Published online Oct 7, 2014. doi: 10.3748/wjg.v20.i37.13325
Revised: June 4, 2014
Accepted: June 26, 2014
Published online: October 7, 2014
Pancreatic cancer (PC) is one of the most aggressive and lethal neoplastic diseases. A valid alternative to the usual invasive diagnostic tools would certainly be the determination of biomarkers in peripheral fluids to provide less invasive tools for early diagnosis. Nowadays, biomarkers are generally investigated mainly in peripheral blood and tissues through high-throughput omics techniques comparing control vs pathological samples. The results can be evaluated by two main strategies: (1) classical methods in which the identification of significant biomarkers is accomplished by monovariate statistical tests where each biomarker is considered as independent from the others; and (2) multivariate methods, taking into consideration the correlations existing among the biomarkers themselves. This last approach is very powerful since it allows the identification of pools of biomarkers with diagnostic and prognostic performances which are superior to single markers in terms of sensitivity, specificity and robustness. Multivariate techniques are usually applied with variable selection procedures to provide a restricted set of biomarkers with the best predictive ability; however, standard selection methods are usually aimed at the identification of the smallest set of variables with the best predictive ability and exhaustivity is usually neglected. The exhaustive search for biomarkers is instead an important alternative to standard variable selection since it can provide information about the etiology of the pathology by producing a comprehensive set of markers. In this review, the most recent applications of the omics techniques (proteomics, genomics and metabolomics) to the identification of exploratory biomarkers for PC will be presented with particular regard to the statistical methods adopted for their identification. The basic theory related to classical and multivariate methods for identification of biomarkers is presented and then, the most recent applications in this field are discussed.
Core tip: Biomarkers are statistically identified as significant by: (1) classical statistical tests where each biomarker is independent from the others; and (2) multivariate methods that take into consideration the correlation among the biomarkers. This last approach provides pools of biomarkers with superior diagnostic and prognostic performances. Multivariate techniques are often applied with variable selection procedures to provide the smallest set of biomarkers with the best predictive ability. The exhaustive identification is instead a valid alternative since it can provide comprehensive information about the etiology of the pathology. The most recent applications of the omics approaches to the identification of biomarkers for PC are presented, with particular regard to the statistical methods adopted.
- Citation: Marengo E, Robotti E. Biomarkers for pancreatic cancer: Recent achievements in proteomics and genomics through classical and multivariate statistical methods. World J Gastroenterol 2014; 20(37): 13325-13342
- URL: https://www.wjgnet.com/1007-9327/full/v20/i37/13325.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i37.13325
Pancreatic cancer (PC) is one of the most aggressive and lethal neoplastic diseases; its early detection is therefore fundamental since surgery at an early disease stage is the preferred and most promising therapy. About 20% of patients can be operated on at time of diagnosis; the 5-year survival rate for not-operable patients is about 1%, while the 5-year survival after surgery is about 20% without an adjuvant therapy and about 25%-30% with the therapy[1-3]. The lack of early symptoms and the high aggressiveness are the main causes of late diagnosis and high mortality of this disease. Therefore, the search for biomarkers of early diagnosis is highly recommended to improve the early diagnostic rate, thus improving patients’ prognosis.
Diagnosis is usually based on invasive techniques [ultrasound endoscopy (EUS), explorative laparoscopy or laparotomy] or on methods that can be at least inconvenient for patients [computed tomography (CT), magnetic resonance imaging (MRI), endoscopic retrograde or magnetic resonance cholangiopancreatography (ERCP and MRCP)][2].
A valid alternative would certainly be the determination of specific biomarkers in peripheral fluids in order to provide less invasive tools for early diagnosis. In this direction, the most recent efforts in the field of biomarker identification for PC are directed. A wide range of serum markers for PC has been reported[2,4] but few of them are exploited in clinical routine since they show low sensitivity and/or specificity in general. Bünger et al[2] reviewed about 43 serum biomarkers for PC divided into four main groups: carbohydrates (CA19-9, CA 50, CA 125, CA195, CA 72-4), carcinoembryonic antigens, other markers and the combination of different markers.
Together with diagnostic markers, great efforts have recently been made to identify predictive and prognostic biomarkers for PC. Tissue biomarkers for the prognosis of pancreatic ductal adenocarcinoma (PDAC) have recently been reviewed by Jamieson et al[4]. The considerations that drive the search for diagnostic biomarkers also apply for predictive and prognostic ones and the identification of markers from peripheral blood should be the best alternative to provide prognostic methods that are less invasive for patients. Nowadays, both diagnostic and prognostic/predictive biomarkers are generally investigated mainly in peripheral blood or tissues through high-throughput omics techniques. The results can be evaluated by two different strategies, namely by: (1) classical statistical methods consisting of the use/identification of significant biomarkers by monovariate statistical tests where each biomarker is considered as independent from the others; and (2) multivariate methods, able to take into consideration the multivariate structure of the data and the correlations among the potential biomarkers. This last approach is very powerful since it allows the identification of pools of biomarkers with diagnostic and/or prognostic performance superior to single markers in terms of sensitivity, specificity and robustness.
It is important to point out that biostatistical methods can usually be defined as multivariate (several endpoints and several predictors) or multivariable (one endpoint, several predictors). In this specific context, the authors will generally apply the term multivariate to identify methods that allow the evaluation of the correlations between the variables, i.e., their synergisms and/or antagonisms.
Multivariate techniques are usually applied with variable selection procedures[5,6] to provide a set of candidate biomarkers with the best predictive ability; however, standard selection tools are aimed at the identification of the smallest set of variables with the best predictive ability. It is the authors’ opinion that exhaustivity should also be addressed[7]. Biomarkers are useful not only for diagnostic/prognostic purposes but also to better understand the etiology of pancreatic cancer. From this point of view, the exploitation of high-throughput methods provides a lot of information that should not be neglected. The exhaustive identification of all possible biomarkers showing large correlations could provide information about the overall mechanism of action of the disease, thus opening the way towards new therapeutic strategies.
In this review, the most recent applications of the omics approaches (proteomics, genomics and metabolomics) for the identification of biomarkers for pancreatic cancer will be presented, with particular regard for the statistical methods adopted for their identification, focusing especially on exploratory biomarkers. High-throughput techniques will probably be the future in the field of searching for exploratory biomarkers due to the great amount of information they convey. Moreover, the possibility of combining the results emerging from proteomic, genomic and metabolomic studies with clinical information can provide exhaustive panels of markers, thus improving their predictive performance with better sensitivity and specificity.
First, the theory of the classical and multivariate methods for identification of biomarkers will be presented, followed by the most recent applications in this field.
The statistical methods presented here can be divided into four main groups: (1) classical methods for identification of biomarkers based on monovariate approaches; (2) tools for biomarkers search based on multivariate approaches; (3) methods for the analysis of survival outcomes; and (4) other methods. Only the tools recently applied to the specific case of pancreatic cancer will be briefly presented as an exhaustive treatment of all multivariate procedures in the field of searching for biomarkers is out of the scope of the present review.
The classical approach to the identification of markers of a specific disease is the evaluation of which variables show a different behavior between two groups of samples (control vs pathological, control vs drug-treated, etc.). The easiest statistical way to solve this problem is the application of classical statistical tests to each biomarker candidate separately and the calculation of the type I error that can be accomplished comparisonwise (for each hypothesis independently) or experimentwise (testing all hypotheses together). The second alternative is preferred since the type I error probability increases as the number of tests increases. The identification of significant markers is therefore accomplished by the Student’s t-test for each variable independently and by applying a correction taking into account the number of multiple tests available: Bonferroni’s method with subsequent modifications[8] or the corrections proposed by Dunn and Sikak and by Dunnet[9-11]. This approach is incorrectly defined as multivariate since it does not take into consideration the correlations eventually existing between the variables. The same approach can also be applied to non-parametric tests to be exploited when the number of samples is too small or when the assumptions at the basis of parametric tests are not verified. Among them, the most widespread in the biomedical field is the Mann-Whitney U-test[8].
An alternative is the exploitation of global tests aimed at demonstrating a global hypothesis (e.g., the effect of a therapy) considering all the tests simultaneously; an example is the Hotelling’s T2 test[8].
Classical procedures also comprise the approach based on the analysis of variance both in its two-way (ANOVA) or multi-way (MANOVA) versions[8]. In this case, it is also possible to compare more than two groups of samples.
An alternative to classical hypothesis testing is the use of the Bayes factors[12], providing a more robust approach. For comparing two hypotheses H1 and H2, this factor may be approximated as the ratio of the marginal likelihood of the data under the two hypotheses and can be interpreted as follows: B ≤ 0.1: strongly against H1; 0.1 < B ≤ 1: against H1; 1 ≤ B < 3: barely worth mentioning for H1; 3 ≤ B << 10: substantially for H1; B > 10: strongly for H1.
Methods that can be properly defined multivariate are based on the comparison of two or more groups of samples taking into account the relationships between the variables, rather than considering them as independent. This approach is certainly more effective since it is fundamental to consider the synergic or antagonistic effects of different factors, i.e., their interactions. Moreover, when independent tests are performed on several factors and a high correlation exists between them, the outcome of the test can be completely wrong[13]. The methods here presented belong to two approaches: (1) unsupervised methods (pattern recognition methods), in which no a priori information is assumed and the evaluation of the existence of groups of samples is suggested by the statistical method itself; and (2) supervised methods (classification tools), in which the a priori information is provided in terms of membership of each sample to a specific class: the statistical method is therefore aimed at the identification of the variables responsible for the separation of the samples in the different classes.
