Published online Feb 28, 2014. doi: 10.3748/wjg.v20.i8.1898
Revised: December 3, 2013
Accepted: January 3, 2014
Published online: February 28, 2014
Colorectal cancer (CRC) remains a highly fatal condition in part due to its resilience to treatment and its propensity to spread beyond the site of primary occurrence. One possible avenue for cancer to escape eradication is via stem-like cancer cells that, through phenotypic heterogeneity, are more resilient than other tumor constituents and are key contributors to cancer growth and metastasis. These proliferative tumor cells are theorized to possess many properties akin to normal intestinal stem cells. Not only do these CRC “stem” cells demonstrate similar restorative ability, they also share many cell pathways and surface markers in common, as well as respond to the same key niche stimuli. With the improvement of techniques for epithelial stem cell identification, our understanding of CRC behavior is also evolving. Emerging evidence about cellular plasticity and epithelial mesenchymal transition are shedding light onto metastatic CRC processes and are also challenging fundamental concepts about unidirectional epithelial proliferation. This review aims to reappraise evidence supporting the existence and behavior of CRC stem cells, their relationship to normal stem cells, and their possible dependence on the stem cell niche.
Core tip: Colorectal (CRC) cancer stem cells are a theorized but poorly characterized cell population believed to be crucial for tumor growth, spread, and tenacity. CRC stem cells share many similar characteristics of normal intestinal stem cells and are hypothesized to originate directly from them. It appears, however, that both the regulation of normal intestinal stem cells and the development of CRC are far more complex than previously imagined. Likely pivotal to the success of both are plasticity pathways able to reverse cellular fate, and stem cell niche signals, ultimately leading to self-replenishment and sometimes also unwanted dissemination.
- Citation: Ong BA, Vega KJ, Houchen CW. Intestinal stem cells and the colorectal cancer microenvironment. World J Gastroenterol 2014; 20(8): 1898-1909
- URL: https://www.wjgnet.com/1007-9327/full/v20/i8/1898.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i8.1898
Colorectal cancer (CRC) remains a highly morbid and fatal disease among both developed nations and globally[1-3]. Based on 2008 world data, CRC is the fourth leading cause of cancer-related mortality behind lung, stomach, and liver cancer, respectively[4,5]. Since Fearon et al[6] introduced a model for colorectal tumorigenesis in 1990, the study of the molecular basis of CRC has been rapidly evolving. While a handful of tumor suppressors and oncogenes (e.g., APC, KRAS, and P53) are commonly found among CRCs, a vast number of low-frequency somatic mutations have since been discovered that are believed to contribute to CRC heterogeneity[7,8]. Given the expanded number of potentially functional mutations, that no CRC therapy is completely curative should come as no surprise[9].
More importantly, individual colorectal cancers can themselves demonstrate phenotypic variability via sub-delegation of constituent cells. Core to this notion are cancer “stem” cells which act as ringleaders that drive CRC proliferation and metastasis[10]. Like normal stem cells, they self-perpetuate and expand in accordance with stem cell hierarchy[10]. Much remains unknown about the origins and regulation of CRC stem cells, though implicated in CRC inception are the signals expressed within the normal intestinal stem cell niche. New light has also been shed onto plasticity pathways that may perhaps be pivotal to CRC metastasis and treatment. The aim of this review is to reappraise current evidence supporting the existence and behavior of CRC stem cells, their relationship to normal stem cells, and their possible dependence on the stem cell microenvironment.
Fearon and Vogelstein’s model for colorectal carcinogenesis illustrates how genetic alterations may allow colorectal cells to escape defined behaviors of the normal intestinal epithelium. By the early 1990s, Fearon et al[11] established three key features about colorectal cancer. First, cells within a colorectal cancer are monoclonal in nature, suggesting that CRC arises from clonal expansion of a small number of cells. Second, Fearon et al[6] surmised that key genetic alterations found commonly among CRC (e.g., RAS, P53, APC) confer functional traits advantageous to the development and expansion of sporadic cancer and are acquired in a sequentially preferred order. For instance, APC mutations often occurred early prior to adenoma formation, whereas P53 mutations frequented tumor phases during the transition of adenomas to overt carcinomas[6]. Finally, based on their own observations and those of others, Fearon et al[6] concluded that the number of accumulated mutations in a tumor was the most consistent feature associated with the clinical and histopathological manifestation of CRC[12].
Fearon and Vogelstein’s original CRC model has since been greatly expounded upon. Numerous low-frequency candidate mutations have been identified among candidate CRC genes, likely contributing to CRC phenotypic heterogeneity[7,8]. Also, carcinogenesis might not rely strictly on Fearon and Vogelstein’s hypothesized mutational gateways. For example, one study found no genetic change between genome-sequenced primary colorectal cancers and their respective metastases, suggesting that insufficient time passed to allow either primary or metastatic lesions to acquire distinguishing mutations[7].
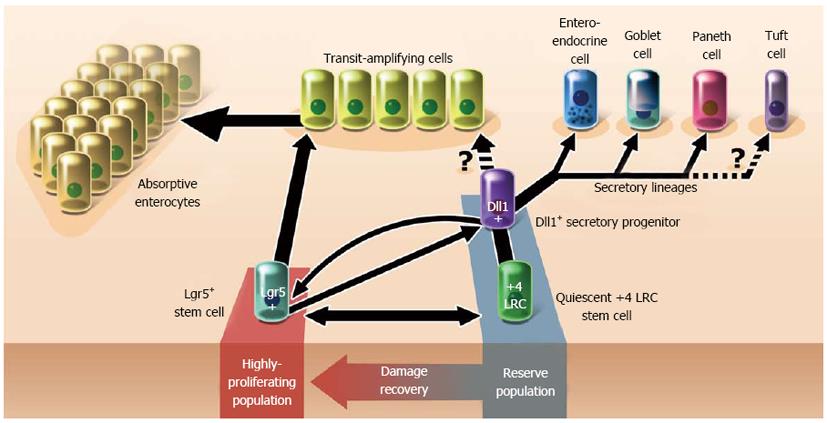
Two functionally distinct populations of putative normal epithelial stem cells have been identified in intestinal crypts of humans and mice: Lgr5+ crypt base columnar stem cells and quiescent label-retaining cells[13-17]. These two cell types replenish and maintain the intestinal epithelium[13].
