosophila), EZH2[102] while miR-29b suppresses tumor angiogenesis, invasion and metastasis by inhibiting expression of matrix metalloproteinase 2 (MMP-2)[103].
Various studies to date have pointed to the miR-106b-25 and the miR-17-92 clusters playing a role in HCC metastasis. Over-expression of miR-17-5p leads to significant cell proliferation and intrahepatic metastasis of HCC. It was postulated and verified that increased miR-17-5p expression led to the down-regulation of its target E2F1 thereby causing the down-regulation of Wip 1 which is an E2F1-regulated gene. Wip 1 is a known suppressor of p38 mitogen-activated protein kinase (MAPK). The down-regulation of Wip 1 can trigger the activation of the p38 (MAPK) pathway, leading to an increase in phosphorylated heat shock protein 27 (HSP27) and as a result, enhances the migration of HCC cells[104]. Increased expression of miR-106b was observed to correlate with high HCC tumor grade. Over-expression of miR-106b was shown to activate the epithelial-mesenchymal transition process leading to cell migration and metastasis[71].
Although these studies suggest that the miR-106b-25 and the miR-17-92 clusters may play a pro-metastatic role, there are also reports which showed that miRNAs encoded by these clusters are anti-metastatic. miRNA-19a was among the 20 metastatic signature miRNAs which were down-regulated in metastatic HCC, suggesting a possible anti-metastatic role[105]. However, it has been documented that the global miRNA expression was down-regulated in the venous metastases compared to non-tumorous tissues[106]. In addition, miR-17 was observed to be down-regulated in metastatic cell lines. This down-regulation is dependent on the action of thyroid hormone on its receptor and the knockdown of miR-17 increased p-AKT expression resulting in enhanced cell invasion. Matrix metalloproteinase 3 (MMP3) was shown to be a direct target of miR-17 in this context[107]. Given the complexity and heterogeneity of HCC, it is possible that the role of these miRNA clusters in regulating metastasis is context-dependent, affecting different targets not only at different stages of HCC but also dependent on the genetic aberrations within the tumor.
Other targets for miR-17-92 and its 2 paralogs, especially the miR-106b-25 cluster, have been identified for other tumors. It is likely that some of these may also be relevant for HCC. Of relevance here is the modulation of the TGF-β signaling pathway by these clusters. Smad7, which is induced by TGF-β to negatively regulate the TGF-β signaling pathway, is targeted by the miR-106b-25 cluster in human breast cancer[108]. TGFBR2 is also targeted by miR-93 and miR-106b[109,110]. The decrease in Smad7 and TGFBR2 expression in HCC could promote the TGF-β-independent JNK-Smad3 pathway and increase intrahepatic metastasis[111]. Another possibility is the tumor protein 53-induced nuclear protein 1 (TP53INP1) which is a stress-induced p53-target gene that enhances p53 stability and transcriptional activity[112]. In gastric cancer, miR-17 and miR-20 were found to target TP53INP1[113]. In HCC, the expression of TP53INP1 in the tumor samples was reduced when compared to the paired non-tumor liver tissues[114]. Thus, although various targets for the miR-106b-25 and the miR-17-92 clusters have already been identified for HCC, chronic liver infections and cirrhosis, it is evident that much more work has to be done to fully dissect out all the targets of these clusters not only for HCC but also for the progression from liver fibrosis to tumorigenesis.
In this review, we focused on the miR-17-92 cluster and its paralog the miR-106b-25 cluster. miRNAs encoded by both clusters are consistently dys-regulated in diseased liver including chronic viral infection, fibrosis, cirrhosis and HCC. Although some work has been done to examine the targets and the interplay of these clusters with key tumor suppressors (PTEN), oncogenes (c-Myc) and signaling pathways that are relevant to HCC (TGF-β and Wnt/β-catenin pathways), more work has to be done to fully understand the regulation and roles of the various members of these clusters, as well as MCM7 which is the host gene for the miR-106b-25 cluster. A thorough understanding of the roles of these clusters could provide new therapeutic, prognostic and diagnostic approaches for HCC.