Here, only the methods most recently applied in the literature to the case of pancreatic cancer will be briefly discussed. The methods presented are listed in Table 1.
| Type of statistical method | Method adopted |
| Classical mono- and multi-variate methods | Student t-test (parametric) |
| Mann-Whitney U-test (non-parametric) | |
| T2 Hotelling | |
| ANOVA and MANOVA | |
| Bayes factors | |
| Unsupervised pattern recognition methods | Principal Component Analysis |
| Cluster Analysis | |
| Multidimensional Scaling | |
| Supervised classification methods | SIMCA |
| Ranking-PCA | |
| O-PLS | |
| CART | |
| Random Forests | |
| Methods for determining survival outcomes | Kaplan Meyer functions |
| Cox Regression | |
| Other methods | PAM |
| Metropolis algorithm and Monte Carlo simulation |
Principal component analysis (PCA)[14,15] represents the objects in a new reference system characterized by new variables called principal components. The first principal component accounts for the maximum variance contained in the original dataset, while subsequent components account for the maximum residual variance. They are calculated hierarchically, so that systematic variations (i.e., information) are explained in the first components while experimental noise and random variations are contained in the last ones. The components are linear combinations of the original variables and are orthogonal to each other, thus containing independent sources of information. They are often used for dimensionality reduction by considering a smaller number of significant components containing only relevant information.
The graphical representation of the scores (the co-ordinates of the samples in the new reference system) in the space of the principal components allows the identification of groups of samples showing a similar behavior (samples close one to the other in the graph) or different characteristics (samples far from each other). The corresponding loading plot (representing the loadings, i.e., the coefficients of the linear combination describing each principal component) identifies the variables that are responsible for the analogies or the differences detected for the samples in the score plot.
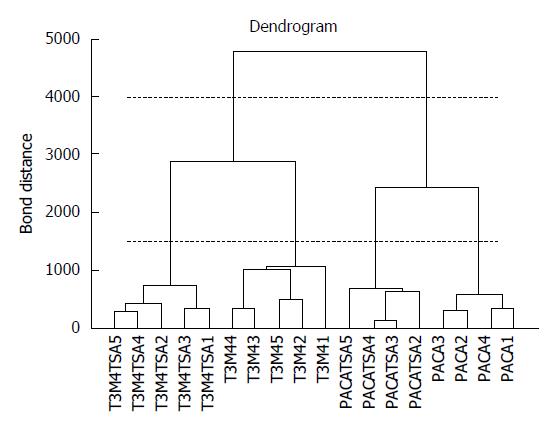
Cluster analysis techniques[14-16] allow the identification of groups of samples or of variables in a dataset by investigating the relationships between the samples or the descriptors. Agglomerative hierarchical methods[14,15] are the most widespread, grouping the samples on the basis of their similarity. The most similar samples or groups are linked first. The final result is a graph, called a dendrogram, where the samples are represented on the X axis and are connected at decreasing levels of similarity along the Y axis. The groups can be identified by applying a horizontal cut of the dendrogram and identifying the number of vertical lines crossed by the horizontal cut. Figure 1 reports a dendrogram where cutting at level 4000 produces only two clusters (the 2 different tumor cell lines), while cutting at level 1500 produces 4 clusters (PACA and T2M4 cell lines, treated and untreated with trichostatin A). The results of hierarchical clustering strongly depend on the measure of similarity and on the linking method adopted. Clustering techniques can be applied to the original variables or to the relevant principal components[16].
Multidimensional scaling (MDS)[17,18] is aimed at dimensionality reduction and graphical representation of the data. Given a set of n objects and a measure of their similarity, MDS searches for a low dimensional space in which the objects are represented by points in the space so that the distances between the points match as much as possible with their original similarities[17]. There are several different approaches to MDS depending on the measure of the similarities matching, on the metrics, on the method used to compute the similarities and on the way the samples configuration is obtained[19,20]. Shepard[21,22] and Kruskal[23] provided an extension of classical MDS to the study of nonparametric similarities.
Classification tools are supervised methods able to separate the objects in the classes present (known a priori, e.g., control vs pathological) and provide the variables most responsible for their belonging to different classes (candidate biomarkers). The final aim of their application in the biomedical field is both the development of diagnostic tools and the identification of the differences existing between the classes to shed light on the etiology of a disease or the effect of a new drug. Here, only the methods already applied to the identification of biomarkers for PC will be described.
The soft independent model of class analogy (SIMCA) method[24-27] is based on the independent modelling of each class by means of PCA. Each class is described by its relevant principal components. The samples belonging to each class are contained in the so-called SIMCA boxes, defined by the relevant components of each class. The classification of each sample with SIMCA is not affected by experimental uncertainty and random variations since each class is modelled only by its relevant components. This method is also useful when more variables than objects are available since it performs a substantial dimensionality reduction.
The identification of the candidate biomarkers by SIMCA can be accomplished by the analysis of the discrimination power (DP), a measure of the ability of each variable to discriminate between two classes at a time. The greater the DP, the more a variable weighs on the classification of an object in one of the two classes compared.
Ranking PCA is a ranking method proposed by Marengo et al[7], Robotti et al[28] and successively applied by Polati et al[29], based on the description of the original data by means of principal components. The use of PCA in the field of identification of biomarkers in the omics sciences is particularly effective since it allows the relationships between the variables to be taken into consideration, providing sets of correlated biomarkers with a similar function and the possibility of solving problems where the number of variables is larger than the number of samples.
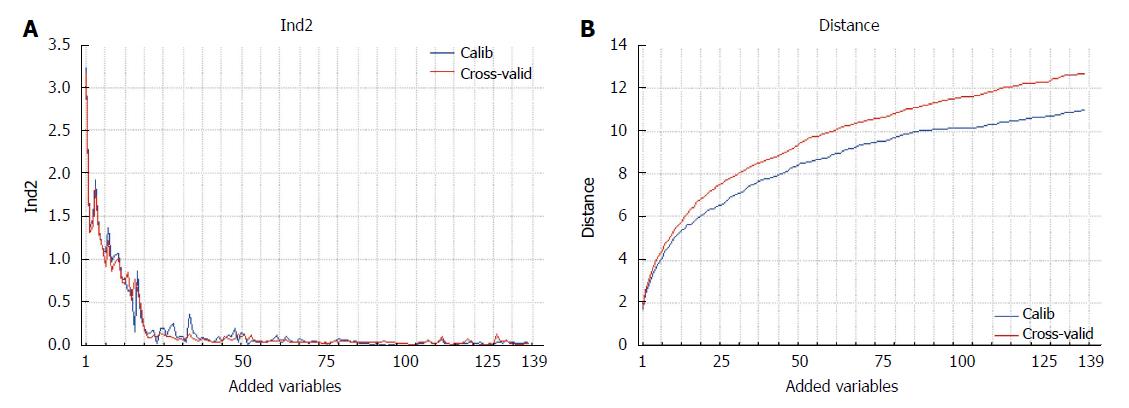
PCA is used here to describe the data coupled to a ranking procedure of the candidate biomarkers in a forward search: one variable is added at each cycle. The first variable selected is the one providing the best separation between the classes on the first principal component. The addition of another discriminating variable further improves the distance between the two classes on the first principal component. If a non-discriminating variable is successively added, the two classes will not be further separated on the same component. Sometimes, more than one component could be necessary for class separation: in this case, different independent sources of information related to the class structure are present in the data and the subsequent principal component accounting for class separation will be included in the model.
The proposed method allows the ranking of the variables according to their discrimination ability, thus assuring the exhaustiveness of the results. The result can be presented in graphical form (Figure 2), where the classification performance or the class distance are reported as a function of the variables added to the model.
Partial least squares (PLS)[14,15,30] establishes a relationship between one or more dependent variables (Y) and a group of descriptors (X). X and Y variables are modeled simultaneously to find the latent variables (LVs) in X that will predict the LVs in Y. These LVs are calculated hierarchically, as for PCA. PLS was originally set up to model continuous responses but it can be applied even for classification purposes (PLS-DA) by establishing an appropriate Y related to the association of each sample to a class. The regression is then carried out between X-block variables and the Y just established. Orthogonal PLS (OPLS)[31] is a modification of PLS developed for highly decorrelated datasets.
Classification trees[24] are built by subsequent divisions (splits) of subgroups of the original data D in two descending subgroups with the aim of classifying the data in homogeneous groups as much as possible, one different from the others. It is possible to derive a tree diagram where, starting from the root node (where the data D are not separated), a series of nodes and branches separate; each node h represents a subgroup of D. Nodes not undergoing a further split are called terminal nodes: a mode for y is associated with each terminal node.
Starting from the root node h1, data are separated in a series of splits: in each node, the split giving the most homogeneous division of the data in the two descendent nodes is selected.
Random forests[32] is an extension of classification trees and is structured to grow many classification trees. To classify a new object, the new object is first classified by each independent tree in the forest. The forest chooses the most recurrent classification (over all the trees in the forest).
The error rate depends on the correlation between any two trees in the forest (increasing the correlation increases the forest error rate) and the strength of each individual tree in the forest (inversely correlated to the tree error rate).
In clinical trials it is usually important to evaluate the time until the participants present with a particular event (endpoint), i.e., a clinical outcome (death, recurrence of a disease, remission etc.). All participants are followed from a certain starting point (operation, starting of a therapy, diagnosis, etc.) up to the moment when the event occurs (the time requested is recorded). However, often the outcome of some participants is unknown: when the study ends before all participants have presented the event or when some participants withdraw from the study. In these cases (censored data), the time of follow-up is recorded.
The Kaplan-Meier method[33] is used to provide survival functions and can also be effectively applied to censored data. It provides a survival curve where time is reported on the X-axis, while the cumulative survival probability is on the Y-axis. The time corresponding to the point where the curve crosses 50% survival is the estimate of median survival. Kaplan-Meier curves can be compared across groups by mainly applying two non-parametric tests (the log rank test[34] and the generalized Wilcoxon test[35]). The generalized Wilcoxon test[35] is a weighted alternative where early time points weigh more than late ones; this is preferred when the effect of an experimental condition vanishes with time. The log rank test instead is preferred to detect differences during all of the follow-up period.
Cox regression[36] is applied to evaluate the effect of several risk factors on survival, defining the hazard as the probability of the endpoint. The hazard is modeled as:
ln [H(t)/H0(t)] = b1X1 + b2X2 + ... + bkXk
where H(t)/H0(t) is the so-called hazard ratio (HR), X1 ... Xk are predictor variables and H0(t) is the baseline hazard at time. In general, the HR may assume only positive values: if it equals 1, the groups show a not statistically different survival; if it is smaller than 1, a subject with a higher value for X has lower risk than a subject with a lower value for X; the opposite behavior is obtained if the HR estimate is larger than 1.