Lgr5+ crypt base columnar cells (CBCs) are multipotent stem cells located in crypts of the small intestine and colon[14]. Lgr5 is an orphan G protein-coupled receptor expressed during embryogenesis and among epithelial stem cell populations in the adult intestine, hair follicles, stomach, mammary glands, and taste buds[18]. CBCs were first characterized in 1974 when an electron microscopy study identified a population of crypt cells that shared common secretory components with all differentiated epithelial cell lineages in the mouse intestine[19]. More recently, Barker et al[14] demonstrated that Lgr5-mediated activation of a permanent cell-labeling gene identified a line of cells originating from the intestinal crypt that yielded three differentiated cell types. The authors surmised that enteroendocrine cells were too rare to be detected among labeled cells[14]. A subsequent in vitro study demonstrated that organoids derived from single Lgr5+ cells form crypt domains containing all lineages of the adult intestinal epithelium including enteroendocrine and crypt paneth cells[20]. Taken together, these findings strongly suggest that multipotent Lgr5+ CBCs are true intestinal epithelial stem cells.
Quite contrary to expected stem cell behavior, evidence suggests that the expansion of Lgr5+ CBCs follows stochastic principles in which cells are equipotent and segregate chromosomes randomly[18,21,22]. Lgr5+ cells are also mitotically-active and demonstrate little asymmetric division[13,21]. Proliferation of these stem cells can at times approximate a square root growth curve, suggesting that they contain potential for rapid, yet very random clonal expansion[13,21,23]. As a likely consequence of their stochastic properties, Lgr5+ stem cells are subject to neutral drift, often resulting in monoclonal or oligoclonal populations in the intestinal crypt[21].
It seems dangerous for a stem cell to propagate in a manner dictated largely by chance. Random chromosomal segregation risks the introduction of genomic errors that can subsequently be passed to both daughters and self-perpetuating clones. Lgr5+ cells also seem to have little control over cell fate, suggesting that they are likely critically regulated by the surrounding milieu.
Quiescent DNA label-retaining intestinal stem cells (LRCs) have remained controversial since the 1970s when these mitotically-inactive cells were found at and around the +4 crypt position[24-26]. Although intestinal LRCs express a number of stem cell markers including Hopx, Tert, Lrig1, and Dclk1, they are widely identified by their expression of Bmi1, a member of chromatin-silencing polycomb-repressing complex 1[13,15,27]. Like Lgr5+ CBCs, Bmi1+ LRCs can form spheroids in vitro containing all differentiated epithelial cell types[13,20]. The multipotency of Bmi1+ LRCs has also been confirmed in vivo through lineage experiments[15]. In contrast to early reports of the radiation sensitivity of +4 position crypt cells, recent evidence suggests that quiescent stem cells are both resistant to and activated by moderate levels of radiation damage, thus suggesting a crucial role in recovery following intestinal injury[13,28]. Notably, Bmi1+ LRCs can single-handedly restore radiation-ablated mouse intestinal epithelium in the total absence of Lgr5+ stem cells[13].
Whether +4 quiescent LRCs are actually stem cells remains a matter of debate. Quiescent stem cells have only been found in the proximal small intestine and to date no presence has yet been found of a corresponding population in the colon[15,29]. Moreover, one study has identified quiescent LRCs not as stem cells, but rather as partially-differentiated secretory precursors[30]. Quiescent stem cell markers (including Bmi1, Tert, Hopx, and Lrig1) have also been found among Lgr5+ stem cells thereby questioning the validity of using such markers to identify a uniquely separate stem cell population[31].
In contrast to current single-lineage stem cell theories, the coexistence of two putative intestinal stem cell types may suggest a more complex pathway for the development of the intestinal epithelium (Figure 1)[10,32]. On one hand, evidence exists supporting the subordinancy of LRCs to LGR5+ cells: LRCs have been characterized as secretory precursors and may not share markers unique from Lgr5+ cells[30,31,33]. On the other hand, evidence also exists conversely that Lgr5+ cells may be subordinate to LRCs: Bmi1+ LRCs restore radiation-ablated Lgr5+ cell populations[13,29]. These findings when taken together suggest that LRCs likely interconvert with Lgr5+ CBCs, regardless of whether LRCs are actually stem cells. Such findings suggest that intestinal epithelial development is neither as hierarchical nor as unidirectional as once thought, though the extent of which is not yet known.
Based on the discussion thus far, perhaps the actions of the stem cell pool as we currently understand it are comprised of the combined properties of Lgr5+ and quiescent stem cells in the crypt (Figure 1). Under normal conditions, Lgr5+ stem cells could function to self-sufficiently maintain epithelial homeostasis through high-output cell production in response to trophic niche signals (e.g., Wnt)[34,35]. However, Lgr5+ CBCs are likely as sensitive to genetic damage as they are to injury. In these situations, the quiescent LRC population may assist with recovery from intestinal injury, either directly or by restoring Lgr5+ stem cells.