P- Reviewers: Grassi G, Romanelli RG S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 2. | Della Corte C, Colombo M. Surveillance for hepatocellular carcinoma. Semin Oncol. 2012;39:384-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Mormone E, George J, Nieto N. Molecular pathogenesis of hepatic fibrosis and current therapeutic approaches. Chem Biol Interact. 2011;193:225-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 4. | Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425-456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1096] [Cited by in F6Publishing: 1229] [Article Influence: 94.5] [Reference Citation Analysis (0)] |
| 5. | Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902-1910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1501] [Cited by in F6Publishing: 1404] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 6. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2902] [Cited by in F6Publishing: 2854] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 7. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3513] [Cited by in F6Publishing: 3505] [Article Influence: 166.9] [Reference Citation Analysis (0)] |
| 8. | Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016-3027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1468] [Cited by in F6Publishing: 1470] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 9. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1918] [Cited by in F6Publishing: 1822] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 10. | Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428-14433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 320] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 11. | Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1990] [Cited by in F6Publishing: 1941] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 12. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1109] [Cited by in F6Publishing: 1066] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 13. | Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979-2990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 283] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 14. | Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056-2060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1474] [Cited by in F6Publishing: 1429] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 15. | Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-596. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1267] [Cited by in F6Publishing: 1263] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 16. | Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 909] [Cited by in F6Publishing: 881] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 17. | Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515-524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1545] [Cited by in F6Publishing: 1568] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 18. | Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3501] [Cited by in F6Publishing: 3443] [Article Influence: 149.7] [Reference Citation Analysis (0)] |
| 19. | Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2408] [Cited by in F6Publishing: 2352] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 20. | Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2066] [Cited by in F6Publishing: 1994] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 21. | Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3675] [Cited by in F6Publishing: 3636] [Article Influence: 165.3] [Reference Citation Analysis (0)] |
| 22. | Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087-3095. [PubMed] [Cited in This Article: ] |
| 23. | He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2729] [Cited by in F6Publishing: 2776] [Article Influence: 146.1] [Reference Citation Analysis (0)] |
| 24. | O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2140] [Cited by in F6Publishing: 2126] [Article Influence: 111.9] [Reference Citation Analysis (0)] |
| 25. | Matsumura I, Tanaka H, Kanakura Y. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2003;2:333-338. [PubMed] [Cited in This Article: ] |
| 26. | Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-9632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1174] [Cited by in F6Publishing: 1187] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 27. | Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720-728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 783] [Cited by in F6Publishing: 784] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 28. | Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175-179. [PubMed] [Cited in This Article: ] |
| 29. | Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 499] [Cited by in F6Publishing: 492] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 30. | Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 452] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 31. | Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1236] [Cited by in F6Publishing: 1268] [Article Influence: 79.3] [Reference Citation Analysis (1)] |
| 32. | Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031-1038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 33. | Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689-1700. [PubMed] [Cited in This Article: ] |
| 34. | Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234-1242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 228] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 35. | Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 343] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 36. | Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672-1681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 376] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 37. | Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465-475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 38. | Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, Zhang X, Tang Q. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335:168-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 39. | Yu XF, Zou J, Bao ZJ, Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol. 2011;17:4711-4717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 97] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2183] [Cited by in F6Publishing: 2337] [Article Influence: 194.8] [Reference Citation Analysis (0)] |
| 41. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 42. | Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, Li RF, Liu YG. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26:1281-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 44. | Jung YJ, Kim JW, Park SJ, Min BY, Jang ES, Kim NY, Jeong SH, Shin CM, Lee SH, Park YS. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol. 2013;85:969-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 45. | Liu Y, Zhang Y, Wen J, Liu L, Zhai X, Liu J, Pan S, Chen J, Shen H, Hu Z. A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One. 2012;7:e32230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 46. | Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 267] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 47. | Kumar V, Yi Lo PH, Sawai H, Kato N, Takahashi A, Deng Z, Urabe Y, Mbarek H, Tokunaga K, Tanaka Y. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV-induced hepatocellular carcinoma. PLoS One. 2012;7:e44743. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 48. | Kishikawa T, Otsuka M, Yoshikawa T, Ohno M, Takata A, Shibata C, Kondo Y, Akanuma M, Yoshida H, Koike K. Regulation of the expression of the liver cancer susceptibility gene MICA by microRNAs. Sci Rep. 2013;3:2739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 49. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1993] [Cited by in F6Publishing: 1929] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 50. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1665] [Cited by in F6Publishing: 1619] [Article Influence: 147.2] [Reference Citation Analysis (0)] |
| 51. | Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, Cheng J, Srinivasan P, Nasheri N, Russell RS. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 52. | Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863-871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 53. | Fleming KM, Aithal GP, Solaymani-Dodaran M, Card TR, West J. Incidence and prevalence of cirrhosis in the United Kingdom, 1992-2001: a general population-based study. J Hepatol. 2008;49:732-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 54. | Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838-851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1446] [Cited by in F6Publishing: 1428] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 55. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 604] [Cited by in F6Publishing: 629] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 56. | Murakami Y, Toyoda H, Tanaka M, Kuroda M, Harada Y, Matsuda F, Tajima A, Kosaka N, Ochiya T, Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6:e16081. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 225] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 57. | Shu J, Kren BT, Xia Z, Wong PY, Li L, Hanse EA, Min MX, Li B, Albrecht JH, Zeng Y. Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology. 2011;54:609-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 58. | Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 164] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 59. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 542] [Cited by in F6Publishing: 605] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 60. | Chen YJ, Zhu JM, Wu H, Fan J, Zhou J, Hu J, Yu Q, Liu TT, Yang L, Wu CL. Circulating microRNAs as a Fingerprint for Liver Cirrhosis. PLoS One. 2013;8:e66577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 61. | Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 616] [Cited by in F6Publishing: 677] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 62. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4162] [Cited by in F6Publishing: 4422] [Article Influence: 245.7] [Reference Citation Analysis (0)] |
| 63. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 876] [Cited by in F6Publishing: 880] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 64. | Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671-678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 492] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 65. | Huang YS, Dai Y, Yu XF, Bao SY, Yin YB, Tang M, Hu CX. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 66. | Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616-1622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 67. | Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 212] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 68. | Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 470] [Cited by in F6Publishing: 499] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 69. | Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25:156-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 70. | Shen G, Jia H, Tai Q, Li Y, Chen D. miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis. 2013;34:211-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 71. | Yau WL, Lam CS, Ng L, Chow AK, Chan ST, Chan JY, Wo JY, Ng KT, Man K, Poon RT. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelial-mesenchymal transition process. PLoS One. 2013;8:e57882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 72. | Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R, Lichter P. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 73. | Nevzorova YA, Hu W, Cubero FJ, Haas U, Freimuth J, Tacke F, Trautwein C, Liedtke C. Overexpression of c-myc in hepatocytes promotes activation of hepatic stellate cells and facilitates the onset of liver fibrosis. Biochim Biophys Acta. 2013;1832:1765-1775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 74. | Yoshida K, Matsuzaki K. Differential Regulation of TGF-β/Smad Signaling in Hepatic Stellate Cells between Acute and Chronic Liver Injuries. Front Physiol. 2012;3:53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 75. | Buendia MA, Bourre L, Cairo S. Myc target miRs and liver cancer: small molecules to get Myc sick. Gastroenterology. 2012;142:214-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 76. | Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162-6170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 571] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 77. | Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 654] [Cited by in F6Publishing: 682] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 78. | Tsuruga H, Yabuta N, Hashizume K, Ikeda M, Endo Y, Nojima H. Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem Biophys Res Commun. 