The Cox model works under the so-called proportional hazards assumption: the ratio between the hazards of two patient groups remains constant over the complete follow-up period. Since a HR is calculated in Cox regression, this estimate should apply to all death times: this simplification is only justified if the group difference remains constant over the whole range of follow-up time.
If the monovariate Cox regression is extended to include more than one X variable (multivariate Cox regression), the effect of the interaction between different factors can be evaluated.
Prediction analysis for microarrays (PAM) performs sample classification from gene expression data and survival outcomes, exploiting the nearest shrunken centroid approach[37]. A standardized centroid for each class is computed. The nearest centroid classification compares the gene expression profile of a new sample to each class centroid: the class whose centroid is closest in squared distance is the predicted class for the new sample.
Nearest shrunken centroid classification[37] is a modification of this approach. When PAM is applied to survival outcomes, supervised principal components analysis[38] is performed, where, instead of using all the genes to perform PCA, only a subset of genes is used, i.e., those highly correlated with survival.
Monte Carlo methods[39] are computational algorithms that rely on repeated random sampling to obtain numerical results; simulations are run several times to calculate probabilities. They are useful for simulating systems with several degrees of freedom and modeling systems characterized by large uncertainty in inputs.
The Metropolis algorithm[40] is used to generate a series of numbers, X1, X2, .., Xn with a distribution fixed a priori. The method is based on the generation of numbers that are accepted or rejected to obtain the selected type of distribution.
The Metropolis algorithm can be implemented in Monte Carlo simulations to perform random sampling. The Monte Carlo optimization can be used in biomarkers search to determine the coefficients of model containing the relevant biomarkers.
The studies based on serum and tissue biomarkers determined by non-omics techniques usually exploit monovariate and multivariate Cox regression to evaluate the effect played by different factors on time to progression (TTP) and overall survival (OS). They will be presented here, divided into prognostic/predictive biomarkers and diagnostic biomarkers.
Recent studies about prognostic and/or predictive biomarkers (Table 2) determined in serum or plasma regard the determination of glycoproteins, both alone or associated with other markers, and the determination of circulating factors of the insulin-like growth factor. Three recent studies have been published based on CA19-9, one of the most debated biomarkers for PC. The first study, by Boeck et al[41], includes 115 patients with histologically confirmed advanced PC treated with first-line therapy. The novelty of this study is the modelling of the effect of CA 19-9 kinetics by treating it as a time-varying covariate. For CA 19-9 kinetics during chemotherapy, data from 69 patients (TTP) and 84 patients (OS) were available. The proposed approach allowed the modeling of the effect of log (CA 19-9) measured during therapy on the event (TTP or OS) rather than modeling the effect of the pretreatment value of log (CA 19-9).
| Ref. | Type of marker | Markers | Sample | Study group | Analytical methods | Statistical methods | Performance |
| 41 | P | CA 19-9 | S | Pretreatment CA 19-9: 115 patients from 5 German centers; 73% treated within prospective clinical trials. Median TTP: 4.4 mo; median OS: 9.4 mo. CA 19-9 kinetics during chemotherapy: 69 patients (TTP) and 84 patients (OS) | Elecsys assay | Cox proportional hazards regression; for CA 19-9 kinetics, CA 19-9 was treated as a time-varying covariate | Univariate analysis: log (CA 19-9) associated with TTP (HR = 1.24; P < 0.001) and OS (HR = 1.16; P = 0.002). Multivariate analysis: results confirmed. Log(CA 19-9) kinetics during chemotherapy: significant predictor for TTP in univariate analyses (HR = 1.48; P < 0.001) and multivariate (HR = 1.45; P < 0.001) and for OS (univariate: HR = 1.34; P < 0.001; multivariate: HR =1.38; P < 0.001) |
| 42 | P | CA 19-9, CEA, CRP, LDH and bilirubin | 291 patients; 253 patients (87 %) received treatment within prospective clinical trials. Median TTP: 5.1 mo. Median OS 9.0 mo | Elecsys assay | Kaplan Meier method and Cox proportional hazards regression | Univariate analysis: pre-treatment CA 19-9 (HR = 1.55), LDH (HR = 2.04) and CEA (HR = 1.89) significantly associated with TTP. Baseline CA 19-9 (HR = 1.46), LDH (HR = 2.07), CRP (HR = 1.69) and bilirubin (HR = 1.62) significant prognostic factors for OS. Multivariate analyses: pre-treatment log (CA 19-9) for TTP and log (bilirubin) and log (CRP) for OS had an independent prognostic value | |
| 44 | P | IGFs | S and P | 80 patients received treatment (40 Ganitumab; 40 placebo) | Immunoassays | Kaplan Meier method and Cox proportional hazards regression | Ganitumab associated with improved OS vs placebo (HR = 0.49; 95%CI: 0.28-0.87) |
| 45 | P | TROP2 | T | 197 patients; subgroup of 134 patients treated surgically | Immunohistochemistry | Kaplan Meier method and Cox proportional hazards regression | TROP2 overexpression observed in 109 (55%) patients and associated with decreased OS (P < 0.01). Univariate Analysis: TROP2 overexpression correlates with lymph node metastasis (P < 0.04) and tumor grade (P < 0.01). In the subgroup of patients treated surgically, TROP2 overexpression correlated with poor progression-free survival (P < 0.01). Multivariate analyses: TROP2 is an independent prognosticator |
| 46 | P | JAM-A | T | 186 patients; subgroup of 83 patients treated surgically | Immunohistochemistry | Kaplan Meier method and Cox proportional hazards regression | Low expression of JAM-A observed in 79 (42 %) patients and associated with poor OS (P < 0.01). Univariate analysis: low expression of JAM-A correlates with positive lymph node status (P = 0.02), the presence of distant metastasis (P = 0.05), and tumor grade (P = 0.04). In the subgroup of patients with surgically resected PC, low expression of JAM-A correlated with decreased progression-free survival (P < 0.01). Multivariate analysis: JAM-A was an independent predictor of poor outcome |
| 47 | P | TBX4 | T | 77 stage II PDAC tumors | Immunohistochemistry | Kaplan Meier method and Cox proportional hazards regression | 48 cases (62.3%) expressed TBX4 at a high level. No significant correlation between TBX4 expression and other clinicopathological parameters, except tumor grade and liver metastasis recurrence. Survival of patients with TBX4-high expression significantly longer than those with TBX4-low expression (P = 0.010). Multivariate analysis: low TBX4 expression independent prognostic factor for OS. TBX4 promoter methylation status frequently observed in PDAC and normal adjacent pancreas |
| 48 | P | HSP27 | T | 86 patients | Tissue microarray (TMA) analysis | Kaplan Meier method and Cox proportional hazards regression | HSP27 expression found in 49% of tumor samples. Univariate analyses: significant correlation between HSP27 expression and survival. Multivariate Cox-regression: HSP27 expression emerged as an independent prognostic factor. HSP27 expression also correlated inversely with nuclear p53 accumulation |
| 49 | P | dCK | T | 45 patients with resected PDAC received adjuvant gemcitabine based-therapy in multicenter phase 2 studies | Immunohistochemistry | Kaplan Meier method and Cox proportional hazards regression | Median follow-up: 19.95 mo (95%CI: 3.3-107.4 mo). Lymph node (LN) ratio and dCK protein expression significant predictors of DFS and OS in univariate analysis. Multivariate analysis: dCK protein expression the only independent prognostic variable (DFS: HR = 3.48, 95%CI: 1.66-7.31, P < 0.001, OS: HR = 3.2, 95%CI: 1.44-7.13, P < 0.004) |
| 50 | P | Notch3 and Hey-1 | T | 42 patients who underwent resection and 50 patients diagnosed with unresectable PDAC | Immunohistochemistry | Mann-Whitney U test, Wilcoxon test, Cox regression analysis, Kaplan-Meier analysis | All 3 Notch family members significantly elevated in tumor tissue. Significantly higher nuclear expression of Notch1, -3 and -4, HES-1, and HEY-1 (all P < 0.001) in locally advanced and metastatic tumors compared to resectable cancers. In survival analyses, nuclear Notch3 and HEY-1 expression significantly associated with reduced OS and DFS following tumor resection with curative intent |
| 51 | D and P | 21 biomarkers | P | clinically defined cohort of 52 locally advanced (Stage II/III) PDAC cases and 43 age-matched controls | Proximity ligation assay | Combination of the PAM algorithm and logistic regression modeling. Biomarkers that were significantly prognostic for survival were determined using univariate and multivariate Cox survival models | CA19-9, OPN and CHI3L1 were found to have superior sensitivity for pancreatic cancer vs CA19-9 alone (93% vs 80%). CEA and CA125 have prognostic significance for survival (P < 0.003) |
| 52 | D | 83 circulating proteins | S | 333 PDAC patients; 144 controls (benign pancreatic conditions); 227 healthy controls. Samples from each group split randomly into training and blinded validation sets. Panels evaluated in validation set and in patients diagnosed with colon (33), lung (62) and breast (108) cancers | bead-based xMAP immunoassays | A Metropolis algorithm with Monte Carlo simulation (MMC) was used to identify discriminatory biomarker panels in the training set | Training set (160 PDAC, 74 Benign, 107 Healthy): panel of CA19–9, ICAM-1, and OPG discriminated PDAC from Healthy controls (SN/SP 88/90%), panel of CA 19–9, CEA, and TIMP-1 discriminated PDAC patients from Benign subjects (SN/SP = 76%/90%). Independent validation set (173 PDAC, 70 Benign, 120 Healthy): panel of CA 19–9, ICAM-1 and OPG demonstrated SN/SP of 78%/94%; panel of CA19–9, CEA, and TIMP-1 demonstrated SN/SP of 71%/89%. The CA19–9, ICAM-1, OPG panel is selective for PDAC and does not recognize breast (SP = 100%), lung (SP = 97%), or colon (SP = 97%) cancer |
| 53 | D and P | YKL-40, IL-6, and CA 19.9 | P | 559 patients with PC from prospective biomarker studies from Denmark (n = 448) and Germany (n = 111) | ELISA and chemiluminescent immunometric assay | Kaplan Meier method and Cox proportional hazards regression | Odds ratios (ORs) for prediction of PC significant for all biomarkers, with CA 19.9 having the highest AUC (CA 19.9: OR = 2.28, 95%CI: 1.97-2.68, P = 0.0001, AUC = 0.94; YKL-40: OR = 4.50, 3.99-5.08, P = 0.0001, AUC = 0.87; IL-6: OR = 3.68, 3.08-4.44, P = 0.0001, AUC = 0.87). Multivariate Cox analysis: high preoperative IL-6 and CA 19.9 independently associated with short OS (CA 19.9: HR = 2.51, 1.22–5.15, P = 0.013; IL-6: HR = 2.03, 1.11-3.70, P = 0.021). Multivariate Cox analysis of non-operable patients: high pre-treatment levels of each biomarker independently associated with short OS (YKL-40: HR = 1.30, 1.03-1.64, P = 0.029; IL-6: HR = 1.71, 1.33-2.20, P = 0.0001; CA 19.9: HR = 1.54, 1.06-2.24, P = 0.022). Patients with preoperative elevation of IL-6 and CA 19.9 had shorter OS (P = 0.005) compared to patients with normal levels (45% vs 92% alive after 12 mo) |
In the second study, Haas et al[42] pooled pre-treatment data on CA 19-9, carcinoembryonic antigen (CEA), C-reactive protein (CRP), lactate dehydrogenase (LDH) and bilirubin from two multicenter randomized phase II trials and prospective patient data. Marker levels were assessed before the start of palliative first-line therapy for advanced PC and during treatment (for CA 19-9 only).
