Published online Jul 7, 2015. doi: 10.3748/wjg.v21.i25.7814
Peer-review started: December 17, 2014
First decision: January 22, 2015
Revised: March 2, 2015
Accepted: April 3, 2015
Article in press: April 3, 2015
Published online: July 7, 2015
AIM: To investigate whether expression of selected miRNAs obtained from fibrotic liver biopsies correlate with fibrosis stage.
METHODS: Altogether, 52 patients were enrolled in the study representing various etiologic backgrounds of fibrosis: 24 cases with chronic hepatitis infections (types B, C), 19 with autoimmune liver diseases (autoimmune hepatitis, primary biliary cirrhosis, primary sclerosing cholangitis, overlapping syndrome cases), and 9 of mixed etiology (alcoholic and nonalcoholic steatosis, cryptogenic cases). Severity of fibrosis was determined by both histologic staging using the METAVIR scoring system and noninvasive transient elastography. Following RNA isolation, expression levels of miR-21, miR-122, miR-214, miR-221, miR-222, and miR-224 were determined using TaqMan MicroRNA Assays applying miR-140 as the reference. Selection of miRNAs was based on their characteristic up- or downregulation observed in hepatocellular carcinoma. Relative expression of miRNAs was correlated with fibrosis stage and liver stiffness (LS) value measured by transient elastography, as well as with serum alanine aminotransferase (ALT) level.
RESULTS: The expression of individual miRNAs showed deregulated patterns in stages F1-F4 as compared with stage F0, but only the reduced level of miR-122 in stage F4 was statistically significant (P < 0.04). When analyzing miRNA expression in relation to fibrosis, levels of miR-122 and miR-221 showed negative correlations with fibrosis stage, and miR-122 was found to correlate negatively and miR-224 positively with LS values (all P < 0.05). ALT levels displayed a positive correlation with miR-21 (P < 0.04). Negative correlations were observed in the fibrosis samples of mixed etiology between miR-122 and fibrosis stage and LS values (P < 0.05), and in the samples of chronic viral hepatitis, between miR-221 and fibrosis stage (P < 0.01), whereas miR-21 showed positive correlation with ALT values in the samples of autoimmune liver diseases (P < 0.03). The results also revealed a strong correlation between fibrosis stage and LS values (P < 0.01) when etiology of fibrosis was not taken into account.
CONCLUSION: Reduced expression of miR-122 in advanced fibrosis and its correlation with fibrosis stage and LS values seem to be characteristic of hepatic fibrosis of various etiologies.
Core tip: In this study, the expression of selected miRNAs was determined in fibrotic liver tissues of various etiologic backgrounds and was correlated with fibrosis stage (METAVIR scores) and liver stiffness as measured by transient elastography. In advanced fibrosis, the level of miR-122 was reduced and showed negative correlations with fibrosis stage and liver stiffness values, indicating that it could be a useful molecule to assess severity of fibrosis regardless of etiology.
- Citation: Halász T, Horváth G, Pár G, Werling K, Kiss A, Schaff Z, Lendvai G. miR-122 negatively correlates with liver fibrosis as detected by histology and FibroScan. World J Gastroenterol 2015; 21(25): 7814-7823
- URL: https://www.wjgnet.com/1007-9327/full/v21/i25/7814.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i25.7814
Hepatic fibrosis develops as a wound-healing response of the liver to cellular injury, reflecting the balance between liver repair and scar formation[1]. Upon injury, the activated hepatic stellate cells (HSC) undergo transition into proliferative, profibrogenic, contractile myofibroblasts, which are responsible for the excess deposition of the extracellular matrix (ECM)[2,3]. The eventual structural abnormalities, which result from the histologic rearrangement of various types of collagens, proteoglycans, and structural glycoproteins and the excess deposition of ECM[2,4-7] cause increased liver stiffness (LS). When injury persists, fibrosis may advance into cirrhosis-the most severe stage, which may then further progress into hepatocellular carcinoma (HCC)[1,2,8]. Liver fibrosis is caused by diverse etiologies that include alcoholic and nonalcoholic steatohepatitis, chronic viral hepatitis, autoimmune disorders, and toxins[1,4,9].
Liver biopsy is the gold standard method for the identification of hepatic fibrosis. As this procedure is invasive, painful, and carries the risk of complications[10-12], other alternatives have been developed, such as noninvasive transient elastography (TE), serum-based aspartate aminotransferase-to-platelet ratio index, and Fibrotest[13,14]. TE is an ultrasound-based examination method that measures the fibrosis-related rigidity of the liver tissue, with the velocity of the share wave being directly related to LS, expressed as the LS value[15]. Using this method, the progress of fibrosis and early asymptotic cirrhosis can be assessed with high sensitivity and specificity. However, not all information needed for the diagnosis of fibrosis can be obtained from TE examination, such as the histologic liver conditions, including necroinflammation.