1997;236:118-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 79. | Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 712] [Cited by in F6Publishing: 734] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 80. | Zhao ZN, Bai JX, Zhou Q, Yan B, Qin WW, Jia LT, Meng YL, Jin BQ, Yao LB, Wang T. TSA suppresses miR-106b-93-25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC. PLoS One. 2012;7:e45133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135-2143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 430] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 82. | Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130-2134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 383] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 83. | Shiraha H, Yamamoto K, Namba M. Human hepatocyte carcinogenesis (review). Int J Oncol. 2013;42:1133-1138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 84. | Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 961] [Cited by in F6Publishing: 971] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 85. | Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Du W, Lu WY, Xuan JW, Deng Z, Yang BB. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci. 2013;126:1517-1530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 86. | Zhao B, Zhu Y, Cui K, Gao J, Yu F, Chen L, Li S. Expression and significance of PTEN and miR-92 in hepatocellular carcinoma. Mol Med Rep. 2013;7:1413-1416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 87. | Peyrou M, Bourgoin L, Foti M. PTEN in liver diseases and cancer. World J Gastroenterol. 2010;16:4627-4633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 61] [Cited by in F6Publishing: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 88. | Watanabe S, Horie Y, Kataoka E, Sato W, Dohmen T, Ohshima S, Goto T, Suzuki A. Non-alcoholic steatohepatitis and hepatocellular carcinoma: lessons from hepatocyte-specific phosphatase and tensin homolog (PTEN)-deficient mice. J Gastroenterol Hepatol. 2007;22 Suppl 1:S96-S100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 89. | Hao LS, Zhang XL, An JY, Karlin J, Tian XP, Dun ZN, Xie SR, Chen S. PTEN expression is down-regulated in liver tissues of rats with hepatic fibrosis induced by biliary stenosis. APMIS. 2009;117:681-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 90. | Takashima M, Parsons CJ, Ikejima K, Watanabe S, White ES, Rippe RA. The tumor suppressor protein PTEN inhibits rat hepatic stellate cell activation. J Gastroenterol. 2009;44:847-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 91. | Cai K, Wang Y, Bao X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J Exp Clin Cancer Res. 2011;30:73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 92. | Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 93. | Miao J, Chen GG, Yun JP, Chun SY, Zheng ZZ, Ho RL, Chak EC, Xia NS, Lai PB. Identification and characterization of BH3 domain protein Bim and its isoforms in human hepatocellular carcinomas. Apoptosis. 2007;12:1691-1701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 94. | Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782-798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 651] [Cited by in F6Publishing: 645] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 95. | Hirano S, Kimoto N, Shimoyama Y, Hirohashi S, Takeichi M. Identification of a neural alpha-catenin as a key regulator of cadherin function and multicellular organization. Cell. 1992;70:293-301. [PubMed] [Cited in This Article: ] |
| 96. | Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J, Tan FW, Ding DP, Xu XH, Zhou F. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725-10734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 97. | Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 98. | Zhai B, Yan HX, Liu SQ, Chen L, Wu MC, Wang HY. Reduced expression of E-cadherin/catenin complex in hepatocellular carcinomas. World J Gastroenterol. 2008;14:5665-5673. [PubMed] [Cited in This Article: ] |
| 99. | Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS One. 2011;6:e19518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 146] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 100. | Liu S, Guo W, Shi J, Li N, Yu X, Xue J, Fu X, Chu K, Lu C, Zhao J. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 133] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 101. | Han ZB, Chen HY, Fan JW, Wu JY, Tang HM, Peng ZH. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol. 2012;138:153-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 102. | Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 103. | Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, Zhang JP, Guan XY, Zhuang SM. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729-1740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 244] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 104. | Yang F, Yin Y, Wang F, Wang Y, Zhang L, Tang Y, Sun S. miR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology. 2010;51:1614-1623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 105. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 543] [Cited by in F6Publishing: 594] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 106. | Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. 2012;55:1453-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 107. | Lin YH, Liao CJ, Huang YH, Wu MH, Chi HC, Wu SM, Chen CY, Tseng YH, Tsai CY, Chung IH. Thyroid hormone receptor represses miR-17 expression to enhance tumor metastasis in human hepatoma cells. Oncogene. 2013;32:4509-4518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 108. | Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162-5171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 109. | Xu D, He XX, Chang Y, Sun SZ, Xu CR, Lin JS. Downregulation of MiR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology. 2012;59:2367-2373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 110. | Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H, Liu Y, Ma C, Huang L, Zhang L. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-β type II receptor. Brain Res. 2010;1357:166-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 111. | Mamiya T, Yamazaki K, Masugi Y, Mori T, Effendi K, Du W, Hibi T, Tanabe M, Ueda M, Takayama T. Reduced transforming growth factor-beta receptor II expression in hepatocellular carcinoma correlates with intrahepatic metastasis. Lab Invest. 2010;90:1339-1345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 112. | Shahbazi J, Lock R, Liu T. Tumor Protein 53-Induced Nuclear Protein 1 Enhances p53 Function and Represses Tumorigenesis. Front Genet. 2013;4:80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 113. | Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, Tao Y, Zhang L, Xu W. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer. 2013;49:2010-2021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 114. | Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 313] [Article Influence: 24.1] [Reference Citation Analysis (0)] |