Published online Jun 7, 2014. doi: 10.3748/wjg.v20.i21.6515
Revised: March 4, 2014
Accepted: April 30, 2014
Published online: June 7, 2014
AIM: To investigate the biological role and underlying mechanism of miR-132 in colorectal cancer (CRC) progression and invasion.
METHODS: Quantitative RT-PCR analysis was used to examine the expression levels of miR-132 in five CRC cell lines (SW480, SW620, HCT116, HT29 and LoVo) and a normal colonic cell line NCM460, as well as in tumor tissues with or without metastases. The Kaplan-Meier method was used to analyze the prognostic significance of miR-132 in CRC patients. The biological effects of miR-132 were assessed in CRC cell lines using the transwell assay. Quantitative RT-PCR and western blot analyses were employed to evaluate the expression of miR-132 targets. The regulation of ZEB2 by miR-132 was confirmed using the luciferase activity assay.
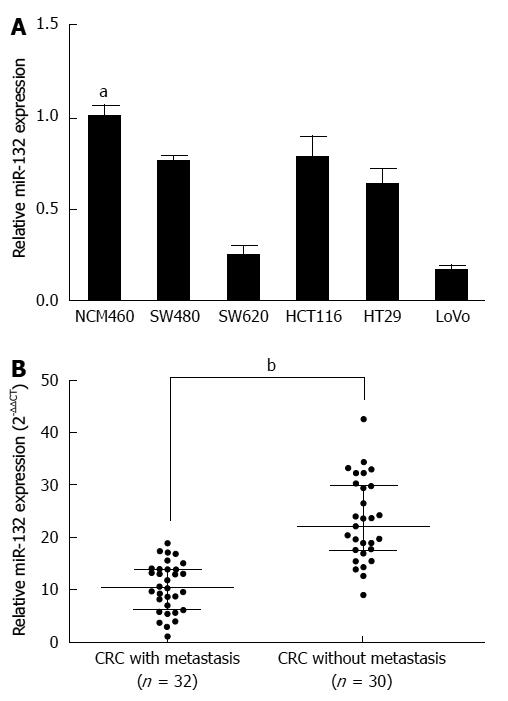
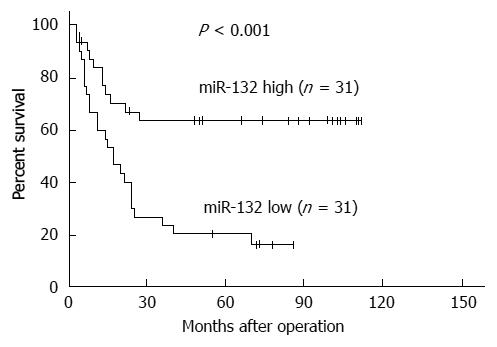
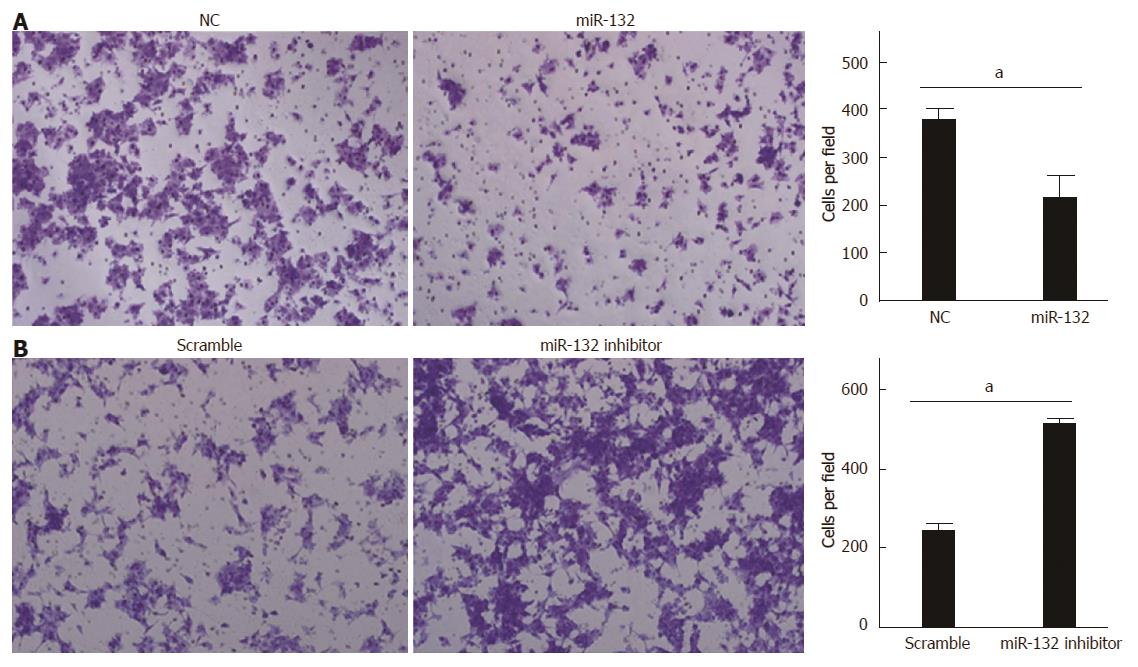
RESULTS: miR-132 was significantly down-regulated in the CRC cell lines compared with the normal colonic cell line (P < 0.05), as well as in the CRC tissues with distant metastases compared with the tissues without metastases (10.52 ± 4.69 vs 23.11 ± 7.84) (P < 0.001). Down-regulation of miR-132 was associated with tumor size (P = 0.016), distant metastasis (P = 0.002), and TNM stage (P = 0.020) in CRC patients. Kaplan-Meier survival curve analysis indicated that patients with low expression of miR-132 tended to have worse disease-free survival than patients with high expression of miR-132 (P < 0.001). Moreover, ectopic expression of miR-132 markedly inhibited cell invasion (P < 0.05) and the epithelial-mesenchymal transition (EMT) in CRC cell lines. Further investigation revealed ZEB2, an EMT regulator, was a downstream target of miR-132.