MicroRNAs (miRNA) are short regulating RNA molecules that interfere with gene expression at the posttranscriptional level by way of inducing translational arrest, which in turn leads to reduced or prevented protein synthesis[3]. As a result of this negative modulating function, miRNAs fine-tune the expression of genes involved predominantly in normal cellular processes, such as development, differentiation, and proliferation[16]. Deregulated miRNA expression in comparison to normal state has been found in many disorders, including liver diseases[17,18]. In hepatic fibrosis, the members of the miR-27, miR-29, and miR-19 families have been reported to show altered expression[5,7,8,19]. These miRNAs either hinder the expression of various ECM components (miR-29) or regulate the signal transduction pathways connected to fibrosis (miR-29)[6,8,20] or the resting state of HSCs (miR-27)[21].
It has been suggested that an imbalance in the normal miRNA pattern can be measured long before the onset of a disease[22]. Therefore, in the present study, the relative expression levels of selected miRNAs that are characteristically up- or downregulated in HCC in comparison to non-tumorous liver tissue[23] were determined in hepatic fibrosis samples of various etiologies. We wished to find out the degree to which these levels were altered in fibrosis and how they correlated with fibrosis stage (analyzed by histology and METAVIR scoring system) and LS values (measured by TE), as well as with serum alanine aminotransferase (ALT) levels.
Biopsy samples of 52 patients were selected from the archives of the First Department of Internal Medicine at the University of Pécs, the Hepatology Center Buda in Budapest and the Second Department of Pathology at the Semmelweis University, Budapest. Selection was based on two criteria: diagnosis of histologically confirmed chronic, diffuse liver disease and LS measurement with intervals no longer than 3 mo between the two examinations. Permission for the retrospective analysis of the samples was obtained from the local Ethical Committee (45727-2/2013/EKU) based on the ethical guidelines of the 1975 Declaration of Helsinki. Patients were aged between 15 and 67 years with an average of 45.18 years. The female/male ratio was 35/17. ALT serum values were also recorded at the time of liver biopsy.
The cases were selected to represent the diverse etiology of fibrosis: 24 cases of chronic viral hepatitis, including 22 hepatitis C virus (HCV) and 2 hepatitis B virus (HBV) infections; 19 autoimmune cases, including 8 autoimmune hepatitis (AIH), 6 primary biliary cirrhosis (PBC), 2 primary sclerosing cholangitis (PSC), and 3 overlapping syndrome cases (AIH/PBC and AIH/PSC); 9 cases of mixed etiology, including 1 case of alcoholic and 2 of nonalcoholic liver diseases (ALD and NAFLD, respectively) and 6 cryptogenic cases (Table 1). Accordingly, three sample groups were formed for analysis: autoimmune (AIH, PBC, PSC, and overlaps), chronic viral hepatitis (HCV, HBV), and mixed etiology (ALD, NAFLD, cryptogenic).
| Etiology | No. of cases | Etiology subgroups(n) | Fibrosis stage2(n) | LS level (kPa) | ALT level (U/L) | HAI |
| Autoimmune | 19 | AIH (8)1 | F0 (1)1 | 6.1 | 452 | 5 |
| PBC (6) | F1 (3) | 5.3-7.6 | 20-904 | 0-4 | ||
| PSC (2) | F2 (4) | 5.1-8.8 | 20-368 | 0-6 | ||
| AIH/PBC (2) | F3 (9) | 5.5-17.1 | 17-558 | 0-12 | ||
| AIH/PSC (1) | F4 (2) | 20.6 - 45.7 | 26-83 | 0-4 | ||
| Chronic viral | 24 | HCV (22) | F0 (3) | 4.6-5.3 | 14-125 | 3-6 |
| HBV (2) | F1 (4) | 3.8-6.8 | 12-35 | 2-6 | ||
| F2 (4) | 5.4-7.6 | 20-88 | 3-4 | |||
| F3 (11) | 5.6-20.4 | 12-257 | 0-8 | |||
| F4 (2) | 18.0-26.3 | 60-108 | 0-10 | |||
| Mixed etiology | 9 | ALD (1) | F0 (1) | 4.9 | 71 | 0 |
| NAFLD (2) | F1 (2) | 3.7-4.1 | 252-272 | 0 | ||
| Cryptogenic (6) | F2 (3) | 5.3-11.9 | 13-101 | 0 | ||
| F3 (1) | 45 | 12 | 0 | |||
| F4 (2) | 75 | 23-32 | 0 | |||
| Total | 52 | - | F0 (5) | 4.6-6.1 | 14-452 | 0-6 |
| F1 (9) | 3.7-7.6 | 12-904 | 0-6 | |||
| F2 (11) | 5.1-11.9 | 13-368 | 0-6 | |||
| F3 (21) | 5.5-45.0 | 12-558 | 0-12 | |||
| F4 (6) | 18.0-75.0 | 23-108 | 0-10 |
Biopsy samples were processed according to routine pathology procedures. In brief, the small, 1-3-cm long samples were submerged in 10% neutral buffered formalin (in PBS, pH 7.0) and fixed for 24 h at room temperature. Following dehydration in a series of ethanols and xylene, the formalin-fixed samples were embedded in paraffin (FFPE samples). These samples were cut into 3-4-μm thick sections and stained with hematoxylin-eosin and picrosyrius red to highlight the connective tissue.
Histologic staging and TE examination were applied to determine the severity of fibrosis. Histologic staging was performed by two pathologists using the METAVIR scoring system from stages F0 to F4, with stage F0 indicating no fibrosis and stage F4 representing cirrhosis[24]. The noninvasive TE was carried out using FibroScan 502 (Echosens, Paris, France), with low LS values reflecting no or mild fibrosis and high LS values implying advanced fibrosis or cirrhosis. The elapsed time between date of histologic sampling and date of LS measurement was a maximum of 3 mo, with an average of 1.5 mo.
RNA was isolated from several 3-4-μm thick sections using the RNeasy FFPE Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions with modifications for copurification of miRNAs[25]. Traces of genomic DNA were eliminated using Turbo DNase digestion (Ambion, Austin, TX, United States).
Expression of individual miRNAs was determined using the following TaqMan MicroRNA Assays (Life Technologies of Thermo Fisher Scientific Inc., Waltham, MA, United States): miR-21 (ID: 000397), miR-122 (ID: 002245), miR-140 (ID: 000462), miR-214 (ID: 002306), miR-221 (ID: 000524), miR-222 (ID: 002276), and miR-224 (ID: 002099). Reverse transcription (RT) and quantitative (q)PCR were performed according to the manufacturer’s instructions. Briefly, RT reaction was carried out using the TaqMan MicroRNA Reverse Transcription Kit in a final volume of 7.5 μL containing 10 ng total RNA. The qPCR was performed using TaqMan Universal PCR Master Mix No AmpErase UNG in a final volume of 10 μL containing 0.65 μL RT product. The amplification reaction was run in triplicate on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems of Thermo Fisher Scientific Inc.). Relative expression was calculated by the 2-∆∆Cq formula, applying miR-140 as the most stable reference determined by the NormFinder application[26] and normalized to the median ∆Cq value of F0 liver samples.
The differences between fibrosis stages F0-F4 were analyzed with a nonparametric Kruskal-Wallis analysis of variance and median test using STATISTICA software, version 9.1 (StatSoft Inc., Tulsa, OK, United States). Correlation analyses between miRNA expression and fibrosis stage, LS values, and ALT levels were performed with a nonparametric Spearman rank order correlation using GraphPad PRISM software, version 5.01 (GraphPad Software Inc, La Jolla, CA, United States). A P value of 0.05 was set as the threshold for statistical significance. The statistical methods of this study were reviewed by Istvan Kenessey from the Second Department of Pathology, Semmelweis University.
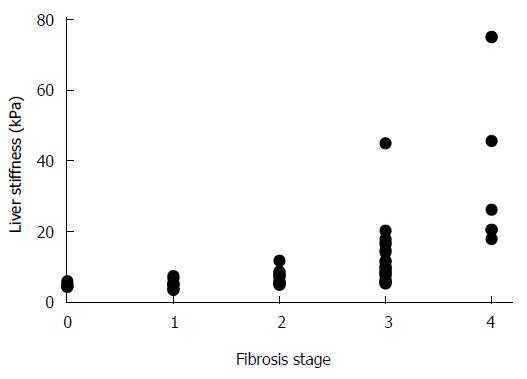
The METAVIR scoring system allowed an unambiguous determination of fibrosis stages in each tissue sample. In contrast, the noninvasive LS measurement showed a wide range of values and when matched with the corresponding METAVIR stages, an overlap between the neighboring ranges was observable (Table 1). This was predominantly manifested in cases of LS value ranges that corresponded to fibrosis stages F0-F3. In addition, the LS value ranges showed slight variances between the various etiology groups. Yet, a highly significant correlation was found between the gradually increasing LS values and fibrosis stage (r = 0.8; P < 0.01), as presented in Figure 1.
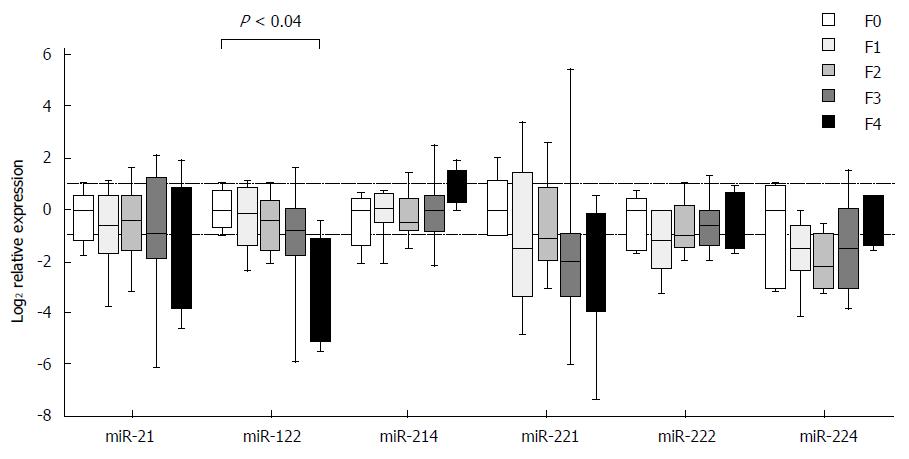
Expression of individual miRNAs showed deregulated patterns in stages F1-F4 in comparison to stage F0, but the observed differences, except for one case, did not reach the set threshold for statistical significance. The exception was miR-122, in which case the expression in stage F4 was decreased as compared with stage F0 (P < 0.04) (Figure 2). The expression differences were close to reaching a statistical significance in two cases: miR-122 between stages F1 and F4 (P = 0.06) and miR-214 between stages F2 and F4 (P = 0.07). When looking at the expressional patterns of individual miRNAs, in general, the levels were lower in stages F1-F4 in comparison to F0, showing an increasing tendency in case of miR-214 from F2 to F4, miR-222 from F1 to F4 and miR-224 from F2 to F4 (Figure 2). Nevertheless, the differences in the three etiologic groups did not reach the set threshold for statistical significance.
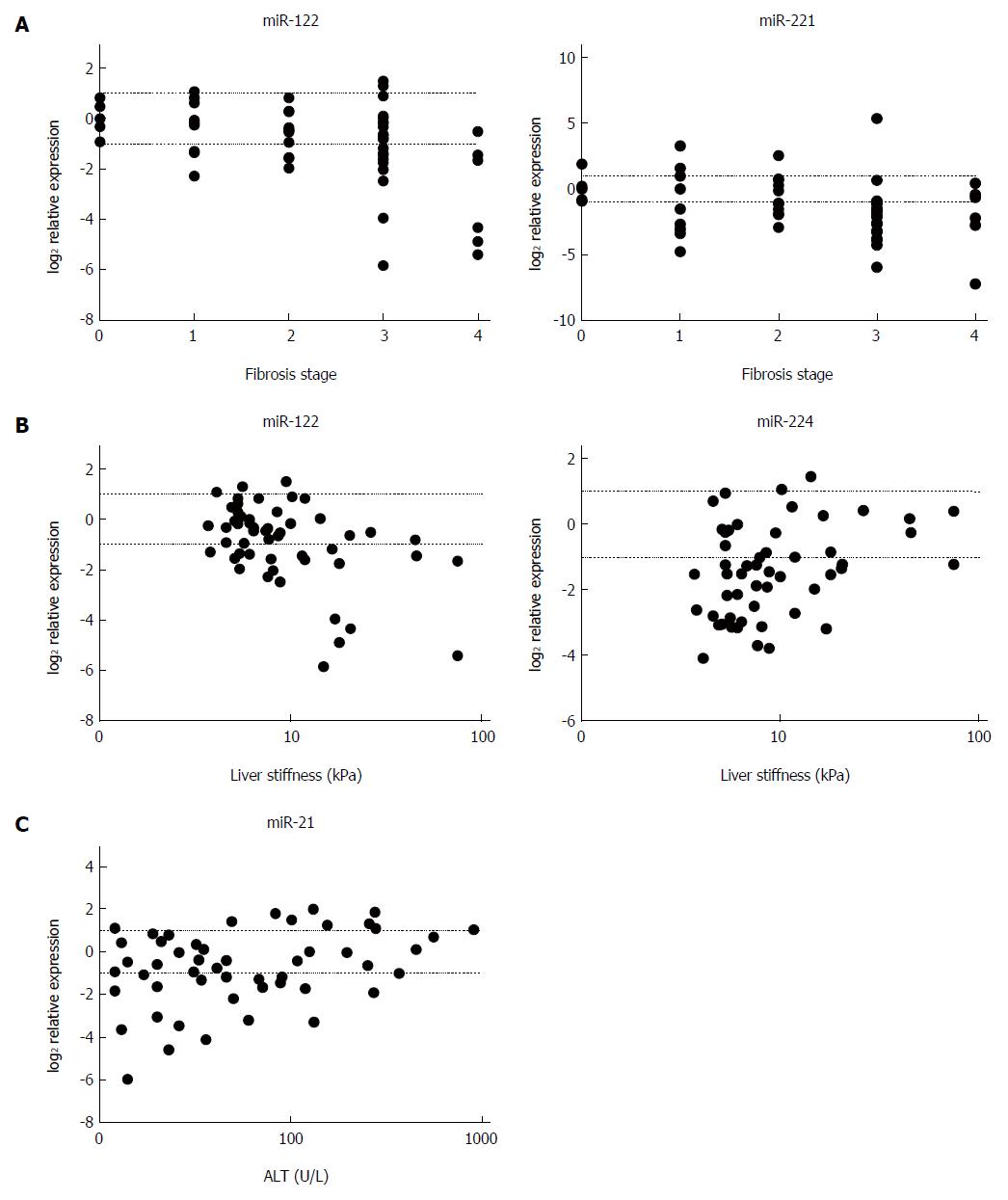
In relation to miRNA expression and fibrosis, miR-122 and miR-221 showed a negative correlation with METAVIR stage (P < 0.01 and P < 0.03, respectively) (Figure 3A), and miR-122 was found to correlate negatively (P < 0.01) and miR-224 positively (P < 0.04) with LS values (Figure 3B). Furthermore, a positive correlation of miR-21 was found between miRNA expression and ALT levels (P < 0.04) (Figure 3C). A summary of the correlation analyses is provided in Table 2. With respect to etiology, miR-122 expression correlated negatively with fibrosis stage and LS values (P < 0.02 and P < 0.05, respectively) in the mixed etiology group, and miR-221 level showed a negative correlation with fibrosis stage (P < 0.01) in the chronic viral hepatitis group, whereas a positive correlation between miR-21 and ALT values (P < 0.03) was found in the autoimmune group.
| miRNA | All samples(n = 52) | Etiology groups | ||
| Autoimmune | Viral | Mixed | ||
| (n = 19) | (n = 24) | (n = 9) | ||
| Fibrosis stage | ||||
| miR-122 | r = -0.4 | r = -0.3 | r = -0.3 | r = -0.8 |
| P < 0.01 | P < 0.20 | P < 0.10 | P < 0.02 | |
| miR-221 | r = -0.3 | r = -0.3 | r = -0.5 | r = -0.2 |
| P < 0.03 | P < 0.20 | P < 0.01 | P < 0.40 | |
| Liver stiffness | ||||
| miR-122 | r = -0.4, | r = -0.4 | r = -0.2 | r = -0.7 |
| P < 0.01 | P < 0.09 | P < 0.30 | P < 0.05 | |
| miR-224 | r = 0.3 | r = 0.3 | r = 0.1 | r = 0.6 |
| P < 0.04 | P < 0.20 | P < 0.70 | P < 0.07 | |
| Alanine aminotransferase | ||||
| miR-21 | r = 0.3 | r = 0.5 | r = 0.4 | r = -0.4 |
| P < 0.04 | P < 0.03 | P < 0.09 | P < 0.30 | |
The diagnosis of liver fibrosis and the decision on therapy are important factors in the treatment of chronic liver diseases, with liver biopsy being the widely used procedure for the accurate determination of fibrosis. Owing to certain limitations of liver biopsy, such as possibility of serious complications, contradictions, sampling as well as intra- and interobserver errors[12], noninvasive approaches, such as TE, are available as alternatives. However, the diagnostic accuracy of TE is not entirely precise over the measuring range. Although the measured values increase with fibrosis stage, the method gives excellent results predominantly from advanced fibrosis (F2) to early and symptom-free cirrhosis (F4), and performs with limitations when early stage fibrosis (F0-F1) is to be determined, and when having to differentiate between F2/F3 stadium or in case of obese patients[27]. Yet, a highly positive correlation between fibrosis stage and LS values was found in the present study, which is supportive of the positive finding reported earlier[28].
There is a general need to find indicators at the molecular level to help predict disease progression. For example, hepatic cirrhosis is characterized by an increased proliferation rate that correlates with a higher tendency to develop HCC[29]. The miRNAs are foreseen to be such indicators based on their altered expression found in liver diseases, fibrosis[7,17], and liver carcinogenesis[23,30]. In addition, deregulated expression of miRNAs may be present long before the onset of a disease[22]. As alterations of miRNA expression in relation to fibrosis stage have predominantly been studied in chronic HCV-infected samples, we aimed, in the present study, to investigate the expression of fibrosis- and hepatocarcinogenesis-related miRNAs in hepatic fibrosis samples of various etiologies, and to correlate the found expression levels with the severity of fibrosis and serum ALT levels.
Our results reveal a reduced level of miR-122 in stage F4 fibrosis as compared with stage F0, and miR-122 showed a negative correlation with fibrosis stage in fibrotic liver samples and, intriguingly, also with LS values. These findings are supported by reports of a negative correlation between miR-122 and fibrosis stage in chronic HCV infection, HCV-based HCC, and cirrhosis[31,32], and also by observations of a decreased level of miR-122 in NAFLD[33,34] and in HCC studies[23]. miR-122 is a liver-characteristic miRNA that composes about 70% of the total miRNAs found in normal hepatocytes[35], most probably due to the fact that it positively regulates the accumulation of cholesterol and triglycerides and the metabolism of fatty acids[16]. Thus, a decreased level of miR-122 in fibrotic liver biopsies may be interpreted as the result of compromised normal hepatocytic activity or as the eliminated suppressive function of miR-122 that hinders fibrogenesis. Namely, miR-122 has been found to suppress the proliferation of HSCs, resulting in decreased maturation of collagen by downregulating the expression of P4HA1, a key enzyme in collagen maturation[36]. miR-122 may also impede carcinogenesis[37], as expression of proteins involved in the cell cycle, differentiation, and proliferation is downregulated by miR-122[38], and loss of miR-122 in HCC is a frequent finding, which correlates with migration, invasion and in vivo tumorigenesis[39]. In association with HCV, miR-122 has been found to protect viral RNA from exonuclease degradation by binding at two positions near the 5′ end of the RNA molecule. However, the capacity of this protection seems to be independent of the promotion of HCV infectivity, indicating that miR-122 has other unknown functions in the viral life cycle[40]. Taken together, downregulation of miR-122 seems to be both a sensitive sign of hepatic injury and a possible step on the path toward liver cancer.
Increased levels of miR-221, miR-224, and miR-21 have been reported in HCC as these oncomiRs inhibit expression of tumor suppressor genes[23]. For example, downregulation of P27 and P57, as targets of miR-221 and key regulators of cell cycle progression, has been found to promote cancer cell proliferation[30,41]. Moreover, miR-221 is observed to be increased in early preneoplastic stage, such as cirrhosis and steatosis or steatohepatitis[42-44], and is also found to be overexpressed in a mouse liver-regeneration model, in which the proliferation of hepatocytes is accelerated by miR-221 in vitro and in vivo in the presence of epidermal or hepatocytes growth factors (EGF or HGF), thereby facilitating a rapid S-phase entry of hepatocytes[45]. In HCV and nonalcoholic steatohepatitis biopsies, miR-221 is observed to increase with fibrosis stage and to correlate positively with expression levels of α1 chain of collagen type I[46]. In contrast, statistical difference in miR-221 expression was not found in the present study; moreover, fibrosis stage showed a negative correlation with miR-221 expression in our samples and the samples of the chronic viral hepatitis group. An explanation for this could be that the representations of the F0-F4 cases, as well as the statistical methods used, were different; furthermore, the extent of regeneration was possibly different in the analyzed samples. Another reason could be the DNA methylation status of the miR-221 locus, as hypomethylation of this locus was found in HCC that contributed to the overexpression of miR-221[47].
miR-224 has been described to promote proliferation, migration, and invasion in HCC by the activation of AKT signaling; thus, miR-224 has been suggested to play a role in liver carcinogenesis and progression[48]. With respect to fibrosis stage, the present study did not reveal any differences or correlations. However, LS values positively correlated with miR-224 expression, suggesting that a gradual increase in miR-224 level may occur in liver tissues prior to malignant transformation. Indeed, miR-224 expression has been found to correlate with fibrosis stage in chronic hepatitis C[49], and elevated levels of miR-224 have also been observed in chronic hepatitis C samples with steatosis and HCV-negative steatotic liver biopsies[50].
It has been reported that miR-21 reduces the expression of fibrogenesis-related tumor suppressor genes, such as SMAD-7, the negative regulator of transforming growth factor-β signaling[8,31], and proliferation-related PTEN, an inhibitor of the AKT pathway[51]. Positive correlation between miR-21 and fibrosis stage is reported in chronic HCV-infected patients and in a CCl4 mouse fibrosis model[31], and stimulation of the fibrogenic effect by miR-21 is also found in a HSC cell line[51]. In contrast, we did not find any difference or correlation between miR-21 and fibrosis stage or LS values in the present study, but positive correlation of miR-21 with serum ALT values was clearly visible. In our chronic viral hepatitis group, this correlation did not reach the set significance level, which is in partial agreement with data reported in chronic hepatitis C patients[31]. In addition, ALT levels have been found to correlate with serum miR-21 levels of chronic HCV patients, suggesting that miR-21 is an indicator of the extent of necroinflammation in the liver[52].
In the present study, the expression of several hepatocarcinogenesis-related miRNAs was assayed in fibrotic liver biopsy samples of various etiologies and correlated with fibrosis stage (measured by METAVIR) and, to the best of our knowledge, for the first time, with LS values (measured by TE). Reduced miR-122 expression was found in advanced fibrosis as compared with stage F0 and a negative correlation was observed not only with fibrosis stage, but with LS values as well. In addition, we detected a positive correlation between miR-224 and LS values, indicating the role of this oncomiR in advanced fibrosis, indicating a link between fibrosis and HCC. Nevertheless, an ideal staging tool should be able to discriminate not only between mild and advanced stages of fibrosis, but also between intermediate stages of fibrosis. Although this is a reasonable demand, the biologic variances may result in overlaps between the observed intermediate values, especially if the intermediate ranges are small. The main focus of the present study was to analyze fibrotic samples of different etiologic backgrounds. The limitation of our analysis is the small sample size in the various etiologic groups. Therefore, further studies are warranted in order to reveal whether the observed miRNA correlations are also characteristic of the various etiology groups or whether these relationships are only summed characteristics of the fibrosis samples by reason of the various etiologies. In conclusion, the observed negative correlation between fibrosis stage and LS values in case of miR-122 indicates that this molecule could be useful in assessing the severity of fibrosis regardless of etiology.
The authors thank Mrs. Elvira Kálé Rigóné for the English proofreading, and Mrs. Magdolna Pekár, Mrs. Csilla Horváth, and Mrs. Violetta Piurkó for their technical assistance.
Hepatic fibrosis is a wound-healing response of the liver to cellular injury that is characterized by deposition of collagen fibers and contributes to the deterioration of normal liver function. When injury persists, fibrosis may advance into cirrhosis (the most severe stage of fibrosis) and further into hepatocellular carcinoma. Assessment of the stage of fibrosis is important for diagnosis and is predominantly based on liver biopsy. Owing to some limitations, other alternatives have been developed such as transient elastography.
The regulatory role of microRNAs (miRNA) is to fine-tune the expression of genes involved in normal cellular processes, such as development, differentiation, and proliferation. For this reason, it may not be surprising that altered miRNA expression can be found in cancers and in several other pathologies, including liver diseases. Moreover, it has been suggested that an imbalance in the normal miRNA pattern is measurable long before the onset of a disease, indicating that miRNAs may be useful molecules in diagnostics.
There is a need to find indicators that will help predict the progression of a disease. As miRNAs have been reported to show altered expression in fibrosis and have also been suggested to play role in liver carcinogenesis, these molecules may be useful candidates. miRNA expression in relation to fibrosis stage has predominantly been investigated in chronic hepatitis C virus-infected samples. Therefore, in the current study, the authors investigated miRNA expression in samples of diverse etiologies, including autoimmune and chronic viral hepatitis and alcoholic and nonalcoholic liver diseases to find out whether miRNA levels become altered in fibrosis and whether they could be correlated with fibrosis stage and liver stiffness values measured by transient elastography.
The reduced expression of miR-122 observed in stage F4 as compared with stage F0 and the negative correlation of miR-122 levels with fibrosis stage and liver stiffness suggest that miR-122 could be a useful molecule to assess fibrosis regardless of etiology. However, further studies are warranted with larger sample sizes. Furthermore, the staging tool should also be able to discriminate between intermediate stages of fibrosis, although biologic variances themselves may result in overlaps between the intermediate values.
miRNAs are regulating molecules that interfere with gene expression upon binding to the untranslated regions of mRNAs and induce translational arrest, the result being reduced or prevented protein synthesis. Transient elastography is an ultrasound-based examination method for the noninvasive assessment of liver stiffness in fibrosis, with the value increasing with the advancement of fibrosis.
The novelty of the study is the correlation analysis of transient elastography and miRNA expression in samples obtained from chronic liver disease of various etiologies. Although the study examined a limited sample size in each subgroup of fibrosis-related liver diseases, the presented results are in agreement with recent reports that support a strong correlation between decrease of miR-122 levels and severity of fibrosis or increase in liver stiffness. The most significant finding of the study is that miR-122 levels are altered independent of the etiologic cause of liver damage. The presented evidences support the usefulness of miR-122 as an indicator of fibrosis progression.
P- Reviewer: Ning Q, Tripodi M S- Editor: Qi Y L- Editor: AmEditor E- Editor: Liu XM
| 1. | Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 618] [Cited by in F6Publishing: 684] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 2. | Mormone E, George J, Nieto N. Molecular pathogenesis of hepatic fibrosis and current therapeutic approaches. Chem Biol Interact. 2011;193:225-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 3. | Chen SL, Zheng MH, Shi KQ, Yang T, Chen YP. A new strategy for treatment of liver fibrosis: letting MicroRNAs do the job. BioDrugs. 2013;27:25-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Gressner OA, Weiskirchen R, Gressner AM. Biomarkers of liver fibrosis: clinical translation of molecular pathogenesis or based on liver-dependent malfunction tests. Clin Chim Acta. 2007;381:107-113. [PubMed] [Cited in This Article: ] |
| 5. | Bandyopadhyay S, Friedman RC, Marquez RT, Keck K, Kong B, Icardi MS, Brown KE, Burge CB, Schmidt WN, Wang Y. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis. 2011;203:1753-1762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 6. | Haybaeck J, Zeller N, Heikenwalder M. The parallel universe: microRNAs and their role in chronic hepatitis, liver tissue damage and hepatocarcinogenesis. Swiss Med Wkly. 2011;141:w13287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | He Y, Huang C, Zhang SP, Sun X, Long XR, Li J. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268-2272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are Central Players in Anti- and Profibrotic Gene Regulation during Liver Fibrosis. Front Physiol. 2012;3:49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 9. | Fowell AJ, Iredale JP. Emerging therapies for liver fibrosis. Dig Dis. 2006;24:174-183. [PubMed] [Cited in This Article: ] |
| 10. | Bedossa P, Dargère D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. 2003;38:1449-1457. [PubMed] [Cited in This Article: ] |
| 11. | Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273-1281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 332] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 12. | El-Kamary SS, Mohamed MM, El-Raziky M, Shardell MD, Shaker OG, ElAkel WA, Esmat G. Liver fibrosis staging through a stepwise analysis of non-invasive markers (FibroSteps) in patients with chronic hepatitis C infection. Liver Int. 2013;33:982-990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Castéra L, Vergniol J, Foucher J, Le Bail B, Chanteloup E, Haaser M, Darriet M, Couzigou P, De Lédinghen V. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. 2005;128:343-350. [PubMed] [Cited in This Article: ] |
| 14. | Castera L, Bedossa P. How to assess liver fibrosis in chronic hepatitis C: serum markers or transient elastography vs. liver biopsy? Liver Int. 2011;31 Suppl 1:13-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Lupsor M, Badea R, Stefanescu H, Grigorescu M, Serban A, Radu C, Crişan D, Sparchez Z, Iancu S, Maniu A. Performance of unidimensional transient elastography in staging non-alcoholic steatohepatitis. J Gastrointestin Liver Dis. 2010;19:53-60. [PubMed] [Cited in This Article: ] |
| 16. | Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. 2012;3:364-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 17. | Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542-552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 428] [Cited by in F6Publishing: 455] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 18. | Gyugos M, Lendvai G, Kenessey I, Schlachter K, Halász J, Nagy P, Garami M, Jakab Z, Schaff Z, Kiss A. MicroRNA expression might predict prognosis of epithelial hepatoblastoma. Virchows Arch. 2014;464:419-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 164] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 20. | Kwiecinski M, Noetel A, Elfimova N, Trebicka J, Schievenbusch S, Strack I, Molnar L, von Brandenstein M, Töx U, Nischt R. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS One. 2011;6:e24568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 21. | Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759-766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 22. | Ciesla M, Skrzypek K, Kozakowska M, Loboda A, Jozkowicz A, Dulak J. MicroRNAs as biomarkers of disease onset. Anal Bioanal Chem. 2011;401:2051-2061. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 24. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [PubMed] [Cited in This Article: ] |
| 25. | Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10:203-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 179] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 26. | Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245-5250. [PubMed] [Cited in This Article: ] |
| 27. | Anastasiou J, Alisa A, Virtue S, Portmann B, Murray-Lyon I, Williams R. Noninvasive markers of fibrosis and inflammation in clinical practice: prospective comparison with liver biopsy. Eur J Gastroenterol Hepatol. 2010;22:474-480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Gómez-Domínguez E, Mendoza J, Rubio S, Moreno-Monteagudo JA, García-Buey L, Moreno-Otero R. Transient elastography: a valid alternative to biopsy in patients with chronic liver disease. Aliment Pharmacol Ther. 2006;24:513-518. [PubMed] [Cited in This Article: ] |
| 29. | Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189-2204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 217] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 30. | Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 252] [Cited by in F6Publishing: 278] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 31. | Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN, McCaffrey AP. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727-1736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 32. | Morita K, Taketomi A, Shirabe K, Umeda K, Kayashima H, Ninomiya M, Uchiyama H, Soejima Y, Maehara Y. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011;31:474-484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 33. | Kerr TA, Korenblat KM, Davidson NO. MicroRNAs and liver disease. Transl Res. 2011;157:241-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: fad or future of liver disease. World J Gastroenterol. 2011;17:2536-2542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 68] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 35. | Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735-739. [PubMed] [Cited in This Article: ] |
| 36. | Li J, Ghazwani M, Zhang Y, Lu J, Li J, Fan J, Gandhi CR, Li S. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J Hepatol. 2013;58:522-528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 146] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 37. | Zeisel MB, Pfeffer S, Baumert TF. miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. J Hepatol. 2013;58:821-823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Boutz DR, Collins PJ, Suresh U, Lu M, Ramírez CM, Fernández-Hernando C, Huang Y, Abreu Rde S, Le SY, Shapiro BA. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem. 2011;286:18066-18078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Scisciani C, Vossio S, Guerrieri F, Schinzari V, De Iaco R, D’Onorio de Meo P, Cervello M, Montalto G, Pollicino T, Raimondo G. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol. 2012;56:855-861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 40. | Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230-4240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 41. | Callegari E, Elamin BK, Giannone F, Milazzo M, Altavilla G, Fornari F, Giacomelli L, D’Abundo L, Ferracin M, Bassi C. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56:1025-1033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 42. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 542] [Cited by in F6Publishing: 605] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 43. | Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152-1161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 44. | Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651-5661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 466] [Cited by in F6Publishing: 465] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 45. | Yuan Q, Loya K, Rani B, Möbus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 46. | Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600-1609. [PubMed] [Cited in This Article: ] |
| 47. | Fornari F, Milazzo M, Galassi M, Callegari E, Veronese A, Miyaaki H, Sabbioni S, Mantovani V, Marasco E, Chieco P. p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol Cancer Res. 2014;12:203-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Ma D, Tao X, Gao F, Fan C, Wu D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett. 2012;4:483-488. [PubMed] [Cited in This Article: ] |
| 49. | Estebaud E, Bieche I, Lapalus M, De Muynck S, Lada O, Martinot-Peignoux M, Duces A, Valla D, Bedossa P, Marcellin P. Differential liver minRNAS expression in chronic hepatitis C patients at early stages of liver fibrosis. J Hepatol. 2011;54:S315. [Cited in This Article: ] |
| 50. | Lendvai G, Jármay K, Karácsony G, Halász T, Kovalszky I, Baghy K, Wittmann T, Schaff Z, Kiss A. Elevated miR-33a and miR-224 in steatotic chronic hepatitis C liver biopsies. World J Gastroenterol. 2014;20:15343-15350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 24] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 52. | Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, Shi Y, Peveling-Oberhag J, Polta A. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |