Published online Jan 7, 2015. doi: 10.3748/wjg.v21.i1.342
Peer-review started: April 11, 2014
First decision: May 29, 2014
Revised: June 24, 2014
Accepted: July 16, 2014
Article in press: July 16, 2014
Published online: January 7, 2015
AIM: To investigate the presence of human papillomavirus (HPV) DNA along with the integration, the quantification and the expression of the HPV16 in colorectal cancers.
METHODS: A prospective series of colorectal tumors were genotyped for HPV DNA. The clinical and pathological variables of the HPV-positive tumors were compared to those of HPV-negative samples. The integration status of HPV16 was evaluated by calculating E2/E6 ng ratios. HPV16-positive tumors were also evaluated for (1) E2, E4, E5, E6 and E7 viral gene ng quantification; (2) relative quantification compared to W12 cells; and (3) viral E2, E4, E5, E6 and E7 mRNA transcripts by real-time polymerase chain reaction.
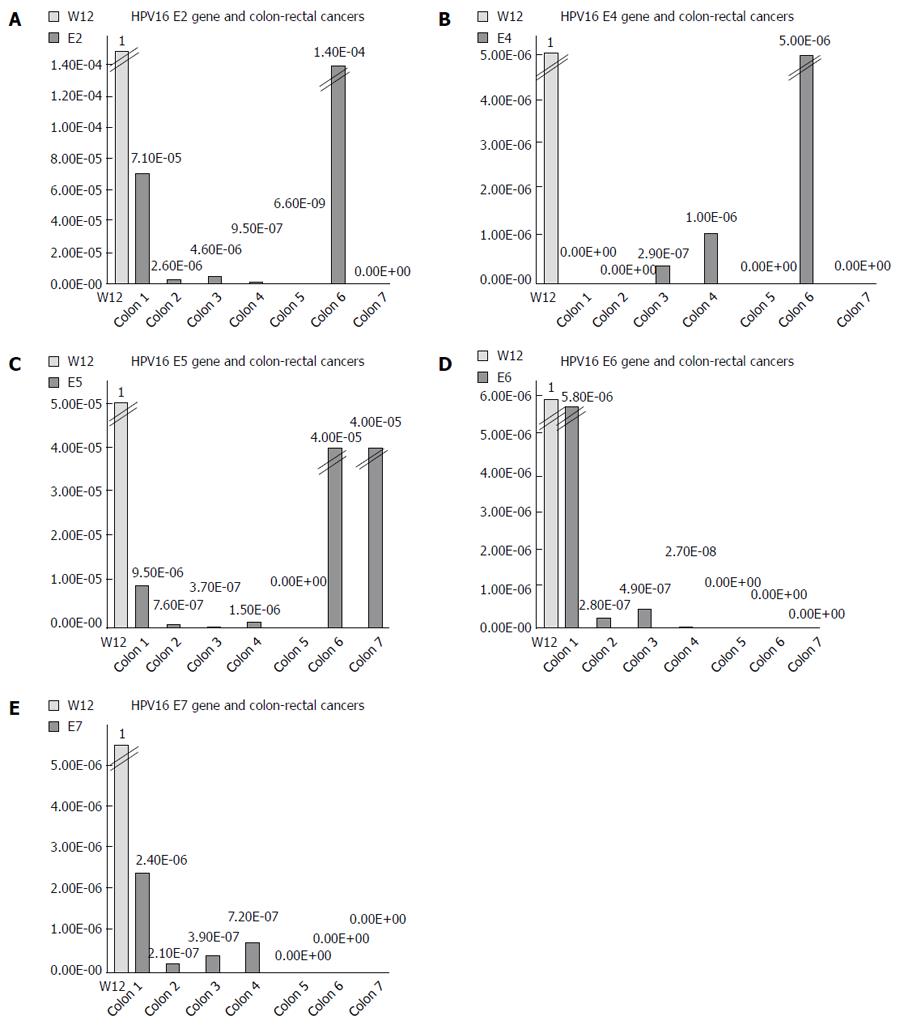
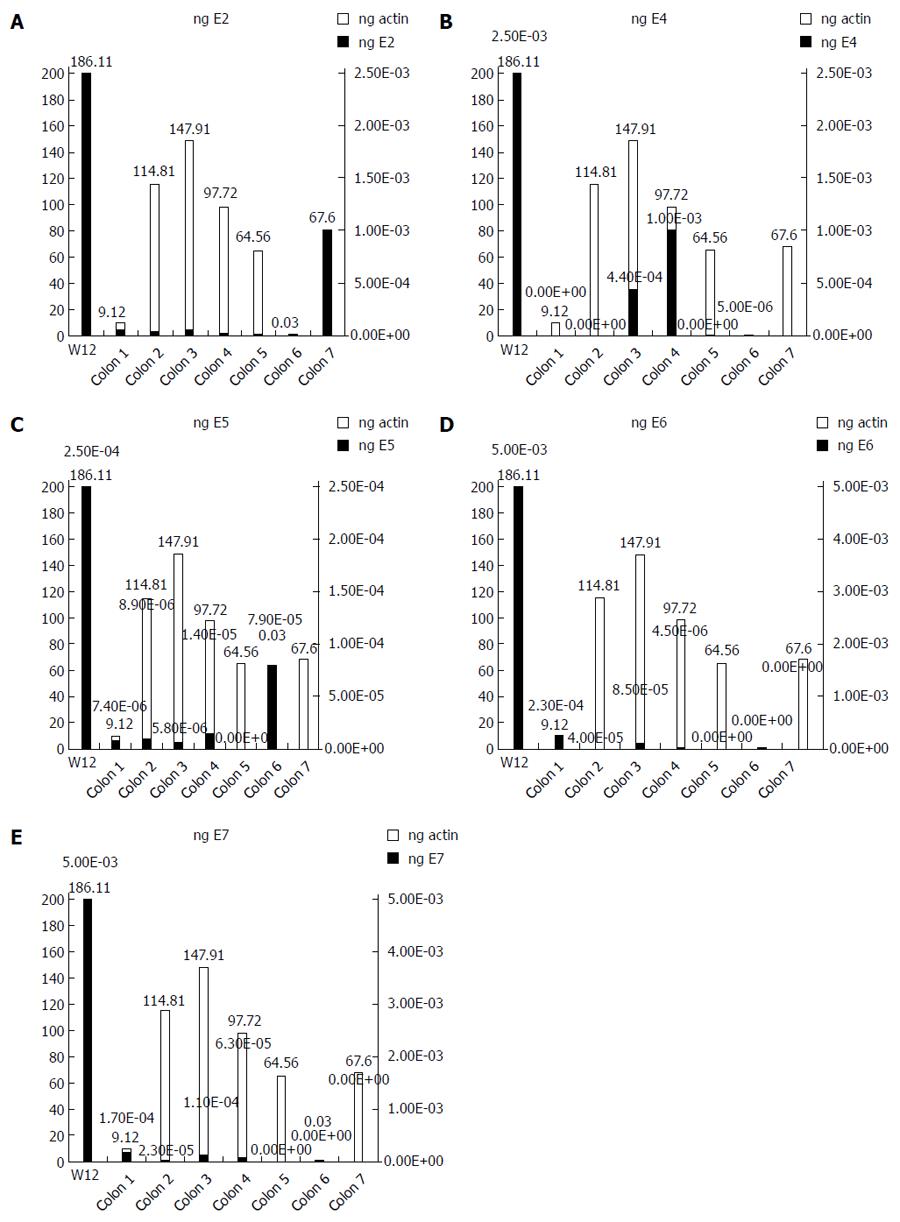
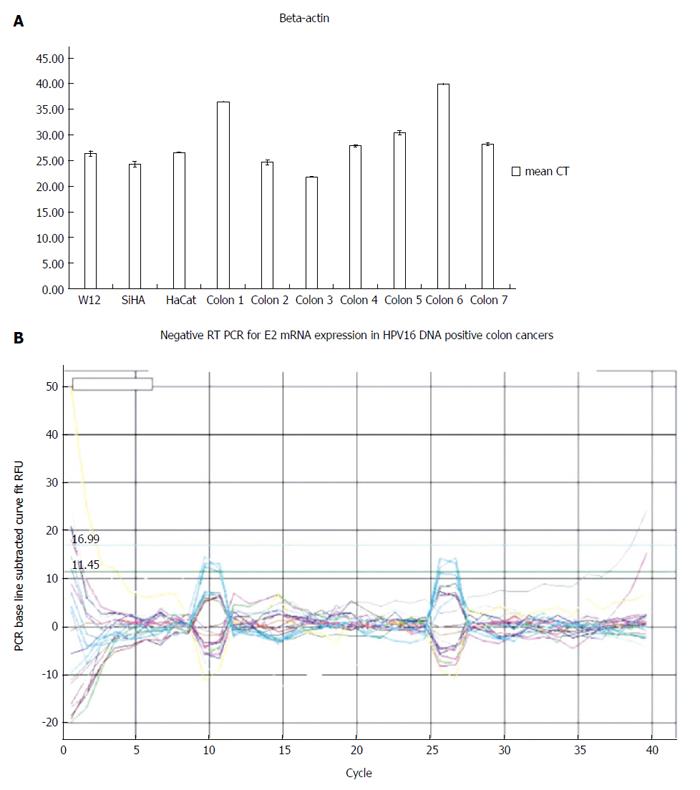
RESULTS: HPV infection was detected in 16.9% of all tumors examined, and HPV16 was the most frequent type detected (63.6% of positive tissues). Notably, the clinical and pathological features of HPV-positive colorectal cancers were not significantly different than those of HPV-negative cancers (χ2 and t-test for all clinical and pathological features of HPV-positive vs HPV-negative colorectal cancers: p ns). HPV16 DNA was present exclusively in episomal form, and the HPV16 E2, E4, E5, E6 and E7 genes were detected in trace nanogram quantities. Furthermore, the HPV16 genes ranged from 10-3 to 10-9 compared to W12 cells at an episomal stage. Although the extractions were validated by housekeeping gene expression, all the HPV16 positive tissues were transcriptionally inactive for the E2, E4, E5, E6 and E7 mRNAs.