In the third study, Boeck et al[43] evaluated pre-treatment (palliative first-line chemotherapy) values and weekly values of cytokeratin 19-fragments (CYFRA 21-1), CA 19-9 and CEA in blood samples from patients with PC. CYFRA 21-1 are biomarkers for different epithelial diseases but their role in PC has not been investigated yet.
In Boeck et al[41] pre-treatment log (CA19-9) proved to be significantly associated with TTP and OS. Moreover, log (CA 19-9) kinetics after the start of treatment was found to be a significant predictor for both TTP and OS. Similar results were found by Haas et al[42] where pre-treatment CA 19-9, LDH and CEA levels were significantly associated with TTP. Regarding OS, baseline CA 19-9, LDH, CRP and bilirubin were significant. Boeck et al[43] found that CYFRA 21-1 and CA 19-9 showed a high correlation with TTP and OS, while in multivariate analysis, only CYFRA 21-1 and performance status were independent predictors for OS.
McCaffery et al[44] assessed the predictive nature of baseline circulating factors of the insulin-like growth factor (IGF) axis on the treatment effect of ganitumab plus gemcitabine in metastatic PDAC. Baseline levels of IGFs/IGF binding proteins were analyzed in serum or plasma while mutations and gene expression were analyzed in archival samples. Ganitumab was associated with improved OS vs placebo. The treatment effect on improved OS was larger in patients with higher levels of IGF-1, IGF-2 or IGFBP-3, or lower levels of IGFBP-2. Interaction between treatment and IGFs/IGFBPs showed predictive potential for IGF-2 and IGFBP-2.
The studies about prognostic and/or predictive biomarkers determined in tissue samples, usually by immunohistochemistry or tissue microarray analysis (TMA), instead include the determination of glycoproteins, other proteins and enzymes.
Fong et al[45] investigated the expression of TROP2 (human trophoblast cell-surface) antigen, a glycoprotein found to be strongly expressed in a variety of human epithelial cancers, and the expression of junctional adhesion molecule A (JAM-A) antigen[46], a type I transmembrane glycoprotein, which has been recently shown to affect the prognosis of several malignancies. The two studies involved 197 and 186 patients with PDAC respectively. TROP2 overexpression was observed in 55% of patients, while low expression of JAM-A was observed in 42% of samples; both markers were significantly associated with decreased OS. They were both correlated with lymph node metastasis and tumor grade. In the subgroup of patients surgically treated with curative intent, TROP2 and low expression of JAM-A correlated with poor progression-free survival.
In the study by Zong et al[47], the expression of the T-box transcription factor 4 (TBX4) was investigated in 77 stage II PDAC tumors. 62.3% of cases expressed TBX4 at a high level. Significant correlation was only detected between TBX4 expression and tumor grade and liver metastasis recurrence. The survival with TBX4-high expression was significantly longer.
Applying tissue microarray (TMA) analysis, Schäfer et al[48] correlated heat shock protein 27 (HSP27) expression status with clinicopathological parameters in PDAC from 86 patients. HSP27 expression was found in 49% of tumor samples. A significant correlation was found with OS. The authors also assessed the impact of HSP27 on chemo- and radio-sensitivity directly in PC cells. HSP27 expression emerged as an independent prognostic factor and correlated inversely with nuclear p53 accumulation, indicating protein interactions between HSP27 and p53 or TP53 mutation-dependent HSP27-regulation. HSP27 overexpression rendered HSP27 low-expressing PL5 PC cells more susceptible to treatment with gemcitabine, while HSP27 protein depletion in HSP27 high-expressing AsPC-1 cells caused increased gemcitabine resistance.
Maréchal et al[49] identified deoxycytidine kinase (dCK), a recombinant enzyme, to be associated with prolonged survival after adjuvant gemcitabine administration for resected PDAC. The study involved 45 patients. The lymph node (LN) ratio and dCK protein expression were significant predictors of DFS and OS in univariate analysis. On multivariate analysis, a step-down procedure based on the likelihood ratio test, dCK protein expression was the only independent prognostic factor.
Having previously reported that Notch3 activation appeared to be associated with more aggressive PC disease, Mann et al[50] examined components of this pathway (Notch1, Notch3, Notch4, HES-1, HEY-1) in resectable and non-resectable tumors compared to uninvolved pancreas. All three Notch family members were significantly increased in tumor tissue, with expression maintained within matched lymph node metastases. Significantly higher nuclear expression of Notch1, -3 and -4, HES-1 and HEY-1 was noted in locally advanced and metastatic tumors compared to resectable cancers. Nuclear Notch3 and HEY-1 expression were significantly associated with reduced OS and DFS following tumor resection.
Regarding diagnostic biomarkers (Table 2), the most recent studies are based on the determination of protein panels in serum or plasma, exploiting ELISA or proximity ligation assay (PLA) for their determination. PLA is a highly sensitive technique for multiplex detection of biomarkers in plasma with little interfering background signal. Some of the studies proposed the determination of both diagnostic and prognostic markers. All studies presented here involve CA19-9 as a potential biomarker in association with other markers.
In the first study, Chang et al[51] applied PLA to the identification of plasma levels of 21 biomarkers in 52 locally advanced PDAC cases and 43 age-matched controls. The optimal diagnostic biomarker panel was computed using a combination of the PAM algorithm and logistic regression modeling.
In the second study, Brand et al[52] investigated 83 circulating proteins in sera of patients diagnosed with PDAC, benign pancreatic conditions and healthy controls. Samples from each group were split randomly into training and blinded validation sets prior to analysis. A Metropolis algorithm with Monte Carlo simulation (MMC) was used to identify discriminatory biomarker panels in the training set. Identified panels were evaluated in the validation set and in patients diagnosed with colon, lung and breast cancers.
In the third study, Schultz et al[53] tested the hypothesis that high plasma YKL-40 and IL-6 are associated with PC and short OS. 559 patients with PC from prospective biomarker studies were studied. Plasma YKL-40 and IL-6 were determined by ELISA and serum CA 19.9 by chemiluminescent immunoassay.
Chang et al[51] found that three markers (CA19-9, OPN and CHI3L1) have superior sensitivity for PC vs CA19-9 alone (93% vs 80%) and two markers (CEA and CA125) proved to have a prognostic significance for survival of PC (P < 0.003) when measured simultaneously. Brand et al[52] found that the panel of CA 19-9, CEA and TIMP-1 discriminated PDAC patients from benign subjects with an SN/SP of 76%/90% in the training set and of 71%/89% in the validation set. The CA19-9, intercellular adhesion molecule 1 (ICAM-1), OPG panel is selective for PDAC and does not recognize breast (SP = 100%), lung (SP = 97%) or colon (SP = 97%) cancer. Schultz et al[53] instead showed that high preoperative IL-6 and CA 19.9 were independently associated with short OS. High pre-treatment levels of each biomarker were independently associated with short OS in non-operable patients.
The most recent studies on identification of biomarkers in proteomics (Table 3) are mainly regarding the determination of diagnostic markers. The studies are presented separating those carried out from serum or tissue samples and those carried out on cell lines or animal models. The studies presented here are mainly devoted to identifying exploratory biomarkers.
