Published online Jun 28, 2014. doi: 10.3748/wjg.v20.i24.7555
Revised: February 18, 2014
Accepted: April 21, 2014
Published online: June 28, 2014
Hepatitis C virus (HCV) is a major cause of liver disease worldwide. HCV is able to evade host defense mechanisms, including both innate and acquired immune responses, to establish persistent infection, which results in a broad spectrum of pathogenicity, such as lipid and glucose metabolism disorders and hepatocellular carcinoma development. The HCV genome is characterized by a high degree of genetic diversity, which can be associated with viral sensitivity or resistance (reflected by different virological responses) to interferon (IFN)-based therapy. In this regard, it is of importance to note that polymorphisms in certain HCV genomic regions have shown a close correlation with treatment outcome. In particular, among the HCV proteins, the core and nonstructural proteins (NS) 5A have been extensively studied for their correlation with responses to IFN-based treatment. This review aims to cover updated information on the impact of major HCV genetic factors, including HCV genotype, mutations in amino acids 70 and 91 of the core protein and sequence heterogeneity in the IFN sensitivity-determining region and IFN/ribavirin resistance-determining region of NS5A, on virological responses to IFN-based therapy.
Core tip: This review aims to cover recent updates on the impact of major hepatitis C virus (HCV) genetic factors, including HCV genotype, mutations in amino acids 70 and 91 of the core protein and sequence heterogeneity in interferon (IFN) sensitivity-determining region and IFN/ribavirin resistance-determining region of Nonstructural proteins 5A, on virological responses to IFN-based therapy.
- Citation: El-Shamy A, Hotta H. Impact of hepatitis C virus heterogeneity on interferon sensitivity: An overview. World J Gastroenterol 2014; 20(24): 7555-7569
- URL: https://www.wjgnet.com/1007-9327/full/v20/i24/7555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i24.7555
Since its discovery in 1989[1,2], hepatitis C virus (HCV) has been the subject of intense research and clinical investigations as its major role in human disease has emerged. Globally, HCV is estimated to infect 180 million people, who represent about 3% of the world’s population. HCV is a major cause of chronic liver disease, such as chronic hepatitis, liver cirrhosis and hepatocellular carcinoma (HCC)[3-6]. HCC is the third most common cause of cancer-related mortality worldwide[7]. In particular, HCV infection accounts for 30%-90% of HCC cases in Western Europe, United States and Asia[8]. Although the treatment of HCV infection is available, it is costly and requires long-term medical support and follow-up. Moreover, current therapies are still impractical for a substantial proportion of HCV-infected patients. The development of a protective vaccine remains a distant prospect.
HCV is an enveloped virus with a positive-strand RNA molecule of approximately 9600 nucleotides. HCV lacks a DNA intermediate; thus, it is incapable of integrating into host chromosomal DNA. Despite this, unlike most RNA viruses, HCV is capable of establishing persistent infection. This ability is central to HCV pathogenesis because it allows chronic infection to occur in 60% to 90% of infected individuals, and virtually all clinically significant HCV-related liver damage takes place during the chronic phase of infection[9,10].
The HCV genome encodes a single open reading frame that encodes a large polyprotein of approximately 3000 amino acids (aa). The polyprotein is processed by host cell peptidases and viral proteases to generate three structural (core, E1, E2) and seven non-structural (p7, NS2 to NS5B) proteins[11]. Both ends of the HCV genome contain highly conserved untranslated regions (5’- and 3’-UTRs) that are critical for genome replication and viral protein translation[12,13]. The 5’-UTR contains the HCV internal ribosome entry site (IRES), an RNA structural element that mediates ribosome binding for translation in a cap-independent manner, while the 3’-UTR is required for HCV RNA replication[14-17].
HCV displays a high nucleotide mutation rate that is estimated to be 1.44 × 10-3 nucleotide changes per site per year over the whole genome[18,19]. This mainly arises from the error-prone nature of its RNA-dependent RNA polymerase, which lacks 3’-to-5’ exonuclease proofreading activity. This has resulted in diversification of HCV into distinct genotypes and subtypes. HCV exists in the host as quasispecies, which are a dynamic distribution of non-identical but closely related genomes[20,21]. This genetic diversity plays a vital role in HCV’s ability to establish persistent infection and to evade the various selective pressures exerted by immune responses and antiviral therapy. Also, different HCV genotypes exhibit different treatment responses and different pathogenicity. Consequently, the impact of sequence heterogeneity within particular regions of the HCV genome, such as the core, E2, NS3 and NS5A, on treatment responses has been a subject of interest for many researchers. In this review, we will discuss the updated information about major HCV genetic factors, including viral genotype and sequence heterogeneity within certain regions of the HCV genome, in particular the core and NS5A regions, that influence the outcome of interferon (IFN)-based therapy.
HCV exhibits genetic variability at several different levels. Most obvious is the genetic divergence of the main genotypes of HCV. Phylogenetic analysis of nucleotide sequences recovered from infected individuals in different geographical regions has classified HCV into seven major genotypes and series of subtypes[22]. HCV genotypes differ in 30%-35% of nucleotide sites over the whole genome, while the subtypes within a given genotype differ in 20%-25% of their nucleotide sites, with more sequence variability concentrated in such regions as the E1 and E2 glycoproteins, while more sequence conservation is found in the 5’- and 3’-UTR, the core gene and some of the nonstructural protein genes, such as NS3[23].
HCV genotypes differ in three major properties that highlight the importance of genetic diversity among the different HCV genotypes: (1) the prevalence of certain HCV genotype is frequently associated with certain geographical ranges; for example, HCV genotype 1 is prevalent in North America and Japan, genotype 3 is most common on the Indian subcontinent, genotype 4 is the most common genotype in Africa and the Middle East, genotype 5 can be found in South Africa and genotype 6 in Southeast Asia[24]; (2) the pathogenicity of HCV infection varies among the different genotypes; for example, HCV genotype 3 infection is associated with a higher degree of liver steatosis[25-27] and genotype 1 infection associated with a higher risk of HCC development[28,29]; and (3) the response rates to IFN-based therapy vary significantly between the different HCV genotypes[30-34].
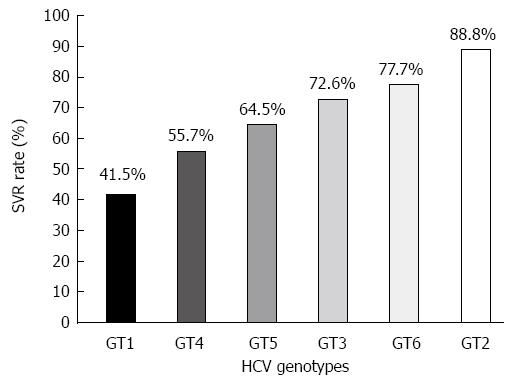
To date, IFN represents the backbone of HCV therapeutic options. Pegylated-IFN and ribavirin (PEG-IFN/RBV) combination therapy is the standard of care for the treatment of chronic hepatitis C infection of different HCV genotypes. Recently, new direct-acting antivirals (DAAs) have been introduced to the treatment of HCV genotype 1 infection[35,36]. HCV genotype is an important determinant of both treatment strategy and outcome. HCV genotype 1 and genotype 4 infections need longer (48 wk) treatment period than genotypes 2 and 3 (24 wk); HCV genotypes 1 and 4 are less responsive to PEG-IFN/RBV treatment, with a sustained virological response (SVR) rate hovering around 50%[30,37-41], while the SVR rate in HCV genotype 2 and genotype 3 infections approaches 80%[30,37]. These differences in the SVR rates observed among different HCV genotypes (Figure 1) have suggested that viral genome variability could play a role in determining treatment outcome. However, it is still unclear which genetic element(s) within the HCV genome accounts for the difference in treatment response rates among different HCV genotypes. Interestingly, a series of detailed phylogenetic analyses have shown that there is a significant correlation between the relative evolutionary age of HCV genotypes and the response rates to IFN-based therapy[42]. In these analyses, HCV genotype 2 branched first, genotypes 1 and 4 branched last, and genotypes 3, 5 and 6 branched in between. Thus, it has been hypothesized that genotypes that emerged earlier exhibit better treatment outcomes, while newly-emerged genotypes have higher rates of resistance to IFN-based therapy than their ancestors. This correlation might be attributable to selective pressures generated by the host immune system, which the IFN-based therapy relies on to a large extent.
As stated above, the response rates to IFN-based therapy vary between different HCV genotypes. More importantly, the sensitivity to IFN also varies between different HCV isolates of a given genotype and subtype. This has further highlighted a possible role for certain viral genetic factors in determining IFN responsiveness. In this context, genetic variations within two genomic regions, the core and NS5A, have been widely discussed for their correlation with treatment outcome. We will cover updated information regarding the impact of sequence heterogeneity within the core and NS5A regions of different HCV genotypes on treatment outcome, as described below.