CONCLUSION: Our study indicated that miR-132 plays an important role in the invasion and metastasis of CRC.
Core tip: In this study, we reported the clinical significance and biological effects of miR-132 in colorectal cancer (CRC). We found that miR-132 was significantly down-regulated in tumor cell lines and CRC tissues with distant metastases. Down-regulation of miR-132 was associated with aggressive tumor phenotypes and adverse prognosis in CRC patients. Moreover, we showed that ectopic expression of miR-132 significantly inhibited cell invasion and the epithelial-mesenchymal transition (EMT), whereas knockdown of miR-132 promoted cell invasion and EMT in CRC cells. Further investigation revealed ZEB2, an EMT regulator, was a downstream target of miR-132.
-
Citation: Zheng YB, Luo HP, Shi Q, Hao ZN, Ding Y, Wang QS, Li SB, Xiao GC, Tong SL. miR-132 inhibits colorectal cancer invasion and metastasis
via directly targeting ZEB2. World J Gastroenterol 2014; 20(21): 6515-6522 - URL: https://www.wjgnet.com/1007-9327/full/v20/i21/6515.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i21.6515
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in western countries[1]. Recent development in the therapeutic strategies has helped to cure many patients with early-stage disease. However, the prognosis of patients with advanced disease and metastasis is still poor. Therefore, further investigation into the underlying molecular mechanisms of CRC progression to identify biomarkers to distinguish CRC patients with or without a high risk of metastasis is of great importance.