| Ref. | Type of marker | Markers | Sample | Study group | Analytical methods | Statistical methods | Performance |
| 54 | D | Among 2393 unique proteins, 104 proteins significantly changed in cancer | T | 5 patients; matched pairs of tumor and non-tumor pancreas | Tissues treated to obtain cytosol, membrane, nucleus and cytoskeletonfractions. Fractions separated and digested underwent LC-MS/MS | PLGEM | 104 proteins significantly changed in cancer. Among these, 4 proteins validated that were up-regulated in cancer: biglycan (BGN), Pigment Epithelium-derived Factor (PEDF) Thrombospondin-2 (THBS-2) and TGF-β induced protein ig-h3 precursor (βIGH3) |
| 57 | D | Serum MALDI-TOF features | S | 15 healthy (H), 24 cancer (Ca), 11 chronic pancreatitis (CP) samples | MALDI-TOF | Nonparametric | 8 serum features: Ca samples differentiated from H (SN = 88%, SP = 93%), Ca from CP (SN = 88%, SP = 30%), and Ca from both H and CP combined (SN = 88%, SP = 66%). 9 features obtained from urine: differentiated Ca from both H and CP combined (SN = 90%, SP = 90%) |
| 59 | D | Serum SELDI-TOF features | S | 96 serum samples from patients undergoing cancer surgery compared with sera from 96 controls | SELDI-TOF | pairwise statistics, MDS, hierarchical analysis Mann-Whitney U test, CART | Data analysis identified 24 differentially expressed protein peaks, 21 of which under-expressed in cancer samples. The best single marker predicts 92% of controls and 89% of cancer samples. Multivariate analysis: best model (3 markers) with SN = 100% and SP = 98% for the training data and SN = 83% and SP = 77% for test data. Apolipoprotein A-II, transthyretin and apolipoprotein A-I identified as markers and decreased at least 2 fold in cancer sera |
| 60 | D | Serum SELDI-TOF features | S | 57 PC samples were compared to 59 controls | SELDI-TOF | Multivariate decorrelation filtering | Improved classification performances when the presented strategy is compared to standard univariate feature selection strategies |
| 61 | D | Proteins | S | Sera from patients diagnosed with PC compared with age- and sex-matched normal subjects | Protein microarrays | Rank-based non-parametric statistical testing | A serum diagnosis of PC was predicted with 86.7% accuracy, with a sensitivity and specificity of 93.3% and 80%. Candidate autoantibody biomarkers studied for their classification power using an independent sample set of 238 sera. Phosphoglycerate kinase-1 and histone H4 noted to elicit a significant differential humoral response in cancer sera compared with age- and sex-matched sera from normal patients and patients with chronic pancreatitis and diabetes |
| 62 | D | Proteins | PDAC cell lines | 435 spots identified from 18 samples from 2 cell lines (Paca44 and T3M4) of control and drug-treated PDAC cells | 2D-PAGE | PCA, SIMCA, Ranking-PCA | Samples were all perfectly classified. Significant proteins were further identified by MS analysis |
| 63 | D | Proteins regulating the conversion of quiescent to activated PaSC cells | rat PaSC cell line | - | SDS-PAGE and GeLC-MS/MS | QSPEC | Qualitative and quantitative proteomic analysis revealed several hundred proteins as differentially abundant between the two cell states. Proteins of greater abundance in activated PaSC: isoforms of actin and ribosomal proteins. Proteins more abundant in non-proliferating PaSC: signaling proteins MAP kinase 3 and Ras-related proteins |
The studies based on proteomic approaches show a great potential for the identification of PC biomarkers; the panels of markers identified by these techniques are characterized by good performance regarding both sensitivity and specificity, showing results at least equal to or in some cases better than classical approaches. It is the authors’ opinion that, in future, the information provided by such high-throughput techniques should be coupled with clinical information to provide exhaustive sets of biomarkers with better predictive ability.
Tissue and serum biomarkers in proteomics are usually determined by: SDS-PAGE followed by LC-MS for identifying the most up- or down-regulated proteins; matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry; and surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) mass spectrometry. The exploitation of instrumental techniques providing a high amount of information makes the application of multivariate methods necessary in order to detect panels of biomarkers with the best predictive ability.
In the first study by McKinney et al[54], matched pairs of tumor and non-tumor pancreas from patients undergoing tumor resection were treated to obtain cytosol, membrane, nucleus and cytoskeleton cellular protein fractions. The fractions were analyzed by SDS-PAGE followed by LC-MS/MS to identify 2393 unique proteins. The spectral count data were compared using a power law global error model (PLGEM) to identify statistically significant protein changes between non-tumor and tumor samples[55,56]. Among the 104 proteins significantly changed in cancers, four (biglycan (BGN), pigment epithelium-derived factor (PEDF), thrombospondin-2 (THBS-2) and TGF-β induced protein ig-h3 precursor (βIGH3)) were further validated and proved to be up-regulated in cancer and have potential for development as minimally-invasive diagnostic markers.
Kojima et al[57] differentiated pancreatic neoplasia from non-neoplastic pancreatic disease. Samples from 50 patients [15 healthy (H), 24 cancer (Ca), 11 chronic pancreatitis (CP)] were collected. A high-throughput method was applied, using high-affinity solid lipophilic extraction resins, enriched low molecular weight proteins for extraction with a high-speed MALDI-MS. Multivariate analysis was carried out by MDS as in Mobley et al[58]. Using eight serum features, Ca were differentiated from H (SN = 88%, SP = 93%), Ca from CP (SN = 88%, SP = 30%) and Ca from both H and CP combined (SN = 88%, SP = 66%). In addition, nine features obtained from urine differentiated Ca from both H and CP, combined with high efficiency (SN = 90%, SP = 90%). Interestingly, the plasma samples did not show significant differences.
Ehmann et al[59] and Hauskrecht et al[60] instead applied SELDI-TOF mass spectrometry. In the first study[59], 96 serum samples from patients undergoing cancer surgery were compared with 96 controls. Samples were fractionated by anion exchange chromatography. Data analysis, involving Mann-Whitney U test and classification and regression tree (CART) analysis, identified 24 differentially expressed protein peaks, 21 of which were under-expressed in cancer samples. The best single marker can predict 92% of controls and 89% of cancer samples. The best model with a set of 3 markers showed a sensitivity of 100% and a specificity of 98% for the training data and a sensitivity of 83% and a specificity of 77% for test data. Apolipoprotein A-II, transthyretin and apolipoprotein A-I were identified as markers (decreased in cancer sera). Hauskrecht et al[60] instead proposed a feature selection method that extracts useful feature panels from high-throughput spectra. 57 PC samples were compared to 59 controls. The results clearly show the improved classification performances when the method is compared to standard strategies.
The last study makes use of protein microarrays to explore whether a humoral response to PC-specific tumor antigens has utility as a biomarker of PC. To determine if such arrays can be used to identify novel autoantibodies in the sera from PC patients, Patwa et al[61] resolved proteins from a PDAC cell line (MIAPACA) by 2-D liquid-based separations and then arrayed them on nitrocellulose slides. The slides were probed with sera from a set of patients diagnosed with PC and compared with age- and sex-matched normal subjects. To account for patient-to-patient variability, a non-parametric Wilcoxon rank-sum test was used in which protein biomarkers were identified. Classification by the PAM algorithm showed 86.7% accuracy, with a SN and SP of 93.3% and 80%, respectively. The identified candidate autoantibody biomarkers were validated using an independent sample set of 238 samples. Phosphoglycerate kinase-1 and histone H4 were noted to elicit a significant differential humoral response in cancer sera.
Diagnostic biomarkers in proteomics have also been recently determined from cell lines (PACA44, T3M4) or on animal models. The most exploited analytical techniques in this case are SDS-PAGE, followed by LC-MS, 2D-PAGE or 2D-LC approaches.
Marengo et al[62] identified the regulatory proteins in human PC treated with trichostatin A by 2D-PAGE maps and multivariate analysis. PCA was applied to a spot quantity dataset comprising 435 spots detected in 18 samples belonging to two different cell lines (Paca44 and T3M4) of control and drug-treated PDAC cells. PCA allowed the identification of the groups of samples present in the dataset; the loadings analysis allowed the identification of the differentially expressed spots, which characterize each group of samples. The treatment of both the cell lines with trichostatin A showed an evident effect on the proteomic pattern of the treated samples. Identification of some of the most relevant spots was also performed by MS analysis. The same authors applied different multivariate statistical tools to the same set of data to provide sets of candidate biomarkers; the first application regards the exploitation of SIMCA classification to evaluate the biomarkers characterized by a significant discriminant power[25], while the second application regards the development of ranking PCA[28]. This second application is particularly interesting for overcoming the limitations of the methods usually adopted as variable selection tools to identify only significant biomarkers: they are usually aimed at the selection of the smallest set of variables (spots) providing the best performances in prediction. This approach does not seems to be the best choice in the identification of potential biomarkers since all the possible candidate biomarkers have to be identified to provide a general picture of the ‘‘pathological state’’; exhaustivity has to be preferred to provide a complete understanding of the mechanisms underlying the pathology. Ranking PCA allowed the exhaustive identification of a complete set of candidate biomarkers.
Paulo et al[63] compared differentially expressed proteins in rat in activated and serum-starved non-proliferating pancreatic stellate cells (PaSC), emerging key mediators in chronic pancreatitis and PC pathogenesis. About 1500 proteins were identified after SDS-PAGE and LC-MS/MS. Qualitative and quantitative proteomic analysis revealed several hundred proteins to be differentially abundant between the two cell states. Significance analysis was performed using QSPEC, a recently published algorithm for determining the statistical significance of differences in spectral counting data from two sample sets[64]. This algorithm exploits the Bayes factor instead of the P-value as a measure of statistical significance[65,66]. Proteins of greater abundance in activated PaSC included isoforms of actin (e.g., smooth muscle actin) and ribosomal proteins. Proteins more abundant in non-proliferating PaSC than in activated PaSC included signaling protein MAPK-3 and Ras-related proteins. The molecular functions and biological pathways for these proteins were also determined by gene ontology analysis and KEGG pathway.
Paulo et al[67] also evaluated the endoscopic pancreatic function test (ePFT) as a method able to safely obtain pancreatic fluid for MS analysis from patients during upper endoscopy and reproducibly identify pancreas-specific proteins. The ePFT-collected pancreatic fluid from 3 individuals without evidence of chronic pancreatitis was analyzed by SDS-PAGE and GeLC-MS/MS. The SDS-PAGE analysis revealed no significant variation in protein concentration during the 1 h collection. The GeLC-MS/MS analysis identified pancreas-specific proteins previously described from endoscopic retrograde cholangiopancreatography and surgical collection methods. Gene ontology further revealed that most of the proteins identified have a molecular function of proteases.