At both the RNA and protein levels, HCV core plays a critical role in the virus life cycle. At the RNA level, a limited region of the first 45 to 60 nucleotides of the core gene, together with the 5’-UTR, is required for IRES function that initiates translation of the HCV polyprotein[14-17,43]. At the protein level, the HCV core is an RNA-binding protein that forms the capsid shell to protect the HCV genome while the virus passes from one cell to another or from one person to another. Furthermore, the core protein plays an important role(s) in the pathogenicity of HCV by modulating host cellular signaling pathways through interaction with a variety of cellular proteins. The core protein has been implicated in IFN resistance, liver steatosis, insulin resistance and hepatocellular carcinoma[44-49].
The HCV core protein shares high homology among different HCV genotypes. There are, however, certain polymorphisms, which are closely associated with the clinical outcome of IFN-based therapy. Akuta et al[50] first reported that aa mutations at positions 70 and 91 of the HCV genotype 1b (HCV-1b) core protein were associated with virological responses to PEG-IFN/RBV treatment. They found a significant correlation between core mutations at aa 70 (Arg70 to Gln70/His70) and/or aa 91 (Leu91 to Met91) and poor treatment response. Gln70/His70 and/or Met91 were found in 100% of ultimate resistance cases, who tested positive for HCV RNA at the end of 48-wk PEG-IFN/RBV treatment, but in only 42% of responders. Subsequently, several clinical studies were conducted by the same investigators and others, including our group, on Japanese patients infected with HCV-1b to follow up this observation. Most of these studies corroborate the initial observation that identified the aa mutation at position 70 of the core protein as a negative predictive marker for resistance to PEG-IFN/RBV treatment[51-60]. The polymorphism at aa 70 is also a useful determinant for the virologic response to extended 72-wk PEG-IFN/RBV treatment[61]. However, it is worth noting that although the significance of the point mutation at aa 70 (Gln70) in prediction of poor treatment response is consistent among most clinical studies, the possible significance of the mutation at aa 91 is contradictory.
Since the discovery of their importance in IFN resistance, mutations at aa 70 and 91 have been the subject of intensive investigation for different aspects of chronic HCV infection including disease progression and pathogenesis. In HCV-1b infection, Gln70/His70 and/or Met91 were significantly associated with severe insulin resistance[62], the severity of liver disease, elevated gamma-glutamyl transpeptidase (γ-GTP) levels, low platelet count and low albumin levels[63]. More importantly, Gln70/His70 and/or Met91 (non-wild core) are significantly associated with an increased risk of HCC development[64-69]. Notably, Gln70 is the only HCV point mutation associated with both an increased risk of HCC and IFN treatment failure in multiple studies. Because increased HCC risk and IFN treatment failure are associated with the same viral point mutation, it is possible that both adverse outcomes result from disruption of the same cellular pathway, specifically, the IFN signaling pathway that is involved in both anti-proliferative and antiviral functions.
It is important to note that the majority of the studies that demonstrated the importance of aa 70 and 91 mutations of the core protein were carried out on Japanese patients infected with HCV-1b. This raised two important questions: (1) is the significance of these viral mutations commonly observed among other HCV genotypes/subtypes; and (2) is the significance of these viral mutations commonly observed among other host ethnicities? To answer the first question, the correlation between the core protein mutations and IFN treatment outcome was investigated in non-HCV-1b infections, such as HCV-1a, -2a, -2b and -4a[70-76]. There was no significant correlation between these mutations and IFN treatment outcome in non-HCV-1b infections, where sequence patterns were quite conserved at these positions. The probability with different aa residues appear at positions 70 and 91 of the core protein of different HCV genotypes is shown in Figure 2. In contrast, point mutations at positions 4 and 110 of the core protein of HCV-2a were significantly associated with responses to PEG-IFN/RBV treatment[70,71]. As for the second question regarding host ethnicity, to the best of our knowledge, there are only three independent studies that were carried out on American[77], Swedish[76] and Saudi patients[75] infected with HCV-1b to investigate the possible correlation between mutations at aa 70 and 91 and IFN treatment outcome. In those studies, the point mutation at aa 70, but not at aa 91, was significantly associated with PEG-IFN/RBV treatment outcome. Overall, the results thus far obtained suggest that the mutation at codon 70 of the HCV core protein can be used as a predictive marker of IFN treatment outcome in only HCV-1b (and possibly HCV-5a) infection, regardless of host ethnicity.
As observed with the standard PEG-IFN/RBV combination therapy, Gln70 of the core protein is also significantly correlated with poor response to the recently approved triple therapy of PEG-IFN/RBV/protease inhibitor (telaprevir) for HCV-1b infection[78-81]. Interestingly, Gln70 showed significant linkage disequilibrium with minor genotypes (T/G and G/G) of the rs8099917 single nucleotide polymorphism (SNP) near the IL28B gene, which has recently been proposed as the strongest host genetic factor that is associated with poor response to PEG-IFN/RBV combination therapy in HCV genotype 1 infection[78,80,82].
Despite accumulated clinical evidence that strongly supports the correlation between HCV-1b core protein mutations and responses to IFN-based treatment, the molecular mechanism underlying this correlation is still obscure. Three experimental attempts aimed primarily to investigate this issue; however these studies produced conflicting results. In two of the studies, there was no significant difference in IFN resistance between the wild-type core protein and mutants at aa 70 and 91[83,84], while in the third study, there was a strong association between the core protein polymorphisms and IFN resistance via IL-6-induced and SOCS3-mediated suppression of IFN signaling[85]. These contradictions may, in part, be attributable to the different experimental models used by each study. Furthermore, Eng et al[86] identified a novel family of HCV core isotypes, referred to as minicores, which contain the C-terminal portion of the classical core protein, but lack the N-terminal portion. Interestingly, the N-termini of two major minicore proteins are at or near aa 70 and 91, and importantly, mutations in aa 70 and 91 regulate the expression levels of 70 and 91 minicores. Accordingly, it was hypothesized that these clinically important mutations at aa 70 and 91 of the core protein alter HCV function through altered expression levels of minicores, which might lead to IFN resistance. Further investigations using biologically relevant experimental models are needed to elucidate the molecular mechanism(s) underlying the role for the core protein mutations in HCV pathogenesis.
The NS5A protein has generated a wide range of interest in HCV research because of its ability to modulate host cell functions, including responses to IFN. NS5A is multifunctional phosphoprotein that is found in a basally phosphorylated form of 56 kDa and a hyperphosphorylated form of 58 kDa[87,88]. NS5A modulates HCV replication through interaction with other viral proteins and certain host proteins to form the HCV replication complex. Moreover, NS5A has been implicated in various forms of viral pathogenesis through interactions with a wide variety of cellular proteins. Thus, NS5A clearly plays multiple roles in mediating viral replication, host-cell interactions, and viral pathogenesis[89]. NS5A is most extensively studied among all the HCV proteins for its relationship with IFN responsiveness. We review recent information regarding the clinical implication of NS5A sequence heterogeneity in predicting treatment outcome of IFN-based therapy.
Initially, in the era of IFN monotherapy, Enomoto et al[90,91] gained a key insight into the clinical influence of NS5A sequence heterogeneity on responses to IFN treatment. In their study, they amplified an NS5A fragment by RT-PCR from sera of HCV-infected patients, determined their sequences at both the nucleotide and amino acid levels, and compared them with a standard (reference) sequence of a given HCV subtype (Figure 3) to determine the number of amino acid substitutions in a sample of a given patient. By comparing the data obtained from patients who successfully responded to IFN therapy and those who did not, the authors identified a region called the IFN sensitivity-determining region (ISDR) spanning from aa 2209 to 2248 of NS5A of HCV-1b, whose sequence heterogeneity was closely associated with IFN treatment outcome. They found that Japanese patients infected with HCV-1b isolates having four or more mutations in the ISDR (ISDR ≥ 4) compared to that of the HCV-1b prototype (HCV-J) successfully responded to IFN therapy whereas patients infected with viral isolates having ISDR ≤ 3 were non-responders. Since this discovery, the ISDR has been the subject of intense clinical and experimental investigations. Although subsequent studies conducted in Japan were consistent with the initial ISDR report[92-95], results obtained from studies conducted in Europe and North America were quite controversial[96-99]. This might be explained in part by the fact that HCV isolates in western countries have lower overall degree of sequence heterogeneity, particularly in the ISDR, than HCV isolates circulating in Japan; the prevalence of HCV isolates with ISDR ≥ 4 is lower in western countries than in Japan. Host genetic differences between western and Japanese populations and the difference in treatment regimens may also contribute, at least partially, to the apparent discrepancy. Despite this, a meta-analysis conducted by Pascu et al[100] on 1230 ISDR sequences obtained from European and Japanese patients infected with HCV-1b clearly confirmed the importance of the ISDR in determining IFN treatment outcome. In this connection, it should be noted that, in the majority of studies dealing with the ISDR heterogeneity, nested RT-PCR followed by direct sequencing of the amplified fragments, without subcloning each sequence of quasispecies, was adopted to obtain the ISDR sequences, which, in general, identifies the most dominant population of quasispecies but may miss minor populations of quasispecies present in the patients.