Tumor invasion and metastasis are parts of a complicated process in which the tumor grows, then detaches from the primary site and metastasizes to a distant organ. Aberrations of protein-coding genes have been widely accepted to play critical roles in the pathology of CRC, including both oncogenes and tumor suppressive genes[2]. Recently, a series of studies have revealed that microRNAs (miRNAs) regulate various genes that play a pivotal role in the process of tumor progression and metastasis.
miRNAs are a class of short (approximately 18-22 nucleotides in length), endogenous, non-coding RNAs that can negatively regulate the expressions of various protein-coding genes by base pairing with their 3’ untranslated region (3’-UTR)[3-5], leading to their post-transcriptional translation inhibition or mRNA degradation[6]. miRNAs have been found to be involved in the regulation of multiple pathological processes that contribute to tumorigenesis and metastasis, such as tumor cell proliferation, differentiation, apoptosis, and invasion[7-9]. In CRC, several reports have indicated the role of miRNAs in regulating tumor invasion and metastasis. For example, miR-93 suppresses the proliferation and colony formation of human colon cancer stem cells by regulating HDAC8 and TLE4[10]. Additionally, miR-21 is a potential biomarker in colon and rectal cancer[11]. Previously, Zhang et al[12] reported that down-regulation of miR-132 by promoter methylation promotes pancreatic cancer development; Formosa et al[13] found that miR-132 is silenced by promoter CpG island methylation, which contributes to prostate cancer progression and metastasis. However, the role of miR-132 in CRC progression and metastasis remains unclear.
In the present study, we first examined the expression level of miR-132 in CRC tissues and cell lines. The results showed that miR-132 was significantly down-regulated in CRC tissues with metastasis compared with tissues without metastasis; the level of miR-132 was lower in CRC cell lines than in the NCM460 cell line (a normal colonic cell line). Ectopic expression of miR-132 markedly inhibited the invasiveness and epithelial-mesenchymal transition (EMT) of CRC cells. Further study indicated that ZEB2 (an EMT regulator) was a direct downstream target of miR-132. Collectively, these results demonstrated that miR-132 inhibited cell invasion and EMT in CRC cells through targeting ZEB2, providing a valuable target for cancer therapy.
CRC tissues with distant metastases (n = 32) and CRC tissues without distant metastases (n = 30) were obtained from 62 CRC patients who underwent initial surgery at Renmin Hospital of Wuhan University between June 2005 and January 2008. All the patients had a histological diagnosis of CRC. Following resection, the specimens were snap-frozen in liquid nitrogen and stored at -80 °C until RNA extraction. All the subjects involved in this study provided written informed consent. This project was approved by the ethics committee of Wuhan University.
The human HT-29, HCT116, SW480, SW620 and LoVo CRC cell lines, the 293T embryonic kidney cell line, and the NCM460 normal colonic epithelial cell line were purchased from the American Type Culture Collection (ATCC). The cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum in a humidified 37 °C incubator supplemented with 5% CO2.
Total RNA from the tissues and cells was extracted using Trizol reagent (Invitrogen, Carlsbad, California, United States). RNA quality and concentration were determined using the Nanodrop 2000 system (Thermo Fisher Scientific, Wilmington, United States). For analysis of the miR-132 level, a TaqMan MicroRNA Assay Kit (Applied Biosystems) was used, and U6 snRNA was used as the reference. For measurement of the ZEB2 mRNA level, a SYBR Premix Ex TaqTM kit (Takara) was used, and β-actin expression was used as endogenous control. Real-time PCR was performed using the Applied Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems, Foster City, California, United States). Data were analyzed using the 2-ΔΔct method.
Western blot analysis was performed as previously described[14]. Briefly, total cellular protein was isolated, and the protein concentration was determined using the Bradford DC protein assay (Bio-Rad, CA, United States). A total of 40 μg of protein was separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. Membranes were then incubated with the following primary antibodies: ZEB2 (1:1000; CST, United States), E-cadherin (1:1000; CST, United States), α-catenin (1:1000; CST, United States), vimentin (1:1000; CST, United States), fibronectin (1:1000; CST, United States) and GAPDH (1:2000; Santa Cruz Biotechnology, United States). Proteins were visualized using the ECL procedure (Amersham Biosciences, United States).