The most recent studies on identification of biomarkers in genomics (Table 4) regard both the determination of prognostic/predictive markers and diagnostic markers and are presented hereafter separated into these two classes. The studies presented here are mainly devoted to identifying exploratory biomarkers.
| Ref. | Type of marker | Markers | Sample | Study group | Analytical methods | Statistical methods | Performance |
| 68 | P | K-ras mutation status and subtypes | endoscopic ultrasound-guided fine-needle aspiration specimens | 242 patients | RT-PCR | Kaplan Meier method and Cox proportional hazards regression | Multivariate analysis: CA19-9 C 1000 U/mL (HR = 1.78, 95 %CI: 1.28-2.46, P < 0.01), metastatic stage (HR 2.26, 95 % CI 1.58-3.24, P < 0.01) and mutant-K-ras (HR 1.76, 95 %CI: 1.03-3.01, P = 0.04) negative prognostic factors. Among patients with K-ras mutation subtypes: CA19-9 C 1000 U/mL (HR 1.65, 95%CI: 1.12-2.37, P < 0.01), metastatic stage (HR 2.12, 95%CI: 1.44-3.14, P < 0.01), and G12D or G12R mutations (HR = 1.60, 95%CI: 1.11-2.28) negative prognostic factors for OS |
| 69 | P | MicroRNA-21 | T | 82 resected Korean PDAC cases. Subgroup of patients treated with adjuvant therapy (n = 52) | Protein expression by immunohistochemistry microRNA expression by qRT-PCR | Cox proportional hazards model | Subgroup with adjuvant therapy: lower than median miR-21 expression associated with lower HR for death (HR = 0.316, 95%CI = 0.166-0.600, P = 0.0004) and recurrence (HR = 0.521, 95%CI = 0.280-0.967, P = 0.04). MiR-21: single most predictive biomarker for treatment outcome. No significant association in patients not treated with adjuvant therapy. Independent validation cohort of 45 frozen PDAC tissues from Italian cases treated with adjuvant therapy: lower than median miR-21 expression confirmed to be correlated with longer OS and DFS |
| 71 | P | 13 putative PDA biomarkers from the public biomarker repository | A survival tissue microarray was constructed comprised of short-term (cancer-specific death ,12 mo, n = 58) and long-term survivors (30 mo, n = 79) who underwent resection for PDA (total, n = 137) | Immunohistochemical analyses; survival tissue microarray (s-TMA) | Wilcoxon rank sum test | Multivariate model: MUC1 (OR = 28.95, 3+ vs negative expression, P = 0.004) and MSLN (OR = 12.47, 3+ vs negative expression, P = 0.01) highly predictive of early cancer-specific death. Pathological factors (size, lymph node metastases, resection margin status, and grade): ORs < 3 and none reached statistical significance. ROC curves used to compare the 4 pathological prognostic features (ROC area = 0.70) to 3 univariate molecular predictors (MUC1, MSLN, MUC2) of survival group (ROC area = 0.80, P = 0.07) | |
| 1 | P | MTA2 mRNA and protein expression | T | 123 PDAC samples and 40 control tissues | qRT-PCR and immunohistochemistry | Kaplan-Meyer curves and Cox analysis | MTA2 mRNA and protein expression levels up-regulated in PC. MTA2 correlated with poor tumor differentiation, TNM stage and lymph node metastasis. Patients with high expression levels of MTA2 showed lower OS. MTA2: independent prognostic factor. |
| 72 | D | Leukocyte DNA Methylation | Blood | Phase I: 132 never-smoker PaC patients and 60 never-smoker healthy controls. Phase II: validation of 88 of 96 phase I selected CpG sites in 240 PaC cases and 240 matched controls | DNA array | Wilcoxon Rank Sum tests and likelihood penalized logistic regression models | Significant differences found in 110 CpG sites (FDR < 0.05). Phase II: 88 of 96 phase I selected CpG sites validated in 240 PaC cases and 240 matched controls (P < 0.05). Prediction model: 5 CpG sites (IL10_P348, LCN2_P86, ZAP70_P220, AIM2_P624, TAL1_P817) discriminated PaC from controls (C = 0.85 in phase I; 0.76 in phase II). One CpG site (LCN2_P86) could discriminate resectable patients from controls (C = 0.78 in phase I; 0.74 in phase II). 3 CpG sites identified (AGXT_P180_F, ALOX12_E85_R, JAK3_P1075_R) where the methylation levels were significantly associated with SNPs (FDR < 0.05) |
| 73 | D | cell-surface targets | T | 28 PC specimens and 4 normal pancreas tissue samples. Expression in normal tissues evaluated by array data from 103 samples representing 28 organ sites as well as mining published data | Complementary assays of mRNA expression. Immunohistochemistry. qRT-PCR | - | 170 unique targets highly expressed in 2 or more PC specimens and not expressed in normal pancreas samples. Two targets (TLR2 and ABCC3) further validated for protein expression by tissue microarray based immunohistochemistry have potential for the development of diagnostic imaging and therapeutic agents for PC |
| 74 | D | Differentially expressed genes | Blood | 25 patients diagnosed with PC and diabetes, 27 patients with PC without diabetes, 25 patients with diabetes mellitus > 5 yr, and 25 healthy controls. Results further validated for 101 blood samples | Microarray and qRT-PCR | - | 58 genes found to be unique in patients with cancer-associated diabetes, including 23 up-regulated and 35 down-regulated genes. 11 up-regulated genes further validated by RT-PCR; 2 of these (VNN1 and MMP9) selected for logistic regression analysis. The combination of VNN1 and MMP9 showed the best discrimination of cancer-associated diabetes from type 2 diabetes. The protein expression of MMP9 and VNN1 was in accordance with the gene expression |
The studies based on genomic approaches have potential to identify PC biomarkers. In future, the information provided by genomic approaches should be coupled to proteomic, metabolomic and clinical information to improve the predictive ability of the panels of identified biomarkers.
Prognostic and predictive biomarkers in genomics (Table 4) are usually determined in tissue samples. Some of the proposed studies include both protein expression and microRNA expression profiles. In these studies, multivariate Cox regression analysis is usually exploited.
Ogura et al[68] studied the K-ras mutation status in 242 patients with unresectable PC. The authors focussed on K-ras mutation subtypes since recent reports indicate that K-ras mutation status acts as a prognostic factor. CA19-9, metastatic stage and mutant-K-ras were negative prognostic factors, indicating a reduced survival. Among the patients who had K-ras mutation subtypes, CA19-9, metastatic stage and the presence of the G12D or G12R mutations were negative prognostic factors for OS.
Hwang et al[69] evaluated whether expression of novel candidate biomarkers, including microRNAs, can predict clinical outcome in PDAC patients treated with adjuvant therapy. 82 resected PDAC cases were analyzed for protein expression by immunohistochemistry and for microRNA expression by quantitative real time PCR (qRT-PCR). Lower than median miR-21 expression was associated with a significantly lower HR for death and recurrence in the subgroup of patients treated with adjuvant therapy. MiR-21 expression status emerged as the single most predictive biomarker for treatment outcome. No significant association was detected in patients not treated with adjuvant therapy. The results were confirmed in an independent validation of 45 PDAC tissues.
One-fifth of patients with seemingly “curable” PDAC experienced an early recurrence and death, while some patients with advanced stage tumors are deemed “unresectable” by conventional staging criteria, yet progress slowly. Effective biomarkers that stratify PDAC based on the biological behavior are therefore needed. Building on a compendium of 2500 published candidate biomarkers in PDAC[70], Winter et al[71] constructed a survival tissue microarray (s-TMA) comprised of short-term (12 mo) and long-term survivors (30 mo) who underwent resection for PDAC. The s-TMA acts as a biological filter to identify prognostic markers. 13 putative PDAC biomarkers were identified from the public biomarker repository and tested against the s-TMA. MUC1 and MSLN were highly predictive of early cancer-specific death. By comparison, no pathological factors (size, lymph node metastases, resection margin status and grade) reached statistical significance.
Chen et al[1] detected metastasis-associated gene 2 (MTA2) expression in PDAC and related it to prognosis. MTA2 mRNA and protein expression were determined by qRT-PCR and immunohistochemistry in primary cancers and their adjacent non-cancerous tissues. MTA2 mRNA and protein expression levels were up-regulated in PC. MTA2 was correlated with poor tumor differentiation, TNM stage and lymph node metastasis. Patients with high expression levels of MTA2 showed lower OS.
Diagnostic biomarkers from genomics-based studies (Table 4) are identified both from tissue and serum samples.
To identify biomarkers for early detection, Pedersen et al[72] examined DNA methylation differences in leukocyte DNA between PC cases and controls. In phase I, methylation levels were measured at 1505 CpG sites in leukocyte DNA from 132 never-smoker PC patients and 60 never-smoker controls. Significant differences were found in 110 CpG sites. In phase II, 88 of 96 phase I selected CpG sites were tested and validated in 240 PC cases and 240 matched controls. Using penalized logistic regression, a prediction model was built consisting of five CpG sites (IL10_P348, LCN2_P86, ZAP70_P220, AIM2_P624, TAL1_P817) that discriminated cancer patients from controls. One CpG site (LCN2_P86) alone could discriminate resectable patients from controls.
Morse et al[73] used complementary assays of mRNA expression profiling of cell-surface genes to determine increased expression in PC vs normal pancreas tissues and validated protein expression by immunohistochemistry on tissue microarrays. This approach was aimed at the identification of targets for potential use in the molecular imaging of cancer, allowing for non-invasive determination of tumor therapeutic response and molecular characterization of the disease, or in the targeted delivery of therapy to tumor cells, decreasing systemic effects. Expression profiles of 2177 cell-surface genes for 28 pancreatic tumor specimens and 4 controls were evaluated. 170 unique targets were highly expressed in 2 or more of the pancreatic tumor specimens and were not expressed in controls. Two targets (TLR2 and ABCC3) were further validated for protein expression and proved to be potential for the development of diagnostic imaging and therapeutic agents for PC.
Huang et al[74] explored specific biomarkers that can differentiate PC-associated diabetes from type 2 diabetes for the early detection of PC. Peripheral blood samples were collected from 25 patients diagnosed with PC and diabetes, 27 patients with PC without diabetes, 25 patients with diabetes mellitus > 5 years, and 25 controls. 32 samples were used in microarray experiments to find differentially expressed genes specific for cancer-associated diabetes. The results were further validated by quantitative qRT-PCR for 101 blood samples. Protein expression of selected genes in serum and tissues was also detected. 58 genes were found to be unique in patients with cancer-associated diabetes (23 up-regulated; 35 down-regulated). 11 up-regulated genes were further validated by RT-PCR and 2 of these, vanin-1 (VNN1) and matrix metalloproteinase 9 (MMP9), showed the best discrimination of cancer-associated from type 2 diabetes.