The predictive value of ISDR initially described in the era of IFN monotherapy continues to be significant also in the era of PEG-IFN/RBV therapy[58,59,101,102]. However, the original criterion of ISDR ≥ 4 to predict SVR was substituted by ISDR ≥ 2. This might result from the selective impact of IFN monotherapy, whereby the prevalence of sensitive isolates with ISDR ≥ 4 was decreased while that of HCV isolates of ISDR ≤ 3 was increased. Subsequently, those IFN monotherapy-resistant isolates with ISDR ≥ 2 were selected by PEG-IFN/RBV as sensitive isolates and those with ISDR ≤ 1 as resistant ones[52,58,102,103]. As for the different genotypes, the degree of sequence heterogeneity in ISDR was significantly correlated with SVR rate in Japanese patients infected with HCV-2a, the second most prevalent genotype in Japan[71,104,105]. In contrast, ISDR sequence heterogeneity did not associate with treatment outcome in patients infected with HCV-2b, -3a or -4a[73,104,106]. Whether the value of ISDR varies with different HCV genotypes and host ethnicity needs further investigation.
The molecular mechanism of ISDR-mediated IFN resistance is still unclear. Some studies have revealed that NS5A binds to and suppresses the function of the IFN-induced double-stranded RNA-activated protein kinase (PKR)[107-109]. PKR is known to inhibit viral replication by inhibiting viral protein synthesis through phosphorylation of eukaryotic initiation factor (eIF)-2. The PKR-binding domain (PKR-BD) of NS5A, spanning from aa 2209 to 2274, consists of the ISDR and its downstream region of 26 aa. The NS5A-PKR interaction was shown to be weakened by the ISDR mutations observed with IFN-sensitive HCV isolates, which would result in weaker suppression of PKR activity. In this context, a significant correlation between sequence variation in PKR-BD and IFN responsiveness was also reported[110-112]. On the other hand, we and other investigators have proposed that NS5A may also play a role(s) in IFN-resistance in an ISDR-independent manner. We have reported that an N-terminal portion of NS5A (aa 1-148) that lacks the ISDR and PKR-BD physically interacts with and inhibits the antiviral activity of 2’,5’-oligoadenylate synthetase in cultured cells[113]. It has also been demonstrated in vitro that NS5A induces the expression of IL-8 at both the mRNA and protein levels. IL-8 is known to inhibit IFN-α signaling pathway. In agreement with this experimental observation, clinical data were reported that IL-8 levels in pretreatment sera were significantly higher in non-responders than in SVR patients[114,115].
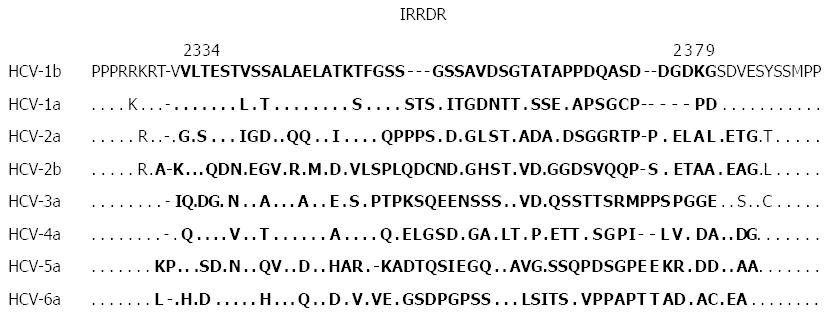
Despite the controversy about the ISDR concept in the era of IFN monotherapy, correlation between NS5A sequence heterogeneity and IFN responsiveness continued to be the subject of interest for many researchers. In this context, by using the same methodology as that for the ISDR analysis, a correlation between sequence heterogeneity of variable region 3 (V3) in a C-terminal portion of NS5A (aa 2356-2379) and responses to IFN-based therapy was also reported[110,112,116-120]. Our group further expanded this observation and gained a key insight when we identified a new region near the C-terminus of NS5A of HCV-1b spanning from aa 2334 to 2379, which we referred to as the IFN/RBV resistance-determining region (IRRDR)[121,122]. The IRRDR consists of the V3 region and its flanking upstream region, pre-V3 (aa 2334-2354). The reference sequences of different HCV subtypes are shown in Figure 4. In our initial study carried out on 47 Japanese patients infected with HCV-1b, we found that a higher degree of IRRDR sequence heterogeneity was closely associated with an early virological response (EVR) at week 16 of the 48-wk PEG-IFN/RBV treatment course[116]. Most importantly, in a follow-up study, the degree of sequence heterogeneity of the IRRDR was significantly associated with SVR. In particular, 16 (76%) of 21 SVR, but only 2 (8%) of 24 Non-SVR, had HCV with 6 or more mutations in the IRRDR (IRRDR ≥ 6). Accordingly, IRRDR ≥ 6 could predict SVR with a positive predictive value of 89% (16/18), while IRRDR ≤ 5 could predict non-SVR with a negative predictive value of 81% (22/27). Thus, we proposed that the degree of sequence heterogeneity of the IRRDR would be a useful positive and negative predictive marker for PEG-IFN/RBV treatment outcomes in HCV-1b infection[121,122]. Following this report, several reports were published by other groups and ours that support the initial idea of the importance of IRRDR in the prediction of treatment outcome[52,53,123]. In a pilot study conducted on patients who underwent liver transplantation, the value of viral genetic factors, including sequence polymorphisms in the core protein, ISDR and IRRDR, in prediction of PEG-IFN/RBV treatment outcome after transplantation was investigated[124]. Interestingly, IRRDR ≥ 6 was the strongest viral genetic factor associated with SVR. Moreover, in a well executed viral genome wide associated study that scanned the whole HCV genome for polymorphisms at certain amino acids or in genomic regions that are significantly associated with PEG-IFN/RBV treatment outcome in HCV-1b infection, a high degree of IRRDR sequence heterogeneity (IRRDR ≥ 4) was selected in multivariate analysis as the strongest factor to predict SVR among various host and viral genetic factors and baseline demographic parameters[125].
Also, it is important to point out that the cutoff number of mutations in IRRDR that is associated with treatment outcome might possibly vary with different geographical regions: In certain geographical regions where HCV isolates with a high degree of sequence heterogeneity are predominant, a higher cutoff number of IRRDR mutations (such as 6 mutations) is applicable[51,53] whereas a lower cutoff number of IRRDR mutations (such as 4 mutations) is better applicable in regions where HCV isolates with a low degree of sequence heterogeneity are predominant[52,125].
While the predictive value of the IRRDR was initially identified in Japanese patients infected with HCV-1b, its predictive value was also confirmed in HCV-2a infection, the second most prevalent HCV genotype in Japan[74]. Furthermore, we investigated for the first time the impact of viral genetics, including the core protein and NS5A polymorphisms, on PEG-IFN/RBV treatment outcome in Egyptian patients with HCV genotype 4 infection. The result clearly demonstrated that the degree of sequence heterogeneity in the IRRDR was the only viral factor that was significantly associated with PEG-IFN/RBV treatment outcome[73]. Therefore, we proposed that the IRRDR would be a useful positive and negative predictive marker for treatment outcome in Egyptian patients infected with HCV genotype 4. Collectively, a series of our studies and others have suggested the significance of IRRDR sequence heterogeneity predicting treatment outcome in different ethnic populations infected with different HCV genotypes.