CONCLUSION: Based on our results, HPV is unlikely involved in colorectal carcinogenesis.
Core tip: The burden of human papillomavirus (HPV)-associated cancers is not limited to the cervix, but includes the oropharynx and the ano-genital area. Moreover, emerging studies have reported the detection of HPV DNA in the tumoral mucosae of several epithelia, including the colorectum. This is the first study aimed to investigate the presence of HPV in specimens using clinically validated methodology, and it is the first to apply the molecular basis of HPV-related carcinogenesis. Because we only detected episomal HPV16s in low quantities, and it was transcriptionally inactive, we conclude that the virus unlikely plays a role in colorectal carcinogenesis.
- Citation: Lorenzon L, Mazzetta F, Pilozzi E, Uggeri G, Torrisi MR, Ferri M, Ziparo V, French D. Human papillomavirus does not have a causal role in colorectal carcinogenesis. World J Gastroenterol 2015; 21(1): 342-350
- URL: https://www.wjgnet.com/1007-9327/full/v21/i1/342.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i1.342
Colorectal carcinoma has been classified as the fourth most frequently diagnosed cancer in Italy and in the United States[1,2].
Recently, a possible correlation between human papillomavirus (HPV) infection and colorectal cancer has been suggested, and several authors detected HPV DNA in cancerous tissues by different techniques[3]. As we recently highlighted, HPV infection rates are significantly different between tumors and controls/tumor-adjacent tissue when HPV DNA is detected, with the most prevalent type reported as HPV16[3]. Similar results have been validated by recent meta-analysis[4].
The literature lacks studies utilizing clinically validated methodologies for HPV genotyping, as well as studies investigating the molecular basis of HPV-associated colorectal carcinogenesis. Thus, a potential role for HPV in colorectal-related carcinogenesis requires further investigation.
The natural history of a high-risk HPV infection in the target epithelium (e.g., the cervix) is a well-described process that involves virus penetration in the basal layer through micro-abrasions and its subsequent replication in episomal form, leading to the expression of six non-structural (E1-E2, E4-E7) and two structural viral proteins (L1- L2)[5-7].
E5, E6 and E7 are viral onco-proteins that cause immortalization and transformation; E6 and E7 inactivate p53 and Rb, respectively[8]. HPV16 E5 also has transforming potential, leading to increased EGFR recycling and signaling[9].
The progression of a persistent and untreated lesion is associated with HPV DNA integration into the host genome, with subsequent disruption of the E2 gene and up-regulation of the E6 and E7 proteins[5,6].
HPV integration has been identified as a critical step in cancer promotion and progression[10]; however, in vivo studies have shown that an integrated, episomal or mixed HPV genome could be detected in pre-cancerous lesions in addition to invasive cancers[11].
Moreover, persistent and progressive HPV infection has been associated with increased viral load[12].
In summary, there are three major issues that scientists face in studying HPV-related carcinogenesis, including: (1) the expression of viral onco-proteins; (2) the integration pattern; and (3) the viral load of a productive/progressive infection.
On the basis of this background, we aimed to translate the molecular basis of HPV-related carcinogenesis in vivo (particularly HPV16), to understand the possible implications of the virus in colorectal carcinogenesis, in relation to: (1) the detection of HPV DNA in a prospective series of colorectal cancer tissues by a clinical validated methodology; (2) HPV16 integration status; (3) the quantification of HPV16; and (4) viral transcript expression.
Sixty-nine cancerous tissues from 66 patients who had undergone surgical resection for colorectal cancer in our Department were prospectively enrolled from 2010 to 2012. The study protocol was approved by the Ethical Board.