Just one paper, by Kaur et al[75], has recently appeared in the literature, reporting for the first time the mass spectrometry-based metabolomic profiling of human pancreas in matched tumor and normal tissues. UPLC coupled with TOF-MS was applied to perform small molecule metabolite profiling of matched normal and PC tissues. The resulting multivariate data matrix was pre-processed for spectral alignment and peak detection, followed by normalization of the data to the feature intensities of the internal standard as well as to the total protein concentration. The normalized data were analyzed first by PCA, followed by OPLS[31]. The authors also exploited random forest clustering[32] to interrogate the top 50 features with significant alterations in the tumor tissue compared to the control. The candidate markers were searched against different databases[76,77] to find compounds that corresponded to the accurate monoisotopic mass measurements detected by UPLC-TOFMS analysis. The authors report a subset of metabolites which were unequivocally identified and found to be significantly de-regulated in PC tissues.
The study reported here proves that metabolomic profiling shows great potential for the identification of biomarkers for PC. Certainly, further characterization and validation with a large sample size is needed and may help establish the utility of such markers as biomarkers of clinical benefit.
This review aimed to present the most recent applications of the omics approaches (proteomics, genomics and metabolomics) to the identification of biomarkers for PC. Particular attention has been paid to the statistical methods adopted for identification of biomarkers, first presenting the main statistical procedures adopted from a theoretical point of view. Then, the most recent applications present in the literature were presented separately for non-omic, proteomic, genomic and metabolomic based studies. Within this distinction, studies were presented separately for diagnostic and prognostic/predictive biomarkers and according to the type of marker.
Different statistical approaches are exploited in the literature for the identification of markers in PC; the methodologies presented here appear to be effective and sound. However, it is the authors’ opinion that multivariate methods have to be preferred. With the term multivariate, the authors refer to methods evaluating the relationships between the variables (both predictors and outcomes if several of both are present) in order to provide a pool of markers highlighting synergistic and antagonistic effects. In fact, the biological effect played by pathology (and PC makes no exception) is the result of a series of different mechanisms independent from each other or showing relevant interactions. Among all strategies, therefore, multivariate ones able to point out these relationships are preferred.
Another hint that must be addressed is the risk of identifying false positives, i.e., markers erroneously identified as such; this risk greatly increases when little information is available (i.e., a small number of cases/patients is investigated). Certainly, this problem is deeply related to the problem of experimental design and sample collection and each study should be carefully designed from a statistical point of view before being performed in order to include all possible sources of biological variation. Of course, this necessity often clashes with the availability of samples, especially when tissue collection is involved. From a statistical point of view, in these cases characterized by little information, it is very important to apply mathematical tools to validate the models built, thus evaluating the predictive ability of the models. In this respect, the use of cross-validation techniques or simulation algorithms is fundamental to identify only statistically significant markers.
Certainly, there is a great gap between the results presented in studies on the identification of candidate markers reported in literature and the actual possibility of exploiting the identified biomarkers at a clinical level. This is due to several aspects, among which the most important are the poor sensitivity/specificity sometimes characterizing the identified pools of markers and the complicated biostatistic design of prospective studies for their validation prior to clinical use.
It is the authors’ opinion that the future perspective in exploratory identification of biomarkers has to be found in the exhaustive search for potential markers. It is impossible to imagine that complex pathology acting on a wide range of individuals characterized by a large biological variability could be reflected in a very restricted panel of markers. We think that the future will rely on high-throughput techniques and the possibility of combining the results emerging from proteomic, genomic, metabolomic studies coupled with clinical information to identify exhaustive panels of markers, thus improving the predictive performance of the panels themselves and providing better sensitivity and specificity. Certainly, great attention has to be paid in such studies to the proper evaluation of experimental and biological variability (i.e., a careful selection of experimental design) to provide sound and robust results and to the evaluation of the results through effective multivariate techniques.
P- Reviewer: Bazhin AV, Duell EJ, Ren CL, Tsuchikawa T S- Editor: Qi Y L- Editor: Roemmele A E- Editor: Wang CH
| 1. | Chen DW, Fan YF, Li J, Jiang XX. MTA2 expression is a novel prognostic marker for pancreatic ductal adenocarcinoma. Tumour Biol. 2013;34:1553-1557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 2. | Bünger S, Laubert T, Roblick UJ, Habermann JK. Serum biomarkers for improved diagnostic of pancreatic cancer: a current overview. J Cancer Res Clin Oncol. 2011;137:375-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8406] [Cited by in F6Publishing: 8902] [Article Influence: 741.8] [Reference Citation Analysis (0)] |
| 4. | Jamieson NB, Carter CR, McKay CJ, Oien KA. Tissue biomarkers for prognosis in pancreatic ductal adenocarcinoma: a systematic review and meta-analysis. Clin Cancer Res. 2011;17:3316-3331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 5. | Negri AS, Robotti E, Prinsi B, Espen L, Marengo E. Proteins involved in biotic and abiotic stress responses as the most significant biomarkers in the ripening of Pinot Noir skins. Funct Integr Genomics. 2011;11:341-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Marengo E, Robotti E, Bobba M, Milli A, Campostrini N, Righetti SC, Cecconi D, Righetti PG. Application of partial least squares discriminant analysis and variable selection procedures: a 2D-PAGE proteomic study. Anal Bioanal Chem. 2008;390:1327-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Marengo E, Robotti E, Bobba M, Gosetti F. The principle of exhaustiveness versus the principle of parsimony: a new approach for the identification of biomarkers from proteomic spot volume datasets based on principal component analysis. Anal Bioanal Chem. 2010;397:25-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Massart DL, Vandeginste BGM, Buydens LMC, De Yong S, Lewi PJ, Smeyers-Verbeke J. Handbook of Chemometrics and Qualimetrics: part A. Amsterdam: Elsevier 1997; . [Cited in This Article: ] |
| 9. | Dunn OJ. Multiple Comparisons Among Means. J Am Stat Assoc. 1961;56:52-64. [Cited in This Article: ] |
| 10. | Dunnett CW. A multiple comparisons procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096-1121. [DOI] [Cited in This Article: ] [Cited by in Crossref: 3083] [Cited by in F6Publishing: 2121] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 11. | Šidák Z. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62:626-633. [Cited in This Article: ] |
| 12. | Kass RE, Raftery AE. Bayes Factors. J Am Stat Assoc. 1995;430:773-795. [Cited in This Article: ] |
| 13. | Box GEP, Hunter WG, Hunter JS. Statistics for experimenters. New York: Wiley 1978; . [Cited in This Article: ] |
| 14. | Massart DL, Vandeginste BGM, Deming SM, Michotte Y, Kaufman L. Chemometrics: A textbook. Amsterdam: Elsevier 1988; . [Cited in This Article: ] |
| 15. | Vandeginste BGM, Massart DL, Buydens LMC, De Yong S, Lewi PJ, Smeyers-Verbeke J. Handbook of Chemometrics and Qualimetrics: Part B. Amsterdam: Elsevier 1998; . [Cited in This Article: ] |
| 16. | Marengo E, Bobba M, Liparota MC, Robotti E, Righetti PG. Use of Legendre moments for the fast comparison of two-dimensional polyacrylamide gel electrophoresis maps images. J Chromatogr A. 2005;1096:86-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Kruskal JB, Wish M. Multidimensional Scaling. Sage University Paper series on Quantitative Application in the Social Sciences. Beverly Hills and London: Sage Publications 1978; . [Cited in This Article: ] |
| 18. | Marengo E, Robotti E, Gianotti V, Righetti PG, Cecconi D, Domenici E. A new integrated statistical approach to the diagnostic use of two-dimensional maps. Electrophoresis. 2003;24:225-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Young G, Householder AS, Discussion of a Set of Points in Terms of Their Mutual Distances. Psychometrica. 1930;3:19-22. [Cited in This Article: ] |
| 20. | Gower JC. Some Distance Properties of Latent Roots and Vector Methods Used in Multivariate Analysis. Biometrika. 1966;53:325-338. [Cited in This Article: ] |
| 21. | Shepard RN. Analysis of proximities: Multidimensional scaling with an unknown distance function I. Psycometrika. 1962;27:125-140. [Cited in This Article: ] |
| 22. | Shepard RN. Analysis of proximities: Multidimensional scaling with an unknown distance function. II. Psycometrika. 1962;27:219-246. [Cited in This Article: ] |
| 23. | Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psycometrika. 1964;29:1-27. [Cited in This Article: ] |
| 24. | Frank IE, Lanteri S. Classification models: Discriminant analysis, SIMCA, CART. Chemometr Intell Lab. 1989;5:247. [DOI] [Cited in This Article: ] |
| 25. | Marengo E, Robotti E, Bobba M, Righetti PG. Evaluation of the variables characterized by significant discriminating power in the application of SIMCA classification method to proteomic studies. J Proteome Res. 2008;7:2789-2796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Marengo E, Robotti E, Bobba M, Liparota MC, Rustichelli C, Zamò A, Chilosi M, Righetti PG. Multivariate statistical tools applied to the characterization of the proteomic profiles of two human lymphoma cell lines by two-dimensional gel electrophoresis. Electrophoresis. 2006;27:484-494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Marengo E, Robotti E, Righetti PG, Campostrini N, Pascali J, Ponzoni M, Hamdan M, Astner H. Study of proteomic changes associated with healthy and tumoral murine samples in neuroblastoma by principal component analysis and classification methods. Clin Chim Acta. 2004;345:55-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Robotti E, Demartini M, Gosetti F, Calabrese G, Marengo E. Development of a classification and ranking method for the identification of possible biomarkers in two-dimensional gel-electrophoresis based on principal component analysis and variable selection procedures. Mol Biosyst. 2011;7:677-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Polati R, Menini M, Robotti E, Millioni R, Marengo E, Novelli E, Balzan S, Cecconi D. Proteomic changes involved in tenderization of bovine Longissimus dorsi muscle during prolonged ageing. Food Chem. 2012;135:2052-2069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 30. | Martens H, Naes T. Multivariate calibration. Wiley: London 1989; . [Cited in This Article: ] |