The clinical correlation between IRRDR sequence heterogeneity and virological responses to IFN-based therapy in HCV infection can be linked to an experimental observation by Tsai et al[126] that an HCV subgenomic RNA replicon containing NS5A of HCV-1b exerted more profound inhibitory effects on IFN activities than the original HCV-2a replicon, and that domain swapping between NS5A sequences of HCV-1b and -2a in the V3 and/or a C-terminal region including the IRRDR resulted in a transfer of their anti-IFN activities. Consistent with this observation, Kumthip et al[127] found that the overexpression of either HCV genotype 1 or genotype 3 NS5A proteins significantly inhibited IFN-induced signaling of IFN-stimulated response element, STAT1 phosphorylation and IFN-stimulated gene expression compared to the respective controls. NS5A of HCV genotype 1 exhibited stronger inhibitory effects on IFN signaling than did that of genotype 3. Furthermore, NS5A of HCV genotype 1 bound to STAT1 with a higher affinity compared to genotype 3. Interestingly, domain mapping revealed that the C-terminal region of NS5A, including ISDR and IRRDR, conferred these inhibitory effects on IFN signaling. Whereas IRRDR is among the most variable sequences across the different genotypes and subtypes of HCV[87], its upstream and downstream sequences show a higher degree of sequence conservation (Figure 4). We speculate that whereas the upstream and downstream sequences are conserved to maintain the capacity of NS5A to participate in RNA replication and virion production across all the HCV genotypes, IRRDR sequences have a genotype-dependent or even a strain-dependent modulatory function(s)[128-130]. Therefore, the sequence heterogeneity of the IRRDR and its significant correlation with IFN responsiveness suggest the possibility that the IRRDR is involved, at least partly, in the viral strategy to evade IFN-mediated antiviral host defense mechanisms. The IRRDR sequence heterogeneity also suggests genetic flexibility of this region and, indeed, a short stretch of sequence in a C-terminal portion of NS5A was shown to tolerate sequence insertions and deletions. This means that the short stretch of sequence is not essential for virus replication in cultured cells. It does not exclude the possibility, however, that the same region might play an important role in modulating the interaction with various host systems, including IFN responsiveness. It is also possible that the genetic flexibility of this region, especially the IRRDR, is accompanied by compensatory changes elsewhere in the viral genome and that these compensatory changes affect overall viral fitness and responses to IFN therapy. We hypothesize that the IRRDR functions as evolution-adaptation machinery for HCV to cope with changes in the surrounding environment. For example, when sequence evolution of NS5A was investigated during IFN treatment, most of the evolutionary mutations were accumulated in the C-terminus, including the IRRDR[119,131]. Furthermore, in a recent retrospective study we investigated sequence evolution of the core protein, NS3 and NS5A (IRRDR and ISDR) during the follow-up period from chronic hepatitis to HCC development by comparing the sequences between pre- and post-HCC isolates[69]. The results showed that IRRDR sequences tended to be more polymorphic at the time of HCC occurrence. The frequency of HCV isolates with IRRDR ≥ 6 was significantly higher in patients with HCC than in those without HCC, and also higher in post-HCC isolates than in pre-HCC isolates. This might imply the possibility that HCV utilizes IRRDR evolution to accommodate certain selective pressures encountered during the course of HCC development. Further studies are needed to elucidate the issue.
E2, one of the two envelope proteins, is the viral component that is required for direct contact with the cell-surface receptors[132]. The first 27 amino acids (aa) of E2 were identified as hypervariable region 1 (HVR1) because of its significant sequence variability and have been reported to be an immunodominant target for neutralization antibodies. Sequence variations in this region might contribute to immune evasion and thereby the persistence of viral infection[133-135]. Also, a region of 12 residues between aa 659-670 of E2, designated as PKR/eIF-2α phosphorylation homology domain (PePHD), has been reported to interact and inhibit the antiviral activity of PKR[136,137]. Accordingly, sequence variations within the PePHD were suspected to influence responses to IFN-based therapy. However, data obtained from clinical studies investigating this issue were controversial[103,138-142].
The NS3 region of the HCV genome is less variable compared to E2 and NS5A, but still displays significant sequence diversity[143]. In a previous study, we demonstrated that polymorphisms in the secondary structure of an N-terminal region of NS3 of HCV-1b were associated with different virological responses to PEG-IFN/RBV therapy and proposed that the viral grouping based on the NS3 polymorphism could be used to predict the outcome of the therapy[144]. In addition, we have recently found that a particular combination of point mutations in aa 1082 and 1112 of NS3 (NS3-Tyr1082/Gln1112) is closely associated with HCC development in HCV-1b infection. We have also noticed that a combination of NS3-Tyr1082/Gln1112 and core-Gln70 is more strongly associated with HCC development than is the mutations of NS3 alone or the core protein alone[69]. Therefore, we propose that NS3-Tyr1082/Gln1112 and core-Gln70 would be independent predictive markers for development of HCV-1b-associated HCC.
Apart from the IFN responsiveness, NS3 has been an intense focus of attention since the introduction of NS3 protease inhibitors as DAAs for treatment of HCV infection. Mutations in four positions in the NS3 protease domain are known to be associated with resistance or reduced sensitivity to telaprevir; R155K/T/S/M, A156T/V/S, V36A/M and T54A[145-148]. Three mutations, T54A, V170A and A156S, conferred low to moderate levels of resistance to boceprevir while variants with the A156T mutation are highly resistant[145-149]. Deep sequencing analysis using “next-generation” sequencers revealed that those DAA-resistant mutations were present even before the initiation of treatment in patients who did not achieve SVR. Thus, the prevalence of the DAA-resistant variants is determined by their replicative fitness and selective pressure of the DAAs[150,151]. In this connection, deep sequencing analysis is also useful to study the possible importance of a viral factor(s) in disease manifestations, including IFN resistance, especially when the target variant(s) constitutes a minor population in the sample and, therefore, undetectable by ordinary direct sequencing.
HCV is an interesting virus to study because of its ability to evade host defense mechanisms, including both innate and acquired immune responses, so as to establish persistent infection, which causes a wide spectrum of pathogenicity, such as lipid and glucose metabolism disorders and HCC development. The HCV genome is characterized by a high degree of genetic diversity that can be associated with the viral sensitivity or resistance (reflected by different virological responses) to IFN-based therapy. In addition to the IL28B SNP as the most important host factor that governs the IFN responsiveness of the patients, a point-mutation at position 70 of the core protein and sequence heterogeneity of the ISDR and IRRDR in NS5A of HCV have significant impact on the outcome of a standard regimen of PEG-IFN/RBV combination therapy. Currently, the HCV therapeutic field is heading towards IFN-free treatment where there are several ongoing clinical trials testing new specific DAAs against HCV. Whether these DAAs can overcome the HCV genetic diversity barrier without the emergence of resistant variants should be carefully monitored and properly assessed. New technologies, such as second and third generations of deep sequencing technologies that are currently available, will open up new doors to further understand the impact of HCV genetics on HCV pathogenesis and treatment responsiveness in more detail.
We thank Adeeb Rahman for editing the manuscript.