The hsa-miR-132 mimic, negative control (NC) oligonucleotides, has-miR-132 inhibitor and scramble oligonucleotides were purchased from Ribobio (Guangzhou, China). The cells were plated in a six-well plate the day before transfection. LoVo cells were transfected with the has-miR-132 mimic or NC (50 nmol/L), and HCT116 cells were infected with the has-miR-132 inhibitor or scramble oligonucleotides (100 nmol/L), using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells were collected, and in vitro assays were performed.
The cell invasive potential was evaluated using specialized transwell chambers (8-μm pore; BD Biosciences). The cells (5 × 104 cells suspended in 500 μL of serum-free medium) were added to the upper chamber of the inserts, which were coated with a Matrigel mix; fetal bovine serum (500 μL) was added to the bottom chamber as a chemoattractant. Twenty-four hours later, the non-invading cells on the upper surface were removed, and the cells that invaded to the bottom side of the membrane were fixed with methanol, stained with 0.1% crystal violet, air dried and subjected to digital image acquisition. The number of invasive cells was evaluated in five independent fields under a microscope. The mean of triplicate assays for each experimental condition was analyzed.
For the luciferase assays, the potential miR-132 binding site in the ZEB2 3′-UTR was predicted using TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org). The 3’-UTR of the ZEB2 mRNA and a mutant ZEB2 mRNA were synthesized and cloned into the Xba I site of a pGL3 basic vector (Promega, United States) downstream from the luciferase stop codon and were designated as pGL3-wt-ZEB2 and pGL3-mt-ZEB2, respectively. Then, 293T cells (1 × 105 cells/well) were cultured in 24-well plates and co-transfected with the pGL3-Control (0.4 mg), pGL3-wt-ZEB2 (0.4 mg) or pGL3-mt-ZEB2 (0.4 mg) plasmid, the pRL-TK luciferase reporters (25 ng/well) and pcDNA-miR-132 (20 nmol/L) or pcDNA-miR-NC (20 nmol/L) using Lipofectamine 2000 (Invitrogen, United States). Forty-eight hours later, the cells were harvested, and luciferase activities were measured using a Dual-Luciferase Reporter Assay kit (Promega, United States).
The data were expressed as the mean ± SD. Statistical significance was analyzed using Student’s t-test (two-tailed). All statistical analyses were performed using SPSS 13.0 or the GraphPad Prism 5.0 software package. The Kaplan-Meier method and log-rank test were performed to analyze the prognostic significance. P < 0.05 was considered to be statistically significant in all tests.
First, we analyzed the expression of miR-132 in five CRC cell lines and the normal colonic cell line NCM460. MiR-132 was significantly down-regulated in CRC cell lines compared with the normal colonic cell line NCM460 (all P < 0.05) (Figure 1A). Among the six CRC cell lines, LoVo possessed the lowest miR-132 level, and HCT116 exhibited the highest miR-132 level (Figure 1A). To explore the expression of miR-132 in CRC tissues with different metastatic characteristics, the miR-132 level was measured in tissues with distant metastases (n = 32) and tissues without distant metastases (n = 30). Interestingly, miR-132 was markedly lower in CRC tissues with distant metastases than that in CRC tissues without distant metastases (10.52 ± 4.69 vs 23.11 ± 7.84) (P < 0.001; Figure 1B). To evaluate the clinical value of miR-132 in CRC patients, we divided the patients into two groups according to the median value (15.40) of the miR-132 level. The correlation between miR-132 and clinicopathological characteristics was then analyzed (Table 1). Low expression of miR-132 was significantly associated with a larger tumor size (P = 0.016), distant metastasis (P = 0.002) and TNM stage (P = 0.020). However, age, gender, CEA level, CA199 level, tumor site and differentiation showed no association with miR-132 expression (Table 1). Kaplan-Meier analysis showed patients with low miR-132 expression had significantly worse disease-free survival than patients with high miR-132 expression (P < 0.001, Figure 2).