| 31. | Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometrics. 2002;16:119-128. [DOI] [Cited in This Article: ] |
| 32. | Breiman L. Random Forests. Machine Learning. 2001;45:5-32. [DOI] [Cited in This Article: ] [Cited by in Crossref: 56052] [Cited by in F6Publishing: 56861] [Article Influence: 5169.2] [Reference Citation Analysis (0)] |
| 33. | Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Amer Statist Assn. 1958;53:457-481. [DOI] [Cited in This Article: ] [Cited by in Crossref: 32610] [Cited by in F6Publishing: 24221] [Article Influence: 367.0] [Reference Citation Analysis (0)] |
| 34. | Berty HP, Shi H, Lyons-Weiler J. Determining the statistical significance of survivorship prediction models. J Eval Clin Pract. 2010;16:155-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Wilcoxin F. Probability tables for individual comparisons by ranking methods. Biometrics. 1947;3:119-122. [PubMed] [Cited in This Article: ] |
| 36. | Cox DR. Regression Models and Life-Tables. J Royal Stat Soc. 1972;34:187-220. [Cited in This Article: ] |
| 37. | Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567-6572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2046] [Cited by in F6Publishing: 1755] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 38. | Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 499] [Cited by in F6Publishing: 414] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 39. | Metropolis N, Ulam S. The Monte Carlo method. J Am Stat Assoc. 1949;44:335-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 679] [Cited by in F6Publishing: 712] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 40. | Hastings WK. Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika. 1970;57: 97-109. [Cited in This Article: ] |
| 41. | Boeck S, Haas M, Laubender RP, Kullmann F, Klose C, Bruns CJ, Wilkowski R, Stieber P, Holdenrieder S, Buchner H. Application of a time-varying covariate model to the analysis of CA 19-9 as serum biomarker in patients with advanced pancreatic cancer. Clin Cancer Res. 2010;16:986-994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Haas M, Heinemann V, Kullmann F, Laubender RP, Klose C, Bruns CJ, Holdenrieder S, Modest DP, Schulz C, Boeck S. Prognostic value of CA 19-9, CEA, CRP, LDH and bilirubin levels in locally advanced and metastatic pancreatic cancer: results from a multicenter, pooled analysis of patients receiving palliative chemotherapy. J Cancer Res Clin Oncol. 2013;139:681-689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 43. | Boeck S, Wittwer C, Heinemann V, Haas M, Kern C, Stieber P, Nagel D, Holdenrieder S. Cytokeratin 19-fragments (CYFRA 21-1) as a novel serum biomarker for response and survival in patients with advanced pancreatic cancer. Br J Cancer. 2013;108:1684-1694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | McCaffery I, Tudor Y, Deng H, Tang R, Suzuki S, Badola S, Kindler HL, Fuchs CS, Loh E, Patterson SD. Putative predictive biomarkers of survival in patients with metastatic pancreatic adenocarcinoma treated with gemcitabine and ganitumab, an IGF1R inhibitor. Clin Cancer Res. 2013;19:4282-4289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Fong D, Moser P, Krammel C, Gostner JM, Margreiter R, Mitterer M, Gastl G, Spizzo G. High expression of TROP2 correlates with poor prognosis in pancreatic cancer. Br J Cancer. 2008;99:1290-1295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 157] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 46. | Fong D, Spizzo G, Mitterer M, Seeber A, Steurer M, Gastl G, Brosch I, Moser P. Low expression of junctional adhesion molecule A is associated with metastasis and poor survival in pancreatic cancer. Ann Surg Oncol. 2012;19:4330-4336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Zong M, Meng M, Li L. Low expression of TBX4 predicts poor prognosis in patients with stage II pancreatic ductal adenocarcinoma. Int J Mol Sci. 2011;12:4953-4963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 48. | Schäfer C, Seeliger H, Bader DC, Assmann G, Buchner D, Guo Y, Ziesch A, Palagyi A, Ochs S, Laubender RP. Heat shock protein 27 as a prognostic and predictive biomarker in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2012;16:1776-1791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 49. | Maréchal R, Mackey JR, Lai R, Demetter P, Peeters M, Polus M, Cass CE, Salmon I, Devière J, Van Laethem JL. Deoxycitidine kinase is associated with prolonged survival after adjuvant gemcitabine for resected pancreatic adenocarcinoma. Cancer. 2010;116:5200-5206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 50. | Mann CD, Bastianpillai C, Neal CP, Masood MM, Jones DJ, Teichert F, Singh R, Karpova E, Berry DP, Manson MM. Notch3 and HEY-1 as prognostic biomarkers in pancreatic adenocarcinoma. PLoS One. 2012;7:e51119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 51. | Chang ST, Zahn JM, Horecka J, Kunz PL, Ford JM, Fisher GA, Le QT, Chang DT, Ji H, Koong AC. Identification of a biomarker panel using a multiplex proximity ligation assay improves accuracy of pancreatic cancer diagnosis. J Transl Med. 2009;7:105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 52. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 175] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 53. | Schultz NA, Christensen IJ, Werner J, Giese N, Jensen BV, Larsen O, Bjerregaard JK, Pfeiffer P, Calatayud D, Nielsen SE. Diagnostic and Prognostic Impact of Circulating YKL-40, IL-6, and CA 19.9 in Patients with Pancreatic Cancer. PLoS One. 2013;8:e67059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | McKinney KQ, Lee YY, Choi HS, Groseclose G, Iannitti DA, Martinie JB, Russo MW, Lundgren DH, Han DK, Bonkovsky HL. Discovery of putative pancreatic cancer biomarkers using subcellular proteomics. J Proteomics. 2011;74:79-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 55. | Pavelka N, Fournier ML, Swanson SK, Pelizzola M, Ricciardi-Castagnoli P, Florens L, Washburn MP. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol Cell Proteomics. 2008;7:631-644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 56. | Pavelka N, Pelizzola M, Vizzardelli C, Capozzoli M, Splendiani A, Granucci F, Ricciardi-Castagnoli P. A power law global error model for the identification of differentially expressed genes in microarray data. BMC Bioinformatics. 2004;5:203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 57. | Kojima K, Asmellash S, Klug CA, Grizzle WE, Mobley JA, Christein JD. Applying proteomic-based biomarker tools for the accurate diagnosis of pancreatic cancer. J Gastrointest Surg. 2008;12:1683-1690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 58. | Mobley JA, Lam YW, Lau KM, Pais VM, L’Esperance JO, Steadman B, Fuster LM, Blute RD, Taplin ME, Ho SM. Monitoring the serological proteome: the latest modality in prostate cancer detection. J Urol. 2004;172:331-337. [PubMed] [Cited in This Article: ] |
| 59. | Ehmann M, Felix K, Hartmann D, Schnölzer M, Nees M, Vorderwülbecke S, Bogumil R, Büchler MW, Friess H. Identification of potential markers for the detection of pancreatic cancer through comparative serum protein expression profiling. Pancreas. 2007;34:205-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 60. | Hauskrecht M, Pelikan R, Malehorn DE, Bigbee WL, Lotze MT, Zeh HJ, Whitcomb DC, Lyons-Weiler J. Feature Selection for Classification of SELDI-TOF-MS Proteomic Profiles. Appl Bioinformatics. 2005;4:227-246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 61. | Patwa TH, Li C, Poisson LM, Kim HY, Pal M, Ghosh D, Simeone DM, Lubman DM. The identification of phosphoglycerate kinase-1 and histone H4 autoantibodies in pancreatic cancer patient serum using a natural protein microarray. Electrophoresis. 2009;30:2215-2226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Marengo E, Robotti E, Cecconi D, Hamdan M, Scarpa A, Righetti PG. Identification of the regulatory proteins in human pancreatic cancers treated with Trichostatin A by 2D-PAGE maps and multivariate statistical analysis. Anal Bioanal Chem. 2004;379:992-1003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 63. | Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of a rat pancreatic stellate cell line using liquid chromatography tandem mass spectrometry (LC-MS/MS). J Proteomics. 2011;75:708-717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 64. | Choi H, Nesvizhskii AI. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J Proteome Res. 2008;7:47-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 161] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 65. | Goodman SN. Toward evidence-based medical statistics. 1: The P value fallacy. Ann Intern Med. 1999;130:995-1004. [PubMed] [Cited in This Article: ] |
| 66. | Goodman SN. Toward evidence-based medical statistics. 2: The Bayes factor. Ann Intern Med. 1999;130:1005-1013. [PubMed] [Cited in This Article: ] |
| 67. | Paulo JA, Lee LS, Wu B, Repas K, Mortele KJ, Banks PA, Steen H, Conwell DL. Identification of pancreas-specific proteins in endoscopically (endoscopic pancreatic function test) collected pancreatic fluid with liquid chromatography--tandem mass spectrometry. Pancreas. 2010;39:889-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Ogura T, Yamao K, Hara K, Mizuno N, Hijioka S, Imaoka H, Sawaki A, Niwa Y, Tajika M, Kondo S. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol. 2013;48:640-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 69. | Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 70. | Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 216] [Cited by in F6Publishing: 210] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 71. | Winter JM, Tang LH, Klimstra DS, Brennan MF, Brody JR, Rocha FG, Jia X, Qin LX, D’Angelica MI, DeMatteo RP. A novel survival-based tissue microarray of pancreatic cancer validates MUC1 and mesothelin as biomarkers. PLoS One. 2012;7:e40157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 72. | Pedersen KS, Bamlet WR, Oberg AL, de Andrade M, Matsumoto ME, Tang H, Thibodeau SN, Petersen GM, Wang L. Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls. PLoS One. 2011;6:e18223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 73. | Morse DL, Balagurunathan Y, Hostetter G, Trissal M, Tafreshi NK, Burke N, Lloyd M, Enkemann S, Coppola D, Hruby VJ. Identification of novel pancreatic adenocarcinoma cell-surface targets by gene expression profiling and tissue microarray. Biochem Pharmacol. 2010;80:748-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 74. | Huang H, Dong X, Kang MX, Xu B, Chen Y, Zhang B, Chen J, Xie QP, Wu YL. Novel blood biomarkers of pancreatic cancer-associated diabetes mellitus identified by peripheral blood-based gene expression profiles. Am J Gastroenterol. 2010;105:1661-1669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 75. | Kaur P, Sheikh K, Kirilyuk A, Kirilyuk K, Singh R, Ressom HW, Cheema AK. Metabolomic profiling for biomarker discovery in pancreatic cancer. Int J Mass Spectrom. 2012;310:44-51. [DOI] [Cited in This Article: ] |
| 76. | Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 511] [Cited by in F6Publishing: 429] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 77. | Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603-D610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1397] [Cited by in F6Publishing: 1387] [Article Influence: 86.7] [Reference Citation Analysis (0)] |