P- Reviewers: Jin B, Kanda T, Rodriguez-Frias F S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4996] [Cited by in F6Publishing: 4592] [Article Influence: 131.2] [Reference Citation Analysis (0)] |
| 2. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2495] [Cited by in F6Publishing: 2324] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 3. | Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S-20S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 481] [Cited by in F6Publishing: 467] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 4. | Di Bisceglie AM. Hepatitis C. Lancet. 1998;351:351-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 271] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 5. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1770] [Cited by in F6Publishing: 1796] [Article Influence: 163.3] [Reference Citation Analysis (0)] |
| 6. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 581] [Cited by in F6Publishing: 599] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 7. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-2150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2571] [Cited by in F6Publishing: 2597] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 8. | Davila JA, Morgan RO, Shaib Y, McGlynn KA, El-Serag HB. Hepatitis C infection and the increasing incidence of hepatocellular carcinoma: a population-based study. Gastroenterology. 2004;127:1372-1380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 385] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 9. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2183] [Cited by in F6Publishing: 2337] [Article Influence: 194.8] [Reference Citation Analysis (0)] |
| 10. | Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 225] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 11. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 416] [Cited by in F6Publishing: 408] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 12. | Okamoto H, Okada S, Sugiyama Y, Yotsumoto S, Tanaka T, Yoshizawa H, Tsuda F, Miyakawa Y, Mayumi M. The 5’-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med. 1990;60:167-177. [PubMed] [Cited in This Article: ] |
| 13. | Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K. Structure of the 3’ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307-3312. [PubMed] [Cited in This Article: ] |
| 14. | Honda M, Ping LH, Rijnbrand RC, Amphlett E, Clarke B, Rowlands D, Lemon SM. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 202] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 15. | Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 278] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 16. | Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959-1962. [PubMed] [Cited in This Article: ] |
| 17. | Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 198] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 18. | Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore EE, Peterson DA, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 274] [Cited by in F6Publishing: 284] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 19. | Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, Yap PL, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321-328. [PubMed] [Cited in This Article: ] |
| 20. | Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26 Suppl 1:83-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Schweitzer CJ, Liang TJ. Impact of host and virus genome variability on HCV replication and response to interferon. Curr Opin Virol. 2013;3:501-507. [PubMed] [Cited in This Article: ] |
| 22. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1070] [Cited by in F6Publishing: 1058] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 23. | Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173-3188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 622] [Cited by in F6Publishing: 575] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 24. | Sy T, Jamal MM. Epidemiology of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:41-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Mihm S, Fayyazi A, Hartmann H, Ramadori G. Analysis of histopathological manifestations of chronic hepatitis C virus infection with respect to virus genotype. Hepatology. 1997;25:735-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 184] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Rubbia-Brandt L, Leandro G, Spahr L, Giostra E, Quadri R, Malé PJ, Negro F. Liver steatosis in chronic hepatitis C: a morphological sign suggesting infection with HCV genotype 3. Histopathology. 2001;39:119-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 94] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 27. | Sharma P, Balan V, Hernandez J, Rosati M, Williams J, Rodriguez-Luna H, Schwartz J, Harrison E, Anderson M, Byrne T. Hepatic steatosis in hepatitis C virus genotype 3 infection: does it correlate with body mass index, fibrosis, and HCV risk factors? Dig Dis Sci. 2004;49:25-29. [PubMed] [Cited in This Article: ] |
| 28. | Bruno S, Crosignani A, Maisonneuve P, Rossi S, Silini E, Mondelli MU. Hepatitis C virus genotype 1b as a major risk factor associated with hepatocellular carcinoma in patients with cirrhosis: a seventeen-year prospective cohort study. Hepatology. 2007;46:1350-1356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 29. | Lee MH, Yang HI, Lu SN, Jen CL, Yeh SH, Liu CJ, Chen PJ, You SL, Wang LY, Chen WJ. Hepatitis C virus seromarkers and subsequent risk of hepatocellular carcinoma: long-term predictors from a community-based cohort study. J Clin Oncol. 2010;28:4587-4593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 30. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4847] [Cited by in F6Publishing: 4689] [Article Influence: 213.1] [Reference Citation Analysis (0)] |
| 31. | McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, Nyberg LM, Lee WM, Ghalib RH, Schiff ER. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580-593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 894] [Cited by in F6Publishing: 872] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 32. | Rumi MG, Aghemo A, Prati GM, D’Ambrosio R, Donato MF, Soffredini R, Del Ninno E, Russo A, Colombo M. Randomized study of peginterferon-alpha2a plus ribavirin vs peginterferon-alpha2b plus ribavirin in chronic hepatitis C. Gastroenterology. 2010;138:108-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 33. | Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat. 2009;16:75-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 188] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2252] [Cited by in F6Publishing: 2192] [Article Influence: 146.1] [Reference Citation Analysis (1)] |
| 35. | Sherman KE, Flamm SL, Afdhal NH, Nelson DR, Sulkowski MS, Everson GT, Fried MW, Adler M, Reesink HW, Martin M. Response-guided telaprevir combination treatment for hepatitis C virus infection. N Engl J Med. 2011;365:1014-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 592] [Cited by in F6Publishing: 628] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 36. | Limaye AR, Draganov PV, Cabrera R. Boceprevir for chronic HCV genotype 1 infection. N Engl J Med. 2011;365:176; author reply 177-178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4736] [Cited by in F6Publishing: 4508] [Article Influence: 196.0] [Reference Citation Analysis (0)] |
| 38. | Alfaleh FZ, Hadad Q, Khuroo MS, Aljumah A, Algamedi A, Alashgar H, Al-Ahdal MN, Mayet I, Khan MQ, Kessie G. Peginterferon alpha-2b plus ribavirin compared with interferon alpha-2b plus ribavirin for initial treatment of chronic hepatitis C in Saudi patients commonly infected with genotype 4. Liver Int. 2004;24:568-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Kamal SM, El Tawil AA, Nakano T, He Q, Rasenack J, Hakam SA, Saleh WA, Ismail A, Aziz AA, Madwar MA. Peginterferon {alpha}-2b and ribavirin therapy in chronic hepatitis C genotype 4: impact of treatment duration and viral kinetics on sustained virological response. Gut. 2005;54:858-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 130] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 40. | Derbala M, Amer A, Bener A, Lopez AC, Omar M, El Ghannam M. Pegylated interferon-alpha 2b-ribavirin combination in Egyptian patients with genotype 4 chronic hepatitis. J Viral Hepat. 2005;12:380-385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 41. | El-Zayadi AR, Attia M, Barakat EM, Badran HM, Hamdy H, El-Tawil A, El-Nakeeb A, Selim O, Saied A. Response of hepatitis C genotype-4 naïve patients to 24 weeks of Peg-interferon-alpha2b/ribavirin or induction-dose interferon-alpha2b/ribavirin/amantadine: a non-randomized controlled study. Am J Gastroenterol. 2005;100:2447-2452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 42. | Pang PS, Planet PJ, Glenn JS. The evolution of the major hepatitis C genotypes correlates with clinical response to interferon therapy. PLoS One. 2009;4:e6579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA. 2007;104:2879-2884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 44. | Kountouras J, Zavos C, Chatzopoulos D. Apoptosis in hepatitis C. J Viral Hepat. 2003;10:335-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 45. | Hahn YS. Subversion of immune responses by hepatitis C virus: immunomodulatory strategies beyond evasion? Curr Opin Immunol. 2003;15:443-449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Mori Y, Moriishi K, Matsuura Y. Hepatitis C virus core protein: its coordinate roles with PA28gamma in metabolic abnormality and carcinogenicity in the liver. Int J Biochem Cell Biol. 2008;40:1437-1442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 47. | Pazienza V, Clément S, Pugnale P, Conzelman S, Foti M, Mangia A, Negro F. The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology. 2007;45:1164-1171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 182] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 48. | Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438-4443. [PubMed] [Cited in This Article: ] |