| Characteristics | n | miR-132 | P value | |
| Low | High | |||
| Age | 0.307 | |||
| < 60 yr | 34 | 19 (55) | 15 (45) | |
| ≥ 60 yr | 28 | 12 (42) | 16 (58) | |
| Gender | 0.437 | |||
| Male | 37 | 20 (54) | 17 (46) | |
| Female | 25 | 11 (44) | 14 (56) | |
| CEA Level | 0.127 | |||
| 0-5 ng/mL | 32 | 13 (41) | 19 (59) | |
| > 5 ng/mL | 30 | 18 (60) | 12 (40) | |
| CA199 Level | 0.082 | |||
| 0-35 μ/mL | 46 | 20 (43) | 26 (57) | |
| > 35 μ/mL | 16 | 11 (69) | 5 (31) | |
| Tumor site | 0.796 | |||
| Colon | 37 | 18 (49) | 19 (51) | |
| Rectum | 25 | 13 (52) | 12 (48) | |
| Tumor size (cm) | 0.016 | |||
| ≤ 5 | 41 | 16 (39) | 25 (61) | |
| > 5 | 21 | 15 (71) | 6 (29) | |
| Differentiation | 0.776 | |||
| Well/moderate | 45 | 23 (51) | 22 (49) | |
| Poor | 17 | 8 (47) | 9 (53) | |
| Distant metastasis | 0.002 | |||
| Yes | 32 | 10 (31) | 22 (69) | |
| No | 30 | 21 (70) | 9 (30) | |
| TNM stage | ||||
| I/II | 11 | 2 (18) | 9 (72) | 0.020 |
| III/IV | 51 | 29 (56) | 22 (44) | |
Because miR-132 was significantly down-regulated in CRC tissues with distant metastases, we hypothesized that miR-132 could inhibit cell invasion. LoVo cells with low miR-132 expression were transfected with the miR-132 mimic to overexpress miR-132, whereas HCT116 cells with high miR-132 expression were transfected with the miR-132 inhibitor to knockdown endogenous miR-132. The efficiency of transfection was confirmed by real-time PCR (data not shown). As expected, ectopic expression of miR-132 reduced LoVo cell invasion by about 40% (P < 0.05; Figure 3A). By contrast, knockdown of miR-132 significantly increased the invasiveness of the HCT116 cells (P < 0.05; Figure 3B).
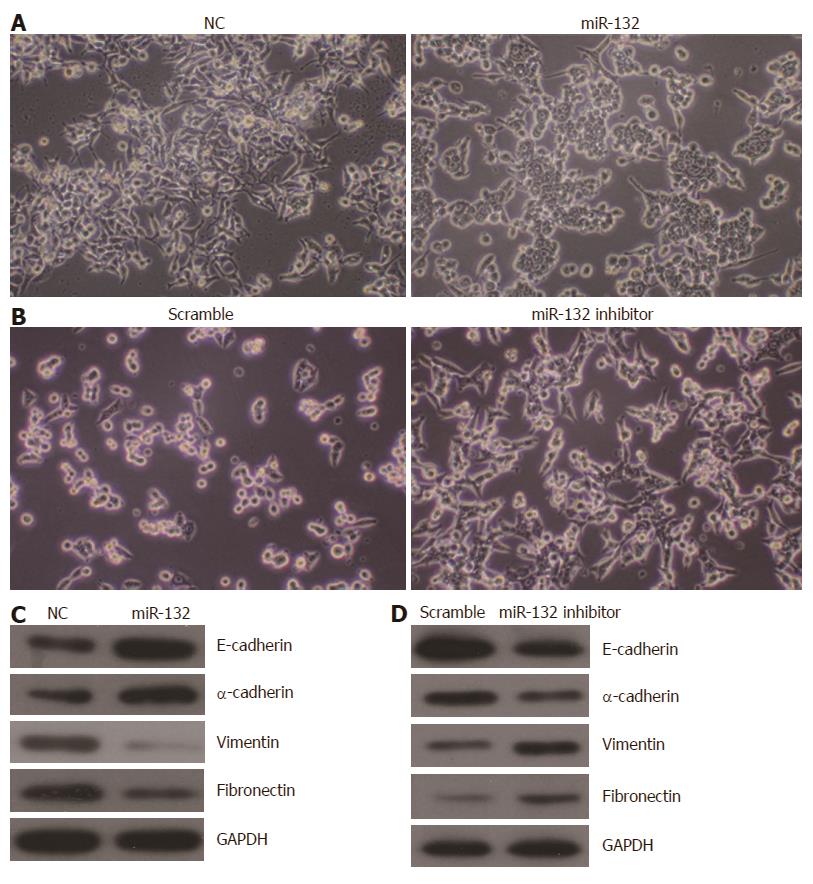