Patients were enrolled if they presented with colorectal cancer independently of sex, age or tumor localization. One exclusion criterion was neo-adjuvant chemo-radiation treatment for rectal cancers, due to HPV infection-related radio-sensitivity[13].
Cancerous tissues were sampled immediately after surgery and frozen. All of the clinical/pathological data were recorded, including patient’s clinical history and demographics (age at the time of surgery, sex), tumor location, and type of surgical procedure.
W12 cells[14] were grown to passage 9 to retain the episomal form of HPV16, and were cultured in M154CF medium plus 1% PSG, 1% HKGS, and calcium (0.07 mmol/L). HaCaT cells (HPV-negative cell line) were cultured in DMEM plus 10% FBS.
HPV genotyping and DNA Real-Time PCR were performed after DNA extraction with the QiAamp DNA extraction kit (Qiagen, Netherlands). HPV detection was performed with the INNO-Lipa HPV Genotyping kit, which can detect 28 different HPVs by DNA amplification followed by reverse hybridization of the SPF10 fragment (Innogenetics, Belgium). The following special attention was paid to avoid cross-contaminations: the personnel were advised to frequently change their disposable gloves and to carefully and singularly open vials. Moreover, amplifications were performed in a separate room.
Real-time polymerase chain reactions (PCRs) were performed using the iCycler IQ™ 5 Multicolor Real Time PCR detection System (Bio-Rad, California, United States) with SYBR Green Supermix (Bio-Rad, California, United States). The HPV16 gene loads was determined in each line by referring to a standard quantification curve generated by diluting from 7 × 106 to 7 × 100 copies of an HPV16 plasmid. Cell lines and cancerous HPV16-positive tissues were then analyzed for DNA ng quantification of the house-keeping gene beta-actin and of the HPV16 E2, E4, E5, E6 and E7 genes. Moreover, a relative quantification of HPV16 genes was obtained for HPV-positive colorectal cancers using the ΔCT method (CT target-CT reference), with to the episomal cell line (W12) as a reference, considered equal to 1.
HPV16 positive tissues were studied using the E2/E6 ng ratio.
RNA extraction was performed with TRIzol Reagent (Invitrogen, California, United States). Retro-transcription was performed with the iScript™ cDNASynthetis kit. The expression of the HPV16 E2, E4, E5, E6 and E7 mRNAs was analyzed by rt real-time PCR. Specimen quality was assessed with beta-actin. Primer sets were the same as those used for qDNA Real Time PCR, with the exception of E2 (E2C: For TTGAAAGCGAAGACAGCGGG; Rev AGTCGTCTGTGTTTCTTCGG) and E4 (E1-E4: For TAATCTACCATGGCTGATCC; Rev AGTCGTCTGTGTTTCTTCGG).
The clinical/pathological features of HPV-positive colorectal cancers were compared with those of HPV-negative cancers. Sub-groups were compared using the Student’s t-test and χ2 test; all test were two-tailed, and P < 0.05 was considered statistical significant. All analyses were obtained with the MedCalc software version 11.4.4.0.
Four samples were excluded from our analysis due to necrotic material, leaving 65 tumors from 62 patients (three patients had synchronous lesions) for data analysis. Table 1 summarizes the features of the population.
| Clinical-pathological features | Value |
| Sex n = 62 | |
| M | 41 (66.1) |
| F | 21 (33.9) |
| M/F | 1.95 |
| Age (yr) n = 62 | |
| Mean ± SD | 70.4 ± 11.3 |
| Median | 72 |
| Range | 38-92 |
| Localizations n = 65 | |
| Rectum | 13 (20.0) |
| Sigmoid | 21 (32.3) |
| Transverse | 6 (9.2) |
| Right | 25 (38.5) |
| Stage-AJCC 9thn = 65 | |
| I | 14 (21.5) |
| II | 22 (33.9) |
| IIa 20-IIb 2 | |
| III | 27 (41.5) |
| IIIa 2-IIIb 20-IIIc 5 | |
| IV | 2 (3.1) |
| T stage n = 65 | |
| T1 | 6 (9.2) |
| T2 | 10 (15.4) |
| T3 | 41 (63.0) |
| T4 | 8 (12.4) |
| N stage n = 65 | |
| N0 | 36 (55.4) |
| N1 | 19 (29.2) |
| N2 | 10 (15.4) |
| Grading n = 65 | |
| G1 | 5 (7.7) |
| G2 | 20 (30.8) |
| G3 | 40 (61.5) |
| V/L/N1 n = 27 | |
| 0 | 4 (14.8) |
| 1 | 22 (81.5) |
| 2 | 1 (3.7) |
| Lymph node harvest n = 65 | |
| Mean ± SD | 21.0 ± 11.5 |
| Median | 19 |
| Range | 6-72 |
| Metastatic nodes n = 65 | |
| Mean ± SD | 2.7 ± 6.1 |
| Median | 0 |
| Range | 0-30 |
| Lymph node ratio n = 65 | |
| Mean ± SD | 0.11 ± 0.2 |
| Median | 0 |
| Range | 0-0.88 |
Eleven tumors tested positive for HPV (16.9%): eight high-risk HPVs (72.7%: seven HPV16-positive tumors, one HPV39), two probable high-risk HPVs (18.2%: one HPV26, one HPV53) and a low-risk HPV6 (9.1%). We most frequently observed the HPV16 viral type (63.6% of the HPV-positive tumors).