| 49. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 931] [Cited by in F6Publishing: 895] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 50. | Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 217] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 51. | El-Shamy A, Kim SR, Ide YH, Sasase N, Imoto S, Deng L, Shoji I, Hotta H. Polymorphisms of hepatitis C virus non-structural protein 5A and core protein and clinical outcome of pegylated-interferon/ribavirin combination therapy. Intervirology. 2012;55:1-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 52. | El-Shamy A, Shoji I, Saito T, Watanabe H, Ide YH, Deng L, Kawata S, Hotta H. Sequence heterogeneity of NS5A and core proteins of hepatitis C virus and virological responses to pegylated-interferon/ribavirin combination therapy. Microbiol Immunol. 2011;55:418-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Kim SR, El-Shamy A, Imoto S, Kim KI, Ide YH, Deng L, Shoji I, Tanaka Y, Hasegawa Y, Ota M. Prediction of response to pegylated interferon/ribavirin combination therapy for chronic hepatitis C genotype 1b and high viral load. J Gastroenterol. 2012;47:1143-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J Hepatol. 2007;46:403-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 216] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 55. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Predictors of viral kinetics to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b. J Med Virol. 2007;79:1686-1695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 56. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Prediction of response to pegylated interferon and ribavirin in hepatitis C by polymorphisms in the viral core protein and very early dynamics of viremia. Intervirology. 2007;50:361-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 57. | Okanoue T, Itoh Y, Hashimoto H, Yasui K, Minami M, Takehara T, Tanaka E, Onji M, Toyota J, Chayama K. Predictive values of amino acid sequences of the core and NS5A regions in antiviral therapy for hepatitis C: a Japanese multi-center study. J Gastroenterol. 2009;44:952-963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 58. | Hayashi K, Katano Y, Ishigami M, Itoh A, Hirooka Y, Nakano I, Urano F, Yoshioka K, Toyoda H, Kumada T. Mutations in the core and NS5A region of hepatitis C virus genotype 1b and correlation with response to pegylated-interferon-alpha 2b and ribavirin combination therapy. J Viral Hepat. 2011;18:280-286. [PubMed] [Cited in This Article: ] |
| 59. | Mori N, Imamura M, Kawakami Y, Saneto H, Kawaoka T, Takaki S, Aikata H, Takahashi S, Chayama K. Randomized trial of high-dose interferon-alpha-2b combined with ribavirin in patients with chronic hepatitis C: Correlation between amino acid substitutions in the core/NS5A region and virological response to interferon therapy. J Med Virol. 2009;81:640-649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 60. | Enomoto N, Maekawa S. HCV genetic elements determining the early response to peginterferon and ribavirin therapy. Intervirology. 2010;53:66-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 61. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. A matched case-controlled study of 48 and 72 weeks of peginterferon plus ribavirin combination therapy in patients infected with HCV genotype 1b in Japan: amino acid substitutions in HCV core region as predictor of sustained virological response. J Med Virol. 2009;81:452-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 62. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitutions in the hepatitis C virus core region of genotype 1b are the important predictor of severe insulin resistance in patients without cirrhosis and diabetes mellitus. J Med Virol. 2009;81:1032-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 63. | Kobayashi M, Akuta N, Suzuki F, Hosaka T, Sezaki H, Kobayashi M, Suzuki Y, Arase Y, Ikeda K, Watahiki S. Influence of amino-acid polymorphism in the core protein on progression of liver disease in patients infected with hepatitis C virus genotype 1b. J Med Virol. 2010;82:41-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 64. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology. 2007;46:1357-1364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 65. | Akuta N, Suzuki F, Seko Y, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Kobayashi M. Complicated relationships of amino acid substitution in hepatitis C virus core region and IL28B genotype influencing hepatocarcinogenesis. Hepatology. 2012;56:2134-2141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 66. | Nakamoto S, Imazeki F, Fukai K, Fujiwara K, Arai M, Kanda T, Yonemitsu Y, Yokosuka O. Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J Hepatol. 2010;52:72-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 67. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S. Amino acid substitutions in hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by antiviral therapy. J Med Virol. 2011;83:1016-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 68. | Seko Y, Akuta N, Suzuki F, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S. Amino acid substitutions in the hepatitis C Virus core region and lipid metabolism are associated with hepatocarcinogenesis in nonresponders to interferon plus ribavirin combination therapy. Intervirology. 2013;56:13-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 69. | El-Shamy A, Shindo M, Shoji I, Deng L, Okuno T, Hotta H. Polymorphisms of the core, NS3, and NS5A proteins of hepatitis C virus genotype 1b associate with development of hepatocellular carcinoma. Hepatology. 2013;58:555-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 70. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 2a high viral load and virological response to interferon-ribavirin combination therapy. Intervirology. 2009;52:301-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 71. | Kadokura M, Maekawa S, Sueki R, Miura M, Komase K, Shindo H, Amemiya F, Uetake T, Inoue T, Sakamoto M. Analysis of the complete open reading frame of hepatitis C virus in genotype 2a infection reveals critical sites influencing the response to peginterferon and ribavirin therapy. Hepatol Int. 2011;5:789-799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 72. | Kadokura M, Maekawa S, Sueki R, Miura M, Komase K, Shindo H, Amemiya F, Uetake T, Inoue T, Sakamoto M. Analysis of the complete open reading frame of genotype 2b hepatitis C virus in association with the response to peginterferon and ribavirin therapy. PLoS One. 2011;6:e24514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 73. | El-Shamy A, Shoji I, El-Akel W, Bilasy SE, Deng L, El-Raziky M, Jiang DP, Esmat G, Hotta H. NS5A sequence heterogeneity of hepatitis C virus genotype 4a predicts clinical outcome of pegylated-interferon-ribavirin therapy in Egyptian patients. J Clin Microbiol. 2012;50:3886-3892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 74. | El-Shamy A, Shoji I, Kim SR, Ide Y, Imoto S, Deng L, Yoon S, Fujisawa T, Tani S, Yano Y. Sequence heterogeneity in NS5A of hepatitis C virus genotypes 2a and 2b and clinical outcome of pegylated-interferon/ribavirin therapy. PLoS One. 2012;7:e30513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 75. | Alhamlan FS, Al-Ahdal MN, Khalaf NZ, Abdo AA, Sanai FM, Al-Ashgar HI, Elhefnawi M, Zaid A, Al-Qahtani AA. Hepatitis C virus genotype 1: how genetic variability of the core protein affects the response to pegylated-interferon and ribavirin therapy. J Med Virol. 2014;86:224-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 76. | Alestig E, Arnholm B, Eilard A, Lagging M, Nilsson S, Norkrans G, Wahlberg T, Wejstål R, Westin J, Lindh M. Core mutations, IL28B polymorphisms and response to peginterferon/ribavirin treatment in Swedish patients with hepatitis C virus genotype 1 infection. BMC Infect Dis. 2011;11:124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 77. | Donlin MJ, Cannon NA, Yao E, Li J, Wahed A, Taylor MW, Belle SH, Di Bisceglie AM, Aurora R, Tavis JE. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol. 2007;81:8211-8224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 100] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 78. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 79. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitutions in the hepatitis C virus core region of genotype 1b affect very early viral dynamics during treatment with telaprevir, peginterferon, and ribavirin. J Med Virol. 2010;82:575-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 80. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitution in HCV core region and genetic variation near the IL28B gene affect viral dynamics during telaprevir, peginterferon and ribavirin treatment. Intervirology. 2012;55:417-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 81. | Akuta N, Suzuki F, Fukushima T, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Kobayashi M. Prediction of treatment efficacy and telaprevir-resistant variants after triple therapy in patients infected with hepatitis C virus genotype 1. J Clin Microbiol. 2013;51:2862-2868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S. Amino acid substitution in HCV core/NS5A region and genetic variation near IL28B gene affect treatment efficacy to interferon plus ribavirin combination therapy. Intervirology. 2012;55:231-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 83. | Ikeda F, Dansako H, Nishimura G, Mori K, Kawai Y, Ariumi Y, Miyake Y, Takaki A, Nouso K, Iwasaki Y. Amino acid substitutions of hepatitis C virus core protein are not associated with intracellular antiviral response to interferon-α in vitro. Liver Int. 2010;30:1324-1331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 84. | Hiraga N, Abe H, Imamura M, Tsuge M, Takahashi S, Hayes CN, Ochi H, Tateno C, Yoshizato K, Nakamura Y. Impact of viral amino acid substitutions and host interleukin-28b polymorphism on replication and susceptibility to interferon of hepatitis C virus. Hepatology. 2011;54:764-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 85. | Funaoka Y, Sakamoto N, Suda G, Itsui Y, Nakagawa M, Kakinuma S, Watanabe T, Mishima K, Ueyama M, Onozuka I. Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91. J Virol. 2011;85:5986-5994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 86. | Eng FJ, Walewski JL, Klepper AL, Fishman SL, Desai SM, McMullan LK, Evans MJ, Rice CM, Branch AD. Internal initiation stimulates production of p8 minicore, a member of a newly discovered family of hepatitis C virus core protein isoforms. J Virol. 2009;83:3104-3114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 87. | Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85:2485-2502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 280] [Cited by in F6Publishing: 280] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 88. | Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980-3986. [PubMed] [Cited in This Article: ] |