EMT is known to be important for tumor metastasis. Thus, we investigated the role of miR-132 in EMT in CRC cells. As shown in Figure 4A, ectopic expression of miR-132 induced a morphological change from the spindle-shaped mesenchymal form to a cobblestone-shaped epithelial-like form in LoVo cells. By contrast, knockdown of miR-132 in HCT116 cells led to more spindle-shaped mesenchymal characteristics (Figure 4B). Moreover, the expression of a set of EMT-related protein markers was also altered along with the morphological changes. Overexpression of miR-132 increased the protein levels of epithelial markers (E-cadherin and α-catenin) but decreased the expression of mesenchymal markers (vimentin and fibronectin) in LoVo cells (Figure 4C). By contrast, knockdown of miR-132 resulted in a decreased level of epithelial markers (E-cadherin and α-catenin) but increased levels of mesenchymal markers (vimentin and fibronectin) in HCT116 cells (Figure 4D).
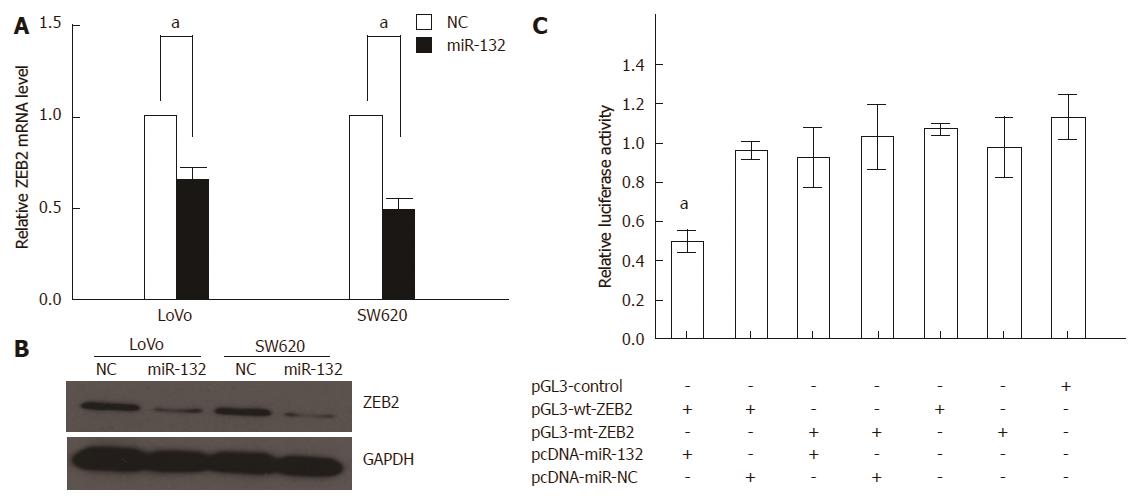
To investigate the underlying mechanism of miR-132 in CRC, we first used TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org) to search for potential targets of miR-132. Because miR-132 could suppress CRC invasion, we focused on genes that could promote CRC metastasis. We found that the 3’-UTR of ZEB2 contains the complementary site for the seed region of miR-132. Additionally, ectopic expression of miR-132 could significantly reduce the ZEB2 mRNA and protein levels in LoVo and SW620 cells (Figure 5A and B). Furthermore, a luciferase activity assay demonstrated that miR-132 could significantly decrease the luciferase activity of the wild-type ZEB2 3’-UTR by 40%, a finding that was not observed for the mutant ZEB2 3’-UTR (Figure 5C).