We thus compared the features of HPV-positive tumors with those of HPV-negative tumors. We did not detect any significantly different clinical/pathological variables between the two groups (P > 0.05; Table 2).
| HPV-positive (11 cancers from 11 patients) | HPV-negative (54 cancers out of 53 patients) | P value | |
| Sex | |||
| M | 9 (81.8) | 34 (64.1) | 0.431 |
| F | 2 (18.2) | 19 (35.9) | |
| M/F | 4.5 | 1.8 | |
| Age (yr) | |||
| Mean ± SD | 68.1 ± 3.1 | 70.6 ± 10.9 | |
| Median | 69 | 72 | 0.52 |
| Range | 38.0-84.0 | 46.0-92 | |
| 95%CI | 52.9-76.9 | 67.5-73.6 | |
| Localization | |||
| Rectum | 2 (18.2) | 11 (20.4) | |
| Sigmoid | 3 (27.3) | 18 (33.3) | 0.731 |
| Transverse | 2 (18.2) | 4 (7.4) | |
| Right | 4 (36.3) | 21 (38.9) | |
| Stage-AJCC 9th | |||
| I | 3 (27.3) | 11 (20.4) | |
| II | 4 (36.3) | 18 (33.3) | 0.481 |
| III | 3 (27.3) | 24 (44.4) | |
| IV | 1 (9.1) | 1 (1.9) | |
| T stage | |||
| T1 | - (-) | 6 (11.1) | |
| T2 | 3 (27.3) | 7 (12.9) | 0.231 |
| T3 | 8 (72.7) | 33 (61.1) | |
| T4 | - (-) | 8 (14.9) | |
| N stage | |||
| N0 | 7 (63.6) | 29 (53.7) | 0.281 |
| N1 | 1 (9.1) | 17 (31.4) | |
| N2 | 3 (27.) | 8 (14.9) | |
| Grading3 | |||
| G1 | - (-) | 5 (9.2) | |
| G2 | 4 (36.36) | 16 (29.7) | 0.551 |
| G3 | 6 (63.64) | 33 (61.1) | |
| V/L/N4 | |||
| V/L/N0 | 2 (33.3) | 2 (9.5) | 0.311 |
| V/L/N1 | 4 (66.7) | 18 (85.7) | |
| V/L/N2 | - (-) | 1 (4.8) | |
| Lymph node harvest | |||
| Mean ± SD | 23.0 ± 11.4 | 20.6 ± 11.6 | |
| Median | 20 | 18 | 0.532 |
| Range | 10-54 | 6-72 | |
| 95%CI | 15.3-30.6 | 17.4-23.7 | |
| Metastatic nodes | |||
| Mean ± SD | 3.1 ± 5.7 | 2.6 ± 6.2 | 0.822 |
| Median | 0 | 0 | |
| Range | 0-15 | 0-30 | |
| 95%CI | 0.7-6.9 | 0.9-4.3 | |
| Lymph node ratio | |||
| Mean ± SD | 0.15 ± 0.3 | 0.1 ± 0.2 | 0.552 |
| Median | 0 | 0 | |
| Range | 0-0.8 | 0-0.8 | |
| 95%CI | 0.04-0.3 | 0.04-0.15 | |
Table 3 shows the genome structure of the HPV16-positive cancers. In 4/7 HPV16-positive tissues, we could map almost the entire genome; in the remaining three patients, we documented fragments of the HPV16 genome. The former four cancers were suitable for integration status analysis, and all patients had an augmented E2/E6 ratio (range: 1.79-20, mean ± SD 8.9 ± 7.8, median 6.9), consistent with episomal forms of HPV DNA.
| E2 | E4 | E5 | E6 | E7 | L1 (SF10) | |
| 1 | + | - | + | + | + | + |
| 2 | + | - | + | + | + | + |
| 3 | + | + | + | + | + | + |
| 4 | + | + | + | + | + | + |
| 5 | + | - | - | - | - | + |
| 6 | + | + | + | - | - | + |
| 7 | - | - | + | - | - | + |
We determined that W12 cells had a full episomal HPV16 genome with an average of 179 copies/cell, which is consistent with previous reports[14].