| 89. | Polyak SJ, Paschal DM, McArdle S, Gale MJ, Moradpour D, Gretch DR. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 114] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 90. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 459] [Cited by in F6Publishing: 447] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 91. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 752] [Cited by in F6Publishing: 727] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 92. | Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750-753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 133] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 93. | Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 160] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 94. | Shen C, Hu T, Shen L, Gao L, Xie W, Zhang J. Mutations in ISDR of NS5A gene influence interferon efficacy in Chinese patients with chronic hepatitis C virus genotype 1b infection. J Gastroenterol Hepatol. 2007;22:1898-1903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 95. | Hayashi K, Katano Y, Honda T, Ishigami M, Itoh A, Hirooka Y, Nakano I, Urano F, Yoshioka K, Toyoda H. Mutations in the interferon sensitivity-determining region of hepatitis C virus genotype 2a correlate with response to pegylated-interferon-alpha 2a monotherapy. J Med Virol. 2009;81:459-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 96. | Hofgärtner WT, Polyak SJ, Sullivan DG, Carithers RL, Gretch DR. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118-126. [PubMed] [Cited in This Article: ] |
| 97. | Frangeul L, Cresta P, Perrin M, Lunel F, Opolon P, Agut H, Huraux JM. Mutations in NS5A region of hepatitis C virus genome correlate with presence of NS5A antibodies and response to interferon therapy for most common European hepatitis C virus genotypes. Hepatology. 1998;28:1674-1679. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 98. | Squadrito G, Orlando ME, Cacciola I, Rumi MG, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to “interferon sensitivity determining region” variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30:1023-1027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 99. | Berg T, Mas Marques A, Höhne M, Wiedenmann B, Hopf U, Schreier E. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology. 2000;32:1386-1395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 100. | Pascu M, Martus P, Höhne M, Wiedenmann B, Hopf U, Schreier E, Berg T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53:1345-1351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 101. | Yen YH, Hung CH, Hu TH, Chen CH, Wu CM, Wang JH, Lu SN, Lee CM. Mutations in the interferon sensitivity-determining region (nonstructural 5A amino acid 2209-2248) in patients with hepatitis C-1b infection and correlating response to combined therapy of pegylated interferon and ribavirin. Aliment Pharmacol Ther. 2008;27:72-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 102. | Shirakawa H, Matsumoto A, Joshita S, Komatsu M, Tanaka N, Umemura T, Ichijo T, Yoshizawa K, Kiyosawa K, Tanaka E. Pretreatment prediction of virological response to peginterferon plus ribavirin therapy in chronic hepatitis C patients using viral and host factors. Hepatology. 2008;48:1753-1760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 103. | Muñoz de Rueda P, Casado J, Patón R, Quintero D, Palacios A, Gila A, Quiles R, León J, Ruiz-Extremera A, Salmerón J. Mutations in E2-PePHD, NS5A-PKRBD, NS5A-ISDR, and NS5A-V3 of hepatitis C virus genotype 1 and their relationships to pegylated interferon-ribavirin treatment responses. J Virol. 2008;82:6644-6653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 104. | Murakami T, Enomoto N, Kurosaki M, Izumi N, Marumo F, Sato C. Mutations in nonstructural protein 5A gene and response to interferon in hepatitis C virus genotype 2 infection. Hepatology. 1999;30:1045-1053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 105. | Akuta N, Suzuki F, Tsubota A, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Arase Y, Ikeda K. Association of amino acid substitution pattern in nonstructural protein 5A of hepatitis C virus genotype2a low viral load and response to interferon monotherapy. J Med Virol. 2003;69:376-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 106. | Bagaglio S, Bruno R, Lodrini S, De Mitri MS, Andreone P, Loggi E, Galli L, Lazzarin A, Morsica G. Genetic heterogeneity of hepatitis C virus (HCV) in clinical strains of HIV positive and HIV negative patients chronically infected with HCV genotype 3a. J Biol Regul Homeost Agents. 2003;17:153-161. [PubMed] [Cited in This Article: ] |
| 107. | Gale MJ, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 631] [Cited by in F6Publishing: 648] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 108. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] [Cited in This Article: ] |
| 109. | Gale MJ, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 127] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 110. | Sarrazin C, Herrmann E, Bruch K, Zeuzem S. Hepatitis C virus nonstructural 5A protein and interferon resistance: a new model for testing the reliability of mutational analyses. J Virol. 2002;76:11079-11090. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 111. | Macquillan GC, Niu X, Speers D, English S, Garas G, Harnett GB, Reed WD, Allan JE, Jeffrey GP. Does sequencing the PKRBD of hepatitis C virus NS5A predict therapeutic response to combination therapy in an Australian population? J Gastroenterol Hepatol. 2004;19:551-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 112. | Nousbaum J, Polyak SJ, Ray SC, Sullivan DG, Larson AM, Carithers RL, Gretch DR. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74:9028-9038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 103] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 113. | Taguchi T, Nagano-Fujii M, Akutsu M, Kadoya H, Ohgimoto S, Ishido S, Hotta H. Hepatitis C virus NS5A protein interacts with 2’,5’-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85:959-969. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 95] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 114. | Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001;75:6095-6106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 248] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 115. | Polyak SJ, Khabar KS, Rezeiq M, Gretch DR. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol. 2001;75:6209-6211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 183] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 116. | Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373-1381. [PubMed] [Cited in This Article: ] |
| 117. | Layden-Almer JE, Kuiken C, Ribeiro RM, Kunstman KJ, Perelson AS, Layden TJ, Wolinsky SM. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J Infect Dis. 2005;192:1078-1087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 118. | Vuillermoz I, Khattab E, Sablon E, Ottevaere I, Durantel D, Vieux C, Trepo C, Zoulim F. Genetic variability of hepatitis C virus in chronically infected patients with viral breakthrough during interferon-ribavirin therapy. J Med Virol. 2004;74:41-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 119. | Puig-Basagoiti F, Forns X, Furcić I, Ampurdanés S, Giménez-Barcons M, Franco S, Sánchez-Tapias JM, Saiz JC. Dynamics of hepatitis C virus NS5A quasispecies during interferon and ribavirin therapy in responder and non-responder patients with genotype 1b chronic hepatitis C. J Gen Virol. 2005;86:1067-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 120. | Murphy MD, Rosen HR, Marousek GI, Chou S. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig Dis Sci. 2002;47:1195-1205. [PubMed] [Cited in This Article: ] |
| 121. | El-Shamy A, Sasayama M, Nagano-Fujii M, Sasase N, Imoto S, Kim SR, Hotta H. Prediction of efficient virological response to pegylated interferon/ribavirin combination therapy by NS5A sequences of hepatitis C virus and anti-NS5A antibodies in pre-treatment sera. Microbiol Immunol. 2007;51:471-482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 122. | El-Shamy A, Nagano-Fujii M, Sasase N, Imoto S, Kim SR, Hotta H. Sequence variation in hepatitis C virus nonstructural protein 5A predicts clinical outcome of pegylated interferon/ribavirin combination therapy. Hepatology. 2008;48:38-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 123. | Sasase N, Kim SR, Kudo M, Kim KI, Taniguchi M, Imoto S, Mita K, Hayashi Y, Shoji I, El-Shamy A. Outcome and early viral dynamics with viral mutation in PEG-IFN/RBV therapy for chronic hepatitis in patients with high viral loads of serum HCV RNA genotype 1b. Intervirology. 2010;53:49-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 124. | Fukuhara T, Taketomi A, Okano S, Ikegami T, Soejima Y, Shirabe K, Maehara Y. Mutations in hepatitis C virus genotype 1b and the sensitivity of interferon-ribavirin therapy after liver transplantation. J Hepatol. 2010;52:672-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 125. | Maekawa S, Sakamoto M, Miura M, Kadokura M, Sueki R, Komase K, Shindo H, Komatsu N, Shindo K, Kanayama A. Comprehensive analysis for viral elements and interleukin-28B polymorphisms in response to pegylated interferon plus ribavirin therapy in hepatitis C virus 1B infection. Hepatology. 2012;56:1611-1621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 126. | Tsai YH, Kuang WF, Lu TY, Kao JH, Lai MY, Liu CJ, Chen PJ, Hwang LH. The non-structural 5A protein of hepatitis C virus exhibits genotypic differences in interferon antagonism. J Hepatol. 2008;49:899-907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 127. | Kumthip K, Chusri P, Jilg N, Zhao L, Fusco DN, Zhao H, Goto K, Cheng D, Schaefer EA, Zhang L. Hepatitis C virus NS5A disrupts STAT1 phosphorylation and suppresses type I interferon signaling. J Virol. 2012;86:8581-8591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 128. | Hughes M, Gretton S, Shelton H, Brown DD, McCormick CJ, Angus AG, Patel AH, Griffin S, Harris M. A conserved proline between domains II and III of hepatitis C virus NS5A influences both RNA replication and virus assembly. J Virol. 2009;83:10788-10796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 129. | Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 313] [Cited by in F6Publishing: 317] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 130. | Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. 2008;82:7964-7976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 280] [Cited by in F6Publishing: 285] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 131. | Yuan HJ, Jain M, Snow KK, Gale M, Lee WM. Evolution of hepatitis C virus NS5A region in breakthrough patients during pegylated interferon and ribavirin therapy*. J Viral Hepat. 2010;17:208-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 132. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] [Cited in This Article: ] |
