Published online Aug 14, 2016. doi: 10.3748/wjg.v22.i30.6800
Peer-review started: March 31, 2016
First decision: May 12, 2016
Revised: June 11, 2016
Accepted: July 6, 2016
Article in press: July 6, 2016
Published online: August 14, 2016
Hepatocellular carcinoma (HCC) is a primary liver cancer, which is one of the most prevalent cancers among humans. Many factors are involved in the liver carcinogenesis as lifestyle and environmental factors. Hepatitis virus infections are now recognized as the chief etiology of HCC; however, the precise mechanism is still enigmatic till now. The inflammation triggered by the cytokine-mediated immune response, was reported to be the closest factor of HCC development. Cytokines are immunoregulatory proteins produced by immune cells, functioning as orchestrators of the immune response. Genes of cytokines and their receptors are known to be polymorphic, which give rise to variations in their genes. These variations have a great impact on the expression levels of the secreted cytokines. Therefore, cytokine gene polymorphisms are involved in the molecular mechanisms of several diseases. This piece of work aims to shed much light on the role of cytokine gene polymorphisms as genetic host factor in hepatitis related HCC.
Core tip: Cytokines have a decisive role in the pathophysiology of many infectious, autoimmune and malignant diseases. Changes in cytokine secretion profile are involved in one way or another in susceptibility to hepatitis B virus (HBV) and hepatitis C virus (HCV) infection and eventualy are thought to influence the disease phenotype and the rapid progression to HCC. Individual variation in cytokine profile might, at least partially, to genetic alteration within the regulatory regions of cytokine genes. One of the most studied phenomena is the allelic polymorphism and its implication in cytokine gene expression, and the susceptibility to infection, clinical severity of diseases and response to treatment. This review concentrates on the cytokine gene polymorphism case-control and meta-analysis studies performed all over the world on hepatocellular carcinoma patients (as a consequence to HBV or HCV infection).
- Citation: Dondeti MF, El-Maadawy EA, Talaat RM. Hepatitis-related hepatocellular carcinoma: Insights into cytokine gene polymorphisms. World J Gastroenterol 2016; 22(30): 6800-6816
- URL: https://www.wjgnet.com/1007-9327/full/v22/i30/6800.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i30.6800
Hepatocellular carcinoma (HCC) is deemed as one of the most life threatening malignancies. It is developed after prolonged and persistent chronic hepatitis infection [hepatitis B virus (HBV) or hepatitis C virus (HCV) infection]. Cytokines represent the key immune-modulatory molecules involved in the defense mechanism against viral infection. Its production is genetically controlled. Variations resulted from single-nucleotide polymorphisms (SNPs) in the cytokine genes’