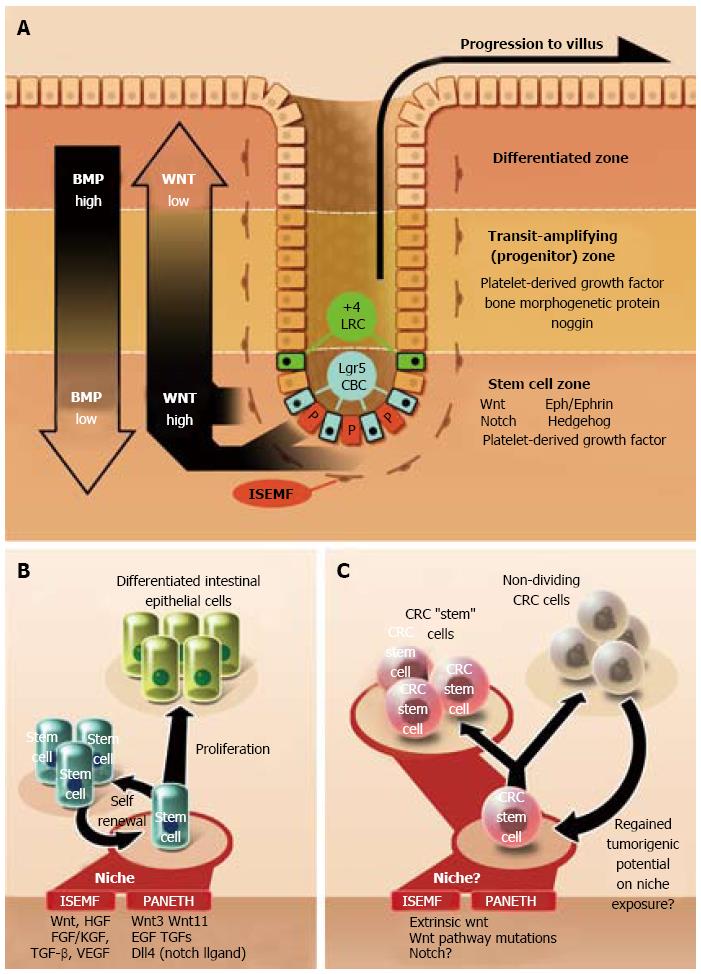
Like other tissues among higher organisms, all intestinal cells reside within a carefully defined construct of chemical signals that directs genetically identical cell populations towards divergent behaviors[36]. Contained in and around the intestinal crypt are a multitude of molecular and cellular effectors that define a unique microenvironment - a “niche”- that directs the optimal function of stem cells[10]. Components of the niche include the subepithelial stroma, adjacent epithelial cells, natural enteric flora, and soluble epithelium-derived factors. Alteration of niche effectors can also lead to aberrant and dysregulated crypt behavior, which in turn may foster neoplasia.
A multitude of signals in the intestinal crypt affect the function and growth of intestinal stem cells (Figure 2A and B)[37]. Of these, Wnt proteins are one of the most crucial for maintaining stem cell homeostasis[34,35,37,38]. Wnt promotes both cellular dedifferentiation and proliferation during embryogenesis and in many adult animal tissues[39-42]. Inhibition of the Wnt pathway results in crypt loss and a marked reduction in epithelial proliferation[43]. Among mice with inducible APC-knockouts, Wnt results in intestinal mucosa populated by undifferentiated cells[44]. Wnt activity is also among the essential signals for the formation of crypt structures from single stem cell cultures as well as for the reprogramming of somatic cells into induced pluripotent stem cells (iPSCs)[20,34,39,41]. Cell-proliferative genes are activated by Wnt via nuclear β-catenin intermediaries and include cell migration controllers (EPH), proliferative signals (c-myc, cyclin D1), and stem and cancer cell markers (Lgr5, Bmi1)[10,14,35,45-47].
The Wnt pathway is also a highly influential mediator of cancer (Figure 2C). APC mutations facilitate Wnt activity by dysregulating β-catenin-mediated gene expression[45,48]. APC mutations are common, occurring in over 80% of sporadic colorectal cancer[48]. Vermeulen et al[49] showed that primary spheroidal cultures derived from human CRCs are regulated by Wnt signals in the surrounding microenvironment, such as those secreted by intestinal myofibroblasts. They also demonstrated that extrinsic Wnt pathway activation was an important determinant in the cellular acquisition of cancer stem cell features (e.g., formation of tumors when injected into immune-deficient mice and in vitro recapitulation of xenograft isolate behavior to that of the original tumor)[49].
Intestinal subepithelial myofibroblasts (ISEMFs), located underneath the basement membrane in the crypt, are stromal cells widely known to promote stem cell self-renewal and differentiation (Figure 2A and B)[20,34,35]. ISEMFs originate from regional intestinal fibroblasts and possibly trans-differentiated bone marrow cells[50]. Intestinal myofibroblasts function as anchors for cell adhesion and provide trophic signals to stem cells via cell-cell interactions and secreted mediators[51]. ISEMFs also contribute to wound healing, mucosal protection, fluid and electrolyte transport, and growth of the basement membrane[50,52]. Secreted myofibroblast mediators are numerous: Wnt proteins, hepatocyte growth factor, fibroblast growth factor, TGF-β, keratinocyte growth factor, matrix metalloproteinases, stem cell factor, VEGF, and numerous interleukins, to name a few[52,53].
ISEMFs have long been implicated in promoting colorectal cancer growth and invasion (Figure 2C)[51]. Little clarity exists regarding whether peri-CRC myofibroblasts are derived from normal ISEMFs. Based on knowledge gleaned from other cancer systems, functional differences between normal and CRC fibroblasts do likely exist[54]. Still, even normal myofibroblasts are capable of facilitating CRC growth. Vermeulen et al[49] found that normal colonic myofibroblasts prevented both the morphological and molecular differentiation of co-cultured colorectal cancer cells. Furthermore, these myofibroblasts were shown to re-induce tumorigenic potential in subpopulations of CRC cells with low degree of proliferative activity[49].
Paneth cells are terminally-differentiated secretory cells intermingled between Lgr5+ CBCs at the base of crypts in the small intestinal mucosa[55]. Though unclear why no Paneth cells have been found elsewhere in the intestine, a population of c-kit+/CD117+ goblet cells in the colon may perhaps function analogously[33,56]. Co-culture of c-kit+ cells with Lgr5+ stem cells promotes the growth of organoids in similar fashion to those produced from Paneth/Lgr5+ cell co-cultures[55,56].
Paneth cells contribute to the preservation of the stem cell compartment through the expression of Wnt proteins and other secreted signals such as epidermal growth factor and Notch ligands, all important in the maintenance of the Lgr5+ CBC population[55]. Paneth cells also secrete antimicrobial peptides[57]. Furthermore, they facilitate epithelial repair by deactivating paneth-specific genes and converting to a phase that promotes Bmi1+ cell proliferation[58].
Paneth cells seemingly serve a redundant role in the intestinal crypt. Wnt proteins released from Paneth cells are also derived from other sources in and around the intestinal crypt[59]. Notably, the complete removal of paneth cells in mouse model systems has not been shown to affect the proliferation of Lgr5+ CBCs[60].
Is there a population of cells in the intestinal epithelium that reliably serves as the source for most, if not all of colorectal cancers? Intestinal stem cells are prime suspects due to their pre-existing proliferative and self-restorative behavior, making them perhaps more sensitive to overt carcinogenesis[10,35]. In support of this notion, Barker et al[61] demonstrated that APC deletions only among Lgr5+ stem cells (6.5% of tumor mass) promoted the formation of adenomas, even in the setting of uniform tumor Wnt target gene activation. Barker and colleagues concluded that Lgr5+ stem cell transformation-especially via loss of APC function-is a highly efficient pathway to neoplasia[61]. Multi-color reporter lineage retracing experiments by Schepers et al[62] have also confirmed that early adenomas are mostly of monoclonal origin, though occasionally oligoclonal. Schepers et al[62] also identified stem-like Lgr5+ tumor origin cells at the base of adenomas that shared organizational resemblances to normal stem cells and were 20-fold more efficient at forming cell colonies in vitro than Lgr5-poor cells derived from the same population.
Still, evidence suggests that colorectal cancer may also arise from non-stem cells, supporting the idea that ultimately any cell harbors the potential to foster neoplasia. Early observations by Cole et al[63] reveal that early adenomatous polyps are positioned at the top of colonic crypts without contact with the stem cell compartment. Schwitalla et al[64] have also demonstrated that Wnt-constitutive intestinal cells can re-acquire stem cell properties in an NF-KB dependent manner and lead to tumor formation. These findings are congruent with iPSC research through which differentiated somatic cells have been reprogrammed back to proliferative stem-like states on account of key genetic alterations[41]. As with other non-intestinal cancers, no clear distinction yet exists identifying which CRCs, if any, are derived from non-stem cells[65].
What are the triggers that stimulate a cell to progress to cancer? Based on the discussion thus far, the neoplastic potential of a cell might be directly correlated with the combined disruptive impact of affected genes. However, One might imagine a situation in which a cell lacking sufficient functional derangement can be driven to cancer in response to external stimuli. Signals may come from cell placement in a Wnt-rich intestinal crypt, or in response to inflammation in light of concurrent genetic Wnt derangements as Schwitalla et al[64] have explored.
Not surprisingly, many normal stem markers such as Lgr5, DCLK1, CD133, CD44, CD24, and ALDH1 have also been found among highly proliferating fractions of colorectal cancers[10,52,66,67]. Given the apparent genetic heterogeneity among CRC[7,8], very few, if any, markers are both specific to CRC stem cells and ubiquitous among all CRCs[9]. Table 1 lists putative CRC stem cell markers as previously covered by other authors[10,68-71]. What remains unclear is whether such markers reflect carry-over from intestinal stem cell precursors as with other cancers (e.g., leukemia)[35] or else a re-activation of stem cell pathways. Regardless of the underlying reason, that CRC and normal intestinal epithelial stem cells express many of the same cell surface markers poses a challenge to the isolation of tumor stem cells.
| Marker | Function |
| ALDH1A1 | Enzyme |
| ALDH1B1 | Enzyme |
| β-catenin | Protein (nuclear) |
| Bmi-1 | Protein (nuclear) |
| CD24 | Cell surface glycoprotein |
| CD26 | Cell surface glycoprotein |
| CD29 | Cell surface glycoprotein |
| CD44 | Cell surface glycoprotein |
| CD133 | Cell surface glycoprotein |
| CD166 (ALCAM) | Cell surface glycoprotein |
| CDX-2 | Transcription factor |
| c-myc | Transcription factor |
| Dclk-1 | Serine-threonine kinase (?) |
| EpCAM | Cell surface glycoprotein |
| Klf-4 | Transcription factor |
| Lgr-5 | Cell surface receptor |
| Lin-28 | Transcription factor |
| Msi-1 | Protein (nuclear) |
| Nanog | Transcription factor |
| 4-Oct | Transcription factor |
| Sox-2 | Transcription factor |
One putative stem cell marker, Doublecortin-like kinase 1 (Dclk1), may be a useful marker for both normal and neoplastic intestinal stem cells. Dclk1 is a complex multi-splicoform transmembrane serine-threonine kinase involved in embryonic neuronal migration through intracellular signaling pathways[72,73]. In the digestive tract, Dclk1+ cells have been found in the stomach and at the +4 position of the intestinal crypt[74,75]. Intestinal Dclk1+ cells are functionally akin to quiescent stem cells via their label retention and radiation-induced activity[74,76]. Some studies contend that Dclk1+ cells are not intestinal stem cells at all. Dclk1 expression may be shared not only by stem cells but also among the enteroendocrine lineage[77]. Alternatively, Gerbe et al[78] propose that Dclk1+ cells are actually novel differentiated tuft cells with unidentified function.
Interestingly, cells aberrantly expressing Dclk1 have been found among both mouse intestinal adenomas and human colorectal cancers, suggesting a potential role for Dclk1 to identify neoplastic stem-like intestinal cells[74,79]. Nakanishi et al[80] recently demonstrated that Dclk1 specifically identifies abnormal intestinal mucosa found among tumors in the small intestine of APCmin/+ mice. Not only did Dclk1+ tumor cells co-express Lgr5, they also demonstrated higher expression of other cancer stem cell markers versus non-tumor cells[80]. Furthermore, ablation of Dclk1+ cells led to regression of the containing polyps without apparent effect to normal intestine[80]. These results concur with findings from our group showing that siRNA-based Dclk1 interference leads to growth arrest of xenoplanted CRC[81,82]. Also notable is a recent study by Li et al[67] demonstrating increased Dclk1+ expression among cell fractions with a higher percentage stem-like HCT116 human CRC cells. Taken together, these findings support the notion that Dclk1+ cells can identify colorectal cancer stem cells and that Dclk1 is critical for tumor growth.
Despite the strong evidence suggesting that only a small fraction of colorectal tumor cells is responsible for maintaining tumor growth, the isolation of “pure” colorectal cancer stem cells has remained an ongoing challenge due to numerous theoretical and practical reasons. In fact, the term “cancer stem cell” may be somewhat of a misnomer. There is no expectation that a dysregulated colorectal cancer cell follows the exact biochemical principles of a normal intestinal epithelial stem cell, even if they share common signaling pathways. So long as the phrase “cancer stem cell” is used loosely to refer to cells in control of the proliferative hierarchy demonstrated by CRC, there is no perceived problem.
The first studies documenting a tumor-initiating CRC subfraction came in 2007 with the identification of CD133+ cells comprising 2.5% of tumor mass[83,84]. However, the significance of CD133 as a specific CRC marker has subsequently been debated[52]. Other markers have further assisted in the enrichment of CRC stem cell fractions (Table 1). Kemper et al[66] found that Lgr5+ cells comprised only 1.9%-11.1% of putative stem cells already marked by Epcam, although admittedly the Lgr5+ fraction was more highly clonogenic. Isolation of DCLK1 among tumor stem cells has been previously discussed, but even Nakanishi et al[80] did not find DCLK1 universally among all tumors in their mouse experiments.
The current methods employed to identify CRC stem cells are derived from non-exclusive properties shared by all intestinal stem cells. These methods include: DNA label retention, in vitro and in vivo proliferation assessments, and detection of cell surface markers[10]. Consequently, the isolation of CRC stem cells is fraught with as much, controversy as normal intestinal stem cells. Not the least of which, subtle differences between humans and animal models may consequently make experimental findings difficult to generalize. The apparent genetic heterogeneity of CRC lends further worry that finding a universal identification standard for CRC stem cells may long remain a daunting task[7,8].
It is becoming increasingly apparent that both the normal intestine and colorectal cancer are subject to “plasticity” processes that convert cells back to less-differentiated forms. Conventional stem cell theory holds that cellular development follows a unidirectional and irreversible hierarchy through semi-differentiated intermediates and concludes with terminal differentiation[85]. The implied goal of such a model is to produce cells capable of specialized organ functions[86]. In the intestine, recent evidence has revealed that short-lived Dll1+ secretory progenitors can readily revert to Lgr5+ stem cells following radiation injury (Figure 1)[87,88]. The apparent conversion of quiescent Bmi1+ LRCs to Lgr5+ stem cells is another clear demonstration of cellular plasticity[30,31]. That differentiated somatic cells, too, can fate-reprogram into iPSCs carries profound implications regarding the exclusivity of stem cell traits and the potential for any cell in an organism to participate in tissue regeneration[41].
Cellular plasticity processes may also depend largely on the cellular microenvironment. For example, extrinsically-derived Wnt signals can sufficiently replace Myc gene mutations during iPSC creation[39]. Also, non-proliferating CRC cells possessing low Wnt activity have been shown to regain proliferative tumorigenic potential when co-cultured with colonic myofibroblasts or the conditioned medium derived from myofibroblast cultures[49]. These results indicate that extrinsic signals -notably activators of the Wnt pathway- are perhaps sufficient to induce behavioral reprogramming, especially in CRC[49].
That fate-reversal occurs in CRC suggests that CRC expansion adheres to a proliferative pattern somewhere in between the classical hierarchical and stochastic growth models[49,85]. Admittedly, however, it is not known to what degree cellular plasticity plays a role in the proliferation of colorectal cancer. Perhaps even among different CRCs there is variation in functional dependence on extrinsic signals, ultimately affecting the growth patterns and behavior of the neoplastic phenotype. In this way, perhaps an extreme disturbance of either genetic derangement or environmental signals alone would also be a sufficient trigger for carcinogenesis[36].
The presence of cancer cells in the lymphatic and systemic circulation have long been known to correlate with poor prognosis, even despite the resection of primary lesions and/or chemotherapy[89-95]. With the apparent monoclonality of colorectal cancer[11], one might infer that circulating cancer stem cells originate from a primary colorectal tumor. Because cell migration brings with it certain constraints on adhesion and cellular interactions, circulating cancer stem cells may be functionally divergent from primary tumor cells.
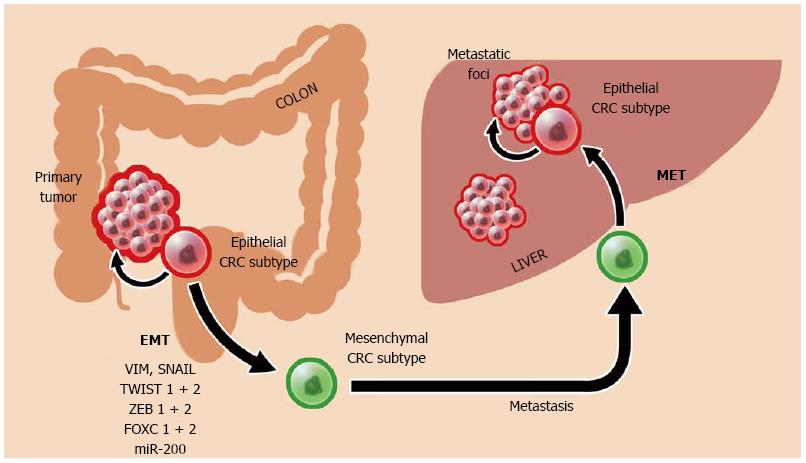
Epithelial-mesenchymal transition (EMT) is a critical extension of cellular plasticity that is believed to govern not only the development of normal tissues but also the growth and spread of colorectal cancer. EMT is defined as the process by which epithelial cells convert to a mesenchymal-like phenotype. Via EMT, a cell relinquishes its native cell-cell interactions, loses tissue-specific polarity, and acquires migratory mesenchymal traits[96]. Important aspects of the EMT process such as the loss of E-cadherin (a hallmark of EMT) is mediated by the Wnt pathway[97]. This process is reversible and plays a key role in normal embryonic development as well as normal wound healing and fibrosis in the adult animal. The opposing process of mesenchymal-epithelial transition (MET) likely occurs through inverse regulation of EMT and is critical for final organ formation once embryonic cells have sufficiently migrated via mesenchymal intermediates[96]. Boundaries demarcating the degree of lineage reprogramming during the EMT process remain vastly gray territory. In fact, cells undergoing EMT may not necessarily have re-written fates, for such changes might only involve alterations to cell mobility.
EMT is likely a dominant mechanism driving colorectal cancer metastasis (Figure 3). In fact, CRC cells that display EMT characteristics have been shown to also possess traits of stem cells[98,99]. Critical to both CRC stem cell formation and EMT induction are Wnt mediators (e.g., nuclear β-catenin), most markedly active at the invasive front of colorectal tumors[97]. Microarray analysis has demonstrated up-regulation of EMT-mediating genes among human CRC (e.g., VIM, TWIST 1 + 2, SNAIL, and FOXC 1 + 2)[100]. EMT is also controlled via the microRNA miR-200 family[100,101]. MicroRNAs are small, non-coding RNAs that regulate post-transcriptional gene expression and serve to activate oncogenes and silence tumor suppressors. The presence of miR-200 family members (notably miR-200c and miR-141) is associated with a gain of epithelial cell characteristics[101]. In contrast, down-regulation of miR-200 family members promotes an invasive mesenchymal phenotype, possibly through the activation of EMT mediators like ZEB1 and ZEB2[96,102,103]. In turn, epigenetic methylation pathways are in control of these miR-200 “switches” that altogether govern the shifting of CRC cells towards either mobile or stationary phases[96,101].
The combined effect of EMT/MET activity is metastatic advancement of a colorectal cancer: EMT enables primary tumor escape and spread by way of mesenchymal intermediates, and MET returns CRC to a highly-proliferative epithelial stem cell phenotype (Figure 3)[101]. In fact, these transitional phases may be the ultimate defining characteristic of CRC and may help direct future CRC therapy. Loboda et al[100] demonstrated that colorectal cancer, despite its vast mutational heterogeneity, can be organized principally as either epithelial or mesenchymal subtypes. Admittedly, the extent that EMT contributes to tumor spread remains unknown.
Interfering with EMT at critical phases of cancer growth is thus seemingly an attractive goal. For instance, anti-EMT therapy could be utilized to prevent primary tumor metastasis in early-stage CRC by forcing cells out of a mesenchymal phenotype or else preventing the entry into EMT (as is apparently the case with cetuximab administration)[96,104,105]. However, one concern regarding EMT/MET exploitation is that the two opposing processes may coexist inseparably. As such, unilaterally-directed therapy might lead to undesirable activity of cells in the opposite transitional phase. For instance, EMT processes are in part responsible for chronic resistance to oxaliplatin[106]. Difficulties in controlling mesenchymal processes may be further complicated by plasticity-mediated recruitment of additional CRC stem cells into the mesenchymal pool. Suffice it to say, our understanding of EMT is still in its infancy.
Much has been learned about the behavior of colorectal cancer stem cells owing to knowledge gained about normal intestinal stem cell behavior. The limitations inherent in our current isolation methods of pure stem cell fractions will likely bear heavily on how we observe and understand CRC as well. Newer developments in the field of stem cell research have provided insight into the vast potential for stem cells to not only be controlled by environmental factors but also be restored by its descendants. Also critical are core pathways such as Wnt that play an integral role in stem cell function, mesenchymal transition, and metastasis. Given the complexity of CRC “homeostasis”, optimal CRC therapy will likely still remain a multi-pronged attack: first by control and/or alteration of trophic niche stimuli, second by the prevention of mesenchymal cell intermediates, and lastly by the elimination of stem cell ringleaders.
P- Reviewers: Herold-Mende C, Li SC, Panciani PP, Sugimura H S- Editor: Cui XM L- Editor: A E- Editor: Liu XM
| 1. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8406] [Cited by in F6Publishing: 8902] [Article Influence: 741.8] [Reference Citation Analysis (0)] |
| 2. | Population Reference Bureau. 2012 World Population Sheet. Available from: http://www.prb.org/pdf12/2012-population-data-sheet_eng.pdf. [Cited in This Article: ] |
| 3. | Cancer research UK. Colorectal Cancer. 2011. Available from: http://www.cancerresearchuk.org/cancer-info/cancerstats/world/colorectal-cancer-world/colorectal-cancer-world. [Cited in This Article: ] |
| 4. | Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374-1403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3526] [Cited by in F6Publishing: 3571] [Article Influence: 324.6] [Reference Citation Analysis (2)] |
| 5. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN v2.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet]. Lyon: International Agency for Research on Cancer 2010; . [Cited in This Article: ] |
| 6. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8087] [Cited by in F6Publishing: 7722] [Article Influence: 227.1] [Reference Citation Analysis (1)] |
| 7. | Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449-2460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1274] [Cited by in F6Publishing: 1304] [Article Influence: 86.9] [Reference Citation Analysis (2)] |
| 8. | Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-1113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2283] [Cited by in F6Publishing: 2213] [Article Influence: 130.2] [Reference Citation Analysis (0)] |
| 9. | Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717-728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 916] [Cited by in F6Publishing: 939] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 10. | Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 11. | Fearon ER, Hamilton SR, Vogelstein B. Clonal analysis of human colorectal tumors. Science. 1987;238:193-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 400] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 12. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4616] [Cited by in F6Publishing: 4374] [Article Influence: 121.5] [Reference Citation Analysis (0)] |
| 13. | Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 573] [Cited by in F6Publishing: 612] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 14. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3854] [Cited by in F6Publishing: 4012] [Article Influence: 236.0] [Reference Citation Analysis (0)] |
| 15. | Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915-920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 928] [Cited by in F6Publishing: 925] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 16. | Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR, Wong MH. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072-2082.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 17. | Reinisch C, Kandutsch S, Uthman A, Pammer J. BMI-1: a protein expressed in stem cells, specialized cells and tumors of the gastrointestinal tract. Histol Histopathol. 2006;21:1143-1149. [PubMed] [Cited in This Article: ] |
| 18. | Barker N, Tan S, Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140:2484-2494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 19. | Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1049] [Cited by in F6Publishing: 1005] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 20. | Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4394] [Cited by in F6Publishing: 4481] [Article Influence: 298.7] [Reference Citation Analysis (0)] |
| 21. | Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1401] [Cited by in F6Publishing: 1179] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 22. | Schepers AG, Vries R, van den Born M, van de Wetering M, Clevers H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 23. | Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822-825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 24. | Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856-1864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 433] [Cited by in F6Publishing: 451] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 25. | Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 370] [Cited by in F6Publishing: 345] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 26. | Potten CS, Kovacs L, Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271-283. [PubMed] [Cited in This Article: ] |
| 27. | Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112:4627-4639. [PubMed] [Cited in This Article: ] |
| 28. | Potten CS. Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature. 1977;269:518-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 340] [Cited by in F6Publishing: 358] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 836] [Cited by in F6Publishing: 869] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 30. | Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 614] [Cited by in F6Publishing: 565] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 31. | Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079-3091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 609] [Cited by in F6Publishing: 562] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 32. | Vooijs M, Liu Z, Kopan R. Notch: architect, landscaper, and guardian of the intestine. Gastroenterology. 2011;141:448-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 33. | Barker N, van Oudenaarden A, Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell. 2012;11:452-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 225] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 34. | Miyamoto S, Rosenberg DW. Role of Notch signaling in colon homeostasis and carcinogenesis. Cancer Sci. 2011;102:1938-1942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895-902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1150] [Cited by in F6Publishing: 1088] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 36. | Bissell MJ, Labarge MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 204] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 37. | Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349-359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 533] [Cited by in F6Publishing: 565] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 38. | Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 345] [Cited by in F6Publishing: 362] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 39. | Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 334] [Cited by in F6Publishing: 353] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 40. | Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol. 2007;18:467-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 41. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13579] [Article Influence: 848.7] [Reference Citation Analysis (0)] |
| 42. | van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1605] [Cited by in F6Publishing: 1578] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 43. | Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709-1713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 758] [Cited by in F6Publishing: 748] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 44. | Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1207] [Cited by in F6Publishing: 1170] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 45. | Burgess AW, Faux MC, Layton MJ, Ramsay RG. Wnt signaling and colon tumorigenesis--a view from the periphery. Exp Cell Res. 2011;317:2748-2758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Cho JH, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406-3418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 47. | Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1147] [Cited by in F6Publishing: 1221] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 48. | Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159-170. [PubMed] [Cited in This Article: ] |
| 49. | Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1301] [Cited by in F6Publishing: 1412] [Article Influence: 100.9] [Reference Citation Analysis (1)] |
| 50. | Andoh A, Bamba S, Brittan M, Fujiyama Y, Wright NA. Role of intestinal subepithelial myofibroblasts in inflammation and regenerative response in the gut. Pharmacol Ther. 2007;114:94-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 51. | De Wever O, Demetter P, Mareel M, Bracke M. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123:2229-2238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 508] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 52. | Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151-2162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 346] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 53. | Yen TH, Wright NA. The gastrointestinal tract stem cell niche. Stem Cell Rev. 2006;2:203-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 54. | Worthley DL, Giraud AS, Wang TC. Stromal fibroblasts in digestive cancer. Cancer Microenviron. 2010;3:117-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 55. | Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1728] [Cited by in F6Publishing: 1784] [Article Influence: 127.4] [Reference Citation Analysis (0)] |
| 56. | Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, Lobo N, Kulkarni S, Sim S, Qian D. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology. 2012;142:1195-1205.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 187] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 57. | Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 725] [Cited by in F6Publishing: 779] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 58. | Roth S, Franken P, Sacchetti A, Kremer A, Anderson K, Sansom O, Fodde R. Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 2012;7:e38965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 59. | Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518-1529.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 475] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 60. | Kim TH, Escudero S, Shivdasani RA. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci USA. 2012;109:3932-3937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 186] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 61. | Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1545] [Cited by in F6Publishing: 1578] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 62. | Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730-735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 831] [Cited by in F6Publishing: 825] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 63. | Cole JW, Mckalen A. Studies on the morphogenesis of adenomatous polyps in the human colon. Cancer. 1963;16:998-1002. [PubMed] [Cited in This Article: ] |
| 64. | Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 821] [Cited by in F6Publishing: 774] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 65. | Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457-472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 379] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 66. | Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378-2386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 67. | Li L, Bellows CF. Doublecortin-like kinase 1 exhibits cancer stem cell-like characteristics in a human colon cancer cell line. Chin J Cancer Res. 2013;25:134-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 17] [Reference Citation Analysis (0)] |
| 68. | Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R. Colon cancer stem cells. J Mol Med (Berl). 2009;87:1097-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 69. | Langan RC, Mullinax JE, Raiji MT, Upham T, Summers T, Stojadinovic A, Avital I. Colorectal cancer biomarkers and the potential role of cancer stem cells. J Cancer. 2013;4:241-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 70. | Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427-13432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 528] [Cited by in F6Publishing: 565] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 71. | Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158-10163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1541] [Cited by in F6Publishing: 1597] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 72. | Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152-9161. [PubMed] [Cited in This Article: ] |
| 73. | Burgess HA, Reiner O. Alternative splice variants of doublecortin-like kinase are differentially expressed and have different kinase activities. J Biol Chem. 2002;277:17696-17705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 74. | May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630-637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 236] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 75. | Zhang Y, Huang X. Investigation of doublecortin and calcium/calmodulin-dependent protein kinase-like-1-expressing cells in the mouse stomach. J Gastroenterol Hepatol. 2010;25:576-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 76. | May R, Sureban SM, Hoang N, Riehl TE, Lightfoot SA, Ramanujam R, Wyche JH, Anant S, Houchen CW. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells. 2009;27:2571-2579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 77. | Bjerknes M, Khandanpour C, Möröy T, Fujiyama T, Hoshino M, Klisch TJ, Ding Q, Gan L, Wang J, Martín MG. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev Biol. 2012;362:194-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 78. | Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer NF, Bourgaux JF, Pignodel C. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192:767-780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 284] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 79. | Gagliardi G, Goswami M, Passera R, Bellows CF. DCLK1 immunoreactivity in colorectal neoplasia. Clin Exp Gastroenterol. 2012;5:35-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 80. | Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 81. | Sureban SM, May R, Ramalingam S, Subramaniam D, Natarajan G, Anant S, Houchen CW. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology. 2009;137:649-659, 659.e1-2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 82. | Sureban SM, May R, Mondalek FG, Qu D, Ponnurangam S, Pantazis P, Anant S, Ramanujam RP, Houchen CW. Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnology. 2011;9:40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 83. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2952] [Cited by in F6Publishing: 2944] [Article Influence: 163.6] [Reference Citation Analysis (0)] |
| 84. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2977] [Cited by in F6Publishing: 2949] [Article Influence: 163.8] [Reference Citation Analysis (0)] |
| 85. | Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338-344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 516] [Cited by in F6Publishing: 525] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 86. | Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6575] [Cited by in F6Publishing: 7362] [Article Influence: 490.8] [Reference Citation Analysis (0)] |
| 87. | van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 552] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 88. | Stamataki D, Holder M, Hodgetts C, Jeffery R, Nye E, Spencer-Dene B, Winton DJ, Lewis J. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS One. 2011;6:e24484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 89. | Yamaguchi K, Takagi Y, Aoki S, Futamura M, Saji S. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Ann Surg. 2000;232:58-65. [PubMed] [Cited in This Article: ] |
| 90. | Steinert G, Schölch S, Koch M, Weitz J. Biology and significance of circulating and disseminated tumour cells in colorectal cancer. Langenbecks Arch Surg. 2012;397:535-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 91. | Joosse SA, Pantel K. Biologic challenges in the detection of circulating tumor cells. Cancer Res. 2013;73:8-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 152] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 92. | Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213-3221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1354] [Cited by in F6Publishing: 1353] [Article Influence: 84.6] [Reference Citation Analysis (0)] |
| 93. | Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, Diener MK, Büchler MW, Koch M, Weitz J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714-1726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 253] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 94. | Liefers GJ, Cleton-Jansen AM, van de Velde CJ, Hermans J, van Krieken JH, Cornelisse CJ, Tollenaar RA. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 419] [Cited by in F6Publishing: 437] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 95. | Hardingham JE, Kotasek D, Sage RE, Eaton MC, Pascoe VH, Dobrovic A. Detection of circulating tumor cells in colorectal cancer by immunobead-PCR is a sensitive prognostic marker for relapse of disease. Mol Med. 1995;1:789-794. [PubMed] [Cited in This Article: ] |
| 96. | Nieto MA, Cano A. The epithelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012;22:361-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 198] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 97. | Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1047] [Cited by in F6Publishing: 1030] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 98. | Han XY, Wei B, Fang JF, Zhang S, Zhang FC, Zhang HB, Lan TY, Lu HQ, Wei HB. Epithelial-mesenchymal transition associates with maintenance of stemness in spheroid-derived stem-like colon cancer cells. PLoS One. 2013;8:e73341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 99. | Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y, Sceusi E, Tozzi F, Ye XC, Mani SA. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 100. | Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, Van’t Veer L, Tollenaar RA, Jackson DB, Agrawal D. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 212] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 101. | Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-1326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 435] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 102. | Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910-14914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1218] [Cited by in F6Publishing: 1280] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 103. | Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1287] [Cited by in F6Publishing: 1351] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 104. | Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1786] [Cited by in F6Publishing: 1832] [Article Influence: 122.1] [Reference Citation Analysis (0)] |
| 105. | Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Queralt B, Báez L, Guardeño R, Hernández-Yagüe X, Martin-Castillo B, Brunet J, Menendez JA. Stem cell property epithelial-to-mesenchymal transition is a core transcriptional network for predicting cetuximab (Erbitux™) efficacy in KRAS wild-type tumor cells. J Cell Biochem. 2011;112:10-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 106. | Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147-4153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 403] [Article Influence: 23.7] [Reference Citation Analysis (0)] |