| 133. | Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 443] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 134. | Smith DB. Evolution of the hypervariable region of hepatitis C virus. J Viral Hepat. 1999;6 Suppl 1:41-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 135. | Alfonso V, Flichman DM, Sookoian S, Mbayed VA, Campos RH. Evolutionary study of HVR1 of E2 in chronic hepatitis C virus infection. J Gen Virol. 2004;85:39-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 136. | Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107-110. [PubMed] [Cited in This Article: ] |
| 137. | Taylor DR, Tian B, Romano PR, Hinnebusch AG, Lai MM, Mathews MB. Hepatitis C virus envelope protein E2 does not inhibit PKR by simple competition with autophosphorylation sites in the RNA-binding domain. J Virol. 2001;75:1265-1273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 138. | Gupta R, Subramani M, Khaja MN, Madhavi C, Roy S, Habibullah CM, Das S. Analysis of mutations within the 5’ untranslated region, interferon sensitivity region, and PePHD region as a function of response to interferon therapy in hepatitis C virus-infected patients in India. J Clin Microbiol. 2006;44:709-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 139. | Saito T, Ito T, Ishiko H, Yonaha M, Morikawa K, Miyokawa A, Mitamura K. Sequence analysis of PePHD within HCV E2 region and correlation with resistance of interferon therapy in Japanese patients infected with HCV genotypes 2a and 2b. Am J Gastroenterol. 2003;98:1377-1383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 140. | Sarrazin C, Bruckner M, Herrmann E, Rüster B, Bruch K, Roth WK, Zeuzem S. Quasispecies heterogeneity of the carboxy-terminal part of the E2 gene including the PePHD and sensitivity of hepatitis C virus 1b isolates to antiviral therapy. Virology. 2001;289:150-163. [PubMed] [Cited in This Article: ] |
| 141. | Gaudy C, Lambelé M, Moreau A, Veillon P, Lunel F, Goudeau A. Mutations within the hepatitis C virus genotype 1b E2-PePHD domain do not correlate with treatment outcome. J Clin Microbiol. 2005;43:750-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 142. | Hung CH, Lee CM, Lu SN, Lee JF, Wang JH, Tung HD, Chen TM, Hu TH, Chen WJ, Changchien CS. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J Viral Hepat. 2003;10:87-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 143. | Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103-126. [PubMed] [Cited in This Article: ] |
| 144. | Sanjo M, Saito T, Ishii R, Nishise Y, Haga H, Okumoto K, Ito J, Watanabe H, Saito K, Togashi H. Secondary structure of the amino-terminal region of HCV NS3 and virological response to pegylated interferon plus ribavirin therapy for chronic hepatitis C. J Med Virol. 2010;82:1364-1370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 145. | Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70:28-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 164] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 146. | Lin C, Gates CA, Rao BG, Brennan DL, Fulghum JR, Luong YP, Frantz JD, Lin K, Ma S, Wei YY. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem. 2005;280:36784-36791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 185] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 147. | Courcambeck J, Bouzidi M, Perbost R, Jouirou B, Amrani N, Cacoub P, Pèpe G, Sabatier JM, Halfon P. Resistance of hepatitis C virus to NS3-4A protease inhibitors: mechanisms of drug resistance induced by R155Q, A156T, D168A and D168V mutations. Antivir Ther. 2006;11:847-855. [PubMed] [Cited in This Article: ] |
| 148. | Hiraga N, Imamura M, Abe H, Hayes CN, Kono T, Onishi M, Tsuge M, Takahashi S, Ochi H, Iwao E. Rapid emergence of telaprevir resistant hepatitis C virus strain from wildtype clone in vivo. Hepatology. 2011;54:781-788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 149. | Yi M, Tong X, Skelton A, Chase R, Chen T, Prongay A, Bogen SL, Saksena AK, Njoroge FG, Veselenak RL. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem. 2006;281:8205-8215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 150. | Adiwijaya BS, Herrmann E, Hare B, Kieffer T, Lin C, Kwong AD, Garg V, Randle JC, Sarrazin C, Zeuzem S. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput Biol. 2010;6:e1000745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 151. | Bartolini B, Giombini E, Zaccaro P, Selleri M, Rozera G, Abbate I, Comandini UV, Ippolito G, Solmone M, Capobianchi MR. Extent of HCV NS3 protease variability and resistance-associated mutations assessed by next generation sequencing in HCV monoinfected and HIV/HCV coinfected patients. Virus Res. 2013;177:205-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 152. | Bruno S, Cammà C, Di Marco V, Rumi M, Vinci M, Camozzi M, Rebucci C, Di Bona D, Colombo M, Craxì A. Peginterferon alfa-2b plus ribavirin for naïve patients with genotype 1 chronic hepatitis C: a randomized controlled trial. J Hepatol. 2004;41:474-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 153. | Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Bernstein D, Rizzetto M. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2216] [Cited by in F6Publishing: 2085] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 154. | Kuboki M, Iino S, Okuno T, Omata M, Kiyosawa K, Kumada H, Hayashi N, Sakai T. Peginterferon alpha-2a (40 KD) plus ribavirin for the treatment of chronic hepatitis C in Japanese patients. J Gastroenterol Hepatol. 2007;22:645-652. [PubMed] [Cited in This Article: ] |
| 155. | Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, Sarrazin C, Harvey J, Brass C, Albrecht J. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993-999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 252] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 156. | Mangia A, Santoro R, Minerva N, Ricci GL, Carretta V, Persico M, Vinelli F, Scotto G, Bacca D, Annese M. Peginterferon alfa-2b and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2005;352:2609-2617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 488] [Cited by in F6Publishing: 509] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 157. | von Wagner M, Huber M, Berg T, Hinrichsen H, Rasenack J, Heintges T, Bergk A, Bernsmeier C, Häussinger D, Herrmann E. Peginterferon-alpha-2a (40KD) and ribavirin for 16 or 24 weeks in patients with genotype 2 or 3 chronic hepatitis C. Gastroenterology. 2005;129:522-527. [PubMed] [DOI] [Cited in This Article: ] |
| 158. | Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 435] [Cited by in F6Publishing: 458] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 159. | Yu ML, Dai CY, Huang JF, Hou NJ, Lee LP, Hsieh MY, Chiu CF, Lin ZY, Chen SC, Hsieh MY. A randomised study of peginterferon and ribavirin for 16 versus 24 weeks in patients with genotype 2 chronic hepatitis C. Gut. 2007;56:553-559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 239] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 160. | Dalgard O, Bjøro K, Ring-Larsen H, Bjornsson E, Holberg-Petersen M, Skovlund E, Reichard O, Myrvang B, Sundelöf B, Ritland S. Pegylated interferon alfa and ribavirin for 14 versus 24 weeks in patients with hepatitis C virus genotype 2 or 3 and rapid virological response. Hepatology. 2008;47:35-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 161. | Hasan F, Asker H, Al-Khaldi J, Siddique I, Al-Ajmi M, Owaid S, Varghese R, Al-Nakib B. Peginterferon alfa-2b plus ribavirin for the treatment of chronic hepatitis C genotype 4. Am J Gastroenterol. 2004;99:1733-1737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 162. | Kamal SM, El Kamary SS, Shardell MD, Hashem M, Ahmed IN, Muhammadi M, Sayed K, Moustafa A, Hakem SA, Ibrahiem A. Pegylated interferon alpha-2b plus ribavirin in patients with genotype 4 chronic hepatitis C: The role of rapid and early virologic response. Hepatology. 2007;46:1732-1740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 118] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 163. | El Makhzangy H, Esmat G, Said M, Elraziky M, Shouman S, Refai R, Rekacewicz C, Gad RR, Vignier N, Abdel-Hamid M. Response to pegylated interferon alfa-2a and ribavirin in chronic hepatitis C genotype 4. J Med Virol. 2009;81:1576-1583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 164. | Bonny CRC, Randl K. Treatment of interferon-naive patients with HCV genotype 5 with interferon (or PEG-interferon) plus ribavirin results in a very high sustained virological response. J Hepatol. 2003;38:738A. [DOI] [Cited in This Article: ] |
| 165. | Legrand-Abravanel F, Sandres-Sauné K, Barange K, Alric L, Moreau J, Desmorat P, Vinel JP, Izopet J. Hepatitis C virus genotype 5: epidemiological characteristics and sensitivity to combination therapy with interferon-alpha plus ribavirin. J Infect Dis. 2004;189:1397-1400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 166. | Dev AT, McCaw R, Sundararajan V, Bowden S, Sievert W. Southeast Asian patients with chronic hepatitis C: the impact of novel genotypes and race on treatment outcome. Hepatology. 2002;36:1259-1265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 167. | Hui CK, Yuen MF, Sablon E, Chan AO, Wong BC, Lai CL. Interferon and ribavirin therapy for chronic hepatitis C virus genotype 6: a comparison with genotype 1. J Infect Dis. 2003;187:1071-1074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 168. | Fung J, Lai CL, Hung I, Young J, Cheng C, Wong D, Yuen MF. Chronic hepatitis C virus genotype 6 infection: response to pegylated interferon and ribavirin. J Infect Dis. 2008;198:808-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 169. | Nguyen NH, VuTien P, Garcia RT, Trinh H, Nguyen H, Nguyen K, Levitt B, Nguyen MH. Response to pegylated interferon and ribavirin in Asian American patients with chronic hepatitis C genotypes 1 vs 2/3 vs 6. J Viral Hepat. 2010;17:691-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 170. | Tsang OT, Zee JS, Chan JM, Li RS, Kan YM, Li FT, Lo FH, Chow DA, Cheung KW, Chan KH. Chronic hepatitis C genotype 6 responds better to pegylated interferon and ribavirin combination therapy than genotype 1. J Gastroenterol Hepatol. 2010;25:766-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 171. | Zhou YQ, Wang XH, Hong GH, Zhu Y, Zhang XQ, Hu YJ, Mao Q. Twenty-four weeks of pegylated interferon plus ribavirin effectively treat patients with HCV genotype 6a. J Viral Hepat. 2011;18:595-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 172. | Tangkijvanich P, Komolmit P, Mahachai V, Poovorawan K, Akkarathamrongsin S, Poovorawan Y. Response-guided therapy for patients with hepatitis C virus genotype 6 infection: a pilot study. J Viral Hepat. 2012;19:423-430. [PubMed] [Cited in This Article: ] |