Metastasis has been widely recognized as an adverse prognostic factor in CRC. miRNAs have been demonstrated as an important regulator of tumor progression and metastasis[15]. In CRC, many miRNAs have been identified that are able to regulate known genes that are involved in the pathology of tumorigenesis and metastasis. For example, miR-21, miR-31 and miR-192 could induce CRC cell resistance to 5-fluorouracil (5-FU), a finding that might shed light on chemotherapy for CRC patients[16-18]; miR-28 inhibits CRC tumor growth and metastasis by CCND1 and NM23-H1[19]. miR-132 has been found to be a tumor suppressor in a series of cancers, such as prostate cancer, hepatocellular carcinoma and ductal carcinoma in situ (DCIS) of the breast[13,20,21]. Recently, a report indicated that miR-132 could regulate apoptosis in non-small cell lung cancer independent of acetylcholinesterase[22]. In the present study, we showed that miR-132 was significantly down-regulated in CRC tissues with distant metastases.
Moreover, miR-132 was associated with tumor size, distant metastasis and TNM stage and could predict survival in CRC patients. These results indicated that miR-132 down-regulation might be a common occurrence in CRC, and miR-132 could be used as a biomarker to predict clinical outcome and metastasis in CRC patients.
EMT is a process in which adherent epithelial cells shed their epithelial traits and acquire mesenchymal properties, including fibroblastoid morphology and increased potential for motility, and, in the case of cancer cells, increased invasion, metastasis and resistance to chemotherapy[23-25]. Although the signaling pathways of EMT are complicated, the hallmark of EMT in tumors is the down-regulation of E-cadherin, which is also considered to be a suppressor of invasion and metastasis. A series of transcription factors have been found to regulate EMT programs in cancer metastasis, including direct transcriptional repressors of E-cadherin-Snail (SNAI1), Slug (SNAI2), ZEB2 (SIP1), ZEB1, FOXC2 and Twist[25]. In our study, we found that ectopic expression of miR-132 inhibited CRC cell invasion and EMT. By contrast, knockdown of miR-132 promoted cell motility and EMT progression. To the best of our knowledge, this is the first study to report the role of miR-132 in CRC metastasis.
MiRNAs are known to function by regulating target genes. Previous reports have found many target genes for miR-132 in cancer. For instance, Lagos et al[26] reported that miR-132 regulated antiviral innate immunity through suppression of the p300 transcriptional co-activator. Alvarez-Saavedra et al[27] showed that genes involved in chromatin remodeling (Mecp2, Ep300, Jarid1a) and translational control (Btg2, Paip2a) were direct targets of miR-132 in the mouse suprachiasmatic nucleus. In our study, we indicated that ZEB2, which is a transcriptional repressor of E-cadherin and plays an important role in EMT, was a direct target of miR-132 in CRC cells. These results showed that one miRNA might target different genes depending on the tumor types and cellular environment. However, further study is needed to investigate the underlying mechanisms of miR-132 in CRC.
In conclusion, we found that miR-132 was significantly down-regulated in CRC with distant metastasis. Moreover, miR-132 could predict disease-free survival and distant metastasis in CRC patients. Further investigation identified that the EMT regulator ZEB2 was a direct target of miR-132. Taken together, these data implicate that miR-132 might be used as a prognostic indicator and therapeutic target in CRC patients.
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in western countries. Although recent developments in therapeutic strategies have helped cure many patients with early stage disease, the prognosis of patients with advanced disease and metastasis remains poor. Further investigation into the underlying molecular mechanisms of CRC progression to identify biomarkers to distinguish CRC patients with or without a high risk of metastasis is of great importance.
MiRNAs have been found to be involved in the regulation of multiple pathological processes that contribute to tumorigenesis and metastasis, such as tumor cell proliferation, differentiation, apoptosis, and invasion. In CRC, studies have indicated that miRNAs play important roles in regulating tumor invasion and metastasis. Previously, Zhang et al reported that down-regulation of miR-132 by promoter methylation promotes pancreatic cancer development. Formosa et al found that miR-132 is silenced by promoter CpG island methylation, a process that contributes to prostate cancer progression and metastasis. However, the role of miR-132 in CRC progression and metastasis remains unclear and needs further exploration.
The authors found that miR-132 was significantly down-regulated in CRC tissues with metastasis compared with tissues without metastasis; the level of miR-132 was lower in CRC cell lines than in NCM460 cells (a normal colonic cell line). Ectopic expression of miR-132 markedly inhibited the invasiveness and epithelial-mesenchymal transition (EMT) in CRC cells. Further study indicated that ZEB2 (an EMT regulator) was a direct downstream target of miR-132. Collectively, these results demonstrated that miR-132 inhibited cell invasion and EMT in CRC cells by targeting ZEB2, thus providing a valuable target for cancer therapy.
The findings in this study indicated that miR-132 was significantly down-regulated in CRC with distant metastases. Moreover, miR-132 could predict disease-free survival and distant metastasis in CRC patients. Further investigation identified that the EMT regulator ZEB2 was a direct target of miR-132. Taken together, these data implicate that miR-132 might be used as a prognostic indicator and therapeutic target in CRC patients.
MiRNA: a small non-coding RNA molecule (approximately 22 nucleotides in length) found in plants, animals, and some viruses that functions in transcriptional and post-transcriptional regulation of gene expression; epithelial-mesenchymal transition: a process by which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory and invasive properties to become mesenchymal stem cells.
The authors report on the biological and clinical significance of miR-132 in colorectal cancer, adding some information on a possible target of that microRNA. The results of the study are interesting and innovative. The sample size of the study is sufficiently large. The methods used are updated and well described, and the statistics are appropriate.
P- Reviewer: Roncucci L S- Editor: Ma YJ L- Editor: O’Neill M E- Editor: Wang CH
| 1. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10002] [Cited by in F6Publishing: 10349] [Article Influence: 739.2] [Reference Citation Analysis (0)] |
| 2. | Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-1113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2283] [Cited by in F6Publishing: 2213] [Article Influence: 130.2] [Reference Citation Analysis (0)] |
| 3. | Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753-1761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 537] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 4. | Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519-1524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 978] [Cited by in F6Publishing: 990] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 5. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [Cited in This Article: ] |
| 6. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5384] [Cited by in F6Publishing: 5479] [Article Influence: 304.4] [Reference Citation Analysis (0)] |
| 7. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5705] [Cited by in F6Publishing: 5849] [Article Influence: 324.9] [Reference Citation Analysis (0)] |
| 8. | Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143-3148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 265] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 618] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 10. | Yu XF, Zou J, Bao ZJ, Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol. 2011;17:4711-4717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 97] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 11. | Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol. 2013;19:5615-5621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 53] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183-1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 13. | Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agrò E. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 14. | Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 15. | Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2843] [Cited by in F6Publishing: 2984] [Article Influence: 186.5] [Reference Citation Analysis (0)] |
| 16. | Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci USA. 2010;107:21098-21103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 281] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 17. | Wang CJ, Stratmann J, Zhou ZG, Sun XF. Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer. 2010;10:616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 18. | Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265-2275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 19. | Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886-896.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 152] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 20. | Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037-1043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu F, Fan Y, Lang R, Fu L, Gu L. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 22. | Zhang B, Lu L, Zhang X, Ye W, Wu J, Xi Q. Hsa-miR-132 Regulates Apoptosis in Non-Small Cell Lung Cancer Independent of Acetylcholinesterase. J Mol Neurosci. 2013;Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449-15454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 747] [Cited by in F6Publishing: 779] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 24. | Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741-4751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1819] [Cited by in F6Publishing: 1976] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 25. | Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2456] [Cited by in F6Publishing: 2486] [Article Influence: 165.7] [Reference Citation Analysis (0)] |
| 26. | Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 27. | Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 150] [Article Influence: 10.7] [Reference Citation Analysis (0)] |