We used the ΔΔCT method to determine the relative amount of HPV16 DNA in each cancer, using the episomal cell line (W12) as a reference. The spread between the HPV16 genes and the housekeeping beta-actin gene in the W12 line was considered to be equal to 1. We detected very low quantities of HPV16 genes, ranging from 10-2 to 10-9 of the amount detected in the W12 cells (Figure 1). We further quantified the ng amounts of HPV16 genes in positive tissues and in the W12 cell line. Even though we sufficiently quantified the beta-actin genes in each sample (range: 0.03-186.11 ng, mean ± SD 85.98 ± 64.27 ng, median 82.66 ng), the HPV16 genes in colorectal cancers were detected in extremely low quantities ng quantities (range: 5.4 × 10-8-0.005 ng, mean ± SD 0.0007 ± 0.0009, median 0.00008), as shown in Figure 2.
Although we successfully validated the efficiency of human RNA extractions from tumors by the expression of beta actin mRNA, all HPV16-positive tissues were negative for E2, E4, E5, E6 and E7 mRNA expressions (Figure 3).
HPV is no longer associated exclusively with cervical cancer[15,16]. Recent studies have detected HPV DNA in the tumoral mucosae of different epithelia[3,17,18].
Burnett-Hartman analyzed the correlation between HPV and colorectal cancer using the Bradford criteria[19] and highlighted evidence for the correlation by analogy, biological plausibility, strength of association, but only weakly provided evidence in consistency, specificity and coherence.
We recently reviewed this association by pooling patients from different studies: the HPV mean detection rate within colorectal carcinomas was 41.7% (range: 0%-84%), compared to a mean detection rate of 32.8% in adjacent colonic mucosae, and 5.8% in disease-free controls (P = 0.001)[20-22]. Moreover, HPV DNA has been reported to be statistically associated with colonic polyp mucosae[3]; conversely, two recent studies provided contrasting results[23,24].
Consistent with the findings of our previous review, Baandrup et al[4] recently reported an HPV prevalence of 36.8% in colorectal adenocarcinomas with an OR of 6.0 (95%CI: 2.0-17.9) for the association between HPV and colorectal cancer.
In our study, we documented an HPV DNA infection in 16.9% of the cancers analyzed. To our knowledge, this is the first study to use a clinically validated methodology. Conversely, the vast majority of past reports employed obsolete methods or used double amplification protocols[3]. According to Burnett-Hartman, prior studies are likely the result of contamination from the anal canal or clinical processing[23].
Despite this, HPV16 was the most frequently identified HPV type in tissues we examined (63.6%), in accordance with previous studies[20-22].
In this study, the presence of HPV showed no difference in distribution of the virus through the colon, as previously reported by other authors[20-22]. The prevalence of right colon localization in our samples may be explained by the fact that we excluded patients with rectal cancers that had previously undergone neo-adjuvant chemo-radiotherapy (which is currently the gold standard treatment for rectal cancers clinically staged T3 or N+), because of the radio-sensitivity related to HPV infection that could bias our results[13].
We thus compared the features of HPV-positive vs HPV-negative cancers, and the sub-groups appeared homogeneous for all clinical/pathological variables observed, in contrast to previous reports by Pérez et al[22] and Cheng et al[25], which highlighted a correlation between virus infection and tumor stage or grading.
Nevertheless, the main objective of our study was to translate the molecular basis of HPV-related carcinogenesis in vivo. Bodaghi was the only author who previously investigated HPV genome integration in colorectal tumors, detecting integrated virus quite frequently in the 31 tumor tissues investigated[21]. Of note, we investigated the entire HPV16 genome, providing an examination of the full viral structure. Indeed, in 57.1% of HPV16-positive tumors, we documented the entire viral genome; in the remaining cases, we detected only viral fragments.
To determine the amount of HPV16 DNA in positive tissues, we generated a relative quantification of the genes comparing to the episomal W12 cells. We detected very low quantities of HPV16 in all positive tissues.
Notably, the reported HPV prevalence of 16.9% in the colorectal cancers we examined is much lower than that of other HPV-associated cancers, as the prevalence of HPV in cervical cancer has been reported to be approximately 96%[26], and approximately 30%-44% in oropharyngeal cancers[27].
Chaturvedi et al[27] recently investigated a large series of oropharyngeal carcinomas using the Inno-Lipa technique; they determined that the HPV prevalence in oropharyngeal tumors was 44.1% (38.8% for HPV16). Among the 102 Inno-LiPA HPV16-positive tissues, 77.5% were positive based on viral load criterion, with a median of 22.1 HPV16 copies/cell, and 84.5% were positive by viral oncogene expression criterion, with a median of 152.9 transcripts/1000 RPLP0 equivalents.
Similar findings were reported in the investigation of HPV16 in cervical cancers; notably, Häfner et al[11] reported viral integration as a key event in malignant lesions: they found integrated HPV16 virus in 51% of cervical cancers tested by APOT assay. Conversely, just 11% of CIN lesions contained HPV16 integration. Furthermore, viral transcript levels were broadly distributed, though they had similar median values irrespective of histo-pathological grading and physical state of the viral genome both in CINs and cancers.
Indeed, we focused this study on viral transcript detection. Although all of the samples’ integrity was validated by housekeeping mRNA expression, all of the tissues were negative for HPV16 mRNAs. Conversely, Chen previously reported E6 protein detection by immunohistochemistry in eight out of eleven HPV16-positive colorectal tissues[28]. Our negative results could be explained by the very low virus quantities in positive tissues or by different transcriptional patterns of the virus in a different target epithelium that could impair viral protein replication and expression (simple cuboidal in colorectum vs squamous stratified in the cervix).
In conclusion, we were able to detect HPV DNA in colorectal cancer tissues; however, the virus was episomal, detectable in barely traceable quantities and transcriptionally inactive. This could be related to the very low amounts of viruses or to a different target epithelium that could bias a productive infection or the transcriptional patterns of the HPV. According to our results, it is unlikely that HPV contributes to colorectal carcinogenesis.
The authors thank Venuti A and Paolini F (Laboratory of Virology, Regina Elena Cancer Institute of Rome, Italy) for the scientific discussions and advice. Moreover, the authors thank Mr. Simone De Michelis for the technical help in the preparation of figures.
Past studies suggested a correlation between human papillomavirus (HPV) infection and colorectal cancer development.
With this authors aimed to translate the most important features of HPV-related carcinogenesis to colorectal cancer.
This is the first study to investigate specimens with a clinically validated methodology and the first one to apply the molecular basis of HPV-related carcinogenesis, investigate viral integration, HPV16 quantification and of HPV transcript expression in colorectal cancerous tissues.
Future studies aiming to correlate the presence of the HPV virus in cancerous tissues other than those widely recognized as part of the HPV-burden of diseases (cervix, ano-genital and head and neck) should refer to the molecular basis of viral carcinogenesis, and should not limit their investigation to the detection of HPV DNA.
This is a unique study with novelty. The authors should be congratulated for this landmark study.
P- Reviewer: Gagliardi G, Shimura T S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
| 1. | AIRTUM.I tumori in Italia: Rapporto 2006. Available from: http://www.registri-tumori.it/incidenza1998-2002/gruppi.html. [Cited in This Article: ] |
| 2. | National Comprehensive Cancer Network. NCCN Guidelines - Colon cancer, version 1.2013. Available from: http://www.nccn.org. [Cited in This Article: ] |
| 3. | Lorenzon L, Ferri M, Pilozzi E, Torrisi MR, Ziparo V, French D. Human papillomavirus and colorectal cancer: evidences and pitfalls of published literature. Int J Colorectal Dis. 2011;26:135-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Baandrup L, Thomsen LT, Olesen TB, Andersen KK, Norrild B, Kjaer SK. The prevalence of human papillomavirus in colorectal adenomas and adenocarcinomas: a systematic review and meta-analysis. Eur J Cancer. 2014;50:1446-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 5. | Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890-907. [PubMed] [Cited in This Article: ] |
| 6. | Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11-22. [PubMed] [Cited in This Article: ] |
| 7. | Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1120] [Cited by in F6Publishing: 1121] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 8. | Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286-2302. [PubMed] [Cited in This Article: ] |
| 9. | Crusius K, Auvinen E, Steuer B, Gaissert H, Alonso A. The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp Cell Res. 1998;241:76-83. [PubMed] [Cited in This Article: ] |
| 10. | Tonon SA, Picconi MA, Bos PD, Zinovich JB, Galuppo J, Alonio LV, Teyssie AR. Physical status of the E2 human papilloma virus 16 viral gene in cervical preneoplastic and neoplastic lesions. J Clin Virol. 2001;21:129-134. [PubMed] [Cited in This Article: ] |
| 11. | Häfner N, Driesch C, Gajda M, Jansen L, Kirchmayr R, Runnebaum IB, Dürst M. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008;27:1610-1617. [PubMed] [Cited in This Article: ] |
| 12. | Hesselink AT, Berkhof J, Heideman DA, Bulkmans NW, van Tellingen JE, Meijer CJ, Snijders PJ. High-risk human papillomavirus DNA load in a population-based cervical screening cohort in relation to the detection of high-grade cervical intraepithelial neoplasia and cervical cancer. Int J Cancer. 2009;124:381-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Lindel K, Rieken S, Daffinger S, Weber KJ, de Villiers EM, Debus J. The transcriptional regulator gene E2 of the Human Papillomavirus (HPV) 16 influences the radiosensitivity of cervical keratinocytes. Radiat Oncol. 2012;7:187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int J Cancer. 1989;43:672-676. [PubMed] [Cited in This Article: ] |
| 15. | Jemal A, Simard EP, Dorell C, Noone AM, Markowitz LE, Kohler B, Eheman C, Saraiya M, Bandi P, Saslow D. Annual Report to the Nation on the Status of Cancer, 1975-2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. J Natl Cancer Inst. 2013;105:175-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 729] [Cited by in F6Publishing: 749] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 16. | Lorenzon L, Benevolo M, Visca P, Venturo I, Filippetti M, Piro FR, Rollo F, Vocaturo A. Human papillomavirus type 16 DNA detected in pulmonary metastases from a penile squamous cell carcinoma: a case study. Int J Surg Pathol. 2013;21:59-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 17. | Zandberg DP, Bhargava R, Badin S, Cullen KJ. The role of human papillomavirus in nongenital cancers. CA Cancer J Clin. 2013;63:57-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 144] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 18. | Frega A, Lorenzon L, Bononi M, De Cesare A, Ciardi A, Lombardi D, Assorgi C, Gentile M, Moscarini M, Torrisi MR. Evaluation of E6 and E7 mRNA expression in HPV DNA positive breast cancer. Eur J Gynaecol Oncol. 2012;33:164-167. [PubMed] [Cited in This Article: ] |
| 19. | Burnett-Hartman AN, Newcomb PA, Potter JD. Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev. 2008;17:2970-2979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 104] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 20. | Damin DC, Caetano MB, Rosito MA, Schwartsmann G, Damin AS, Frazzon AP, Ruppenthal RD, Alexandre CO. Evidence for an association of human papillomavirus infection and colorectal cancer. Eur J Surg Oncol. 2007;33:569-574. [PubMed] [Cited in This Article: ] |
| 21. | Bodaghi S, Yamanegi K, Xiao SY, Da Costa M, Palefsky JM, Zheng ZM. Colorectal papillomavirus infection in patients with colorectal cancer. Clin Cancer Res. 2005;11:2862-2867. [PubMed] [Cited in This Article: ] |
| 22. | Pérez LO, Abba MC, Laguens RM, Golijow CD. Analysis of adenocarcinoma of the colon and rectum: detection of human papillomavirus (HPV) DNA by polymerase chain reaction. Colorectal Dis. 2005;7:492-495. [PubMed] [Cited in This Article: ] |
| 23. | Burnett-Hartman AN, Feng Q, Popov V, Kalidindi A, Newcomb PA. Human papillomavirus DNA is rarely detected in colorectal carcinomas and not associated with microsatellite instability: the Seattle colon cancer family registry. Cancer Epidemiol Biomarkers Prev. 2013;22:317-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Burnett-Hartman AN, Newcomb PA, Mandelson MT, Galloway DA, Madeleine MM, Wurscher MA, Carter JJ, Makar KW, Potter JD, Schwartz SM. No evidence for human papillomavirus in the etiology of colorectal polyps. Cancer Epidemiol Biomarkers Prev. 2011;20:2288-2297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Cheng JY, Sheu LF, Meng CL, Lee WH, Lin JC. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut. 1995;37:87-90. [PubMed] [Cited in This Article: ] |
| 26. | Muñoz N, Bosch FX, Castellsagué X, Díaz M, de Sanjose S, Hammouda D, Shah KV, Meijer CJ. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278-285. [PubMed] [Cited in This Article: ] |
| 27. | Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294-4301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2465] [Cited by in F6Publishing: 2572] [Article Influence: 197.8] [Reference Citation Analysis (0)] |
| 28. | Chen TH, Huang CC, Yeh KT, Chang SH, Chang SW, Sung WW, Cheng YW, Lee H. Human papilloma virus 16 E6 oncoprotein associated with p53 inactivation in colorectal cancer. World J Gastroenterol. 2012;18:4051-4058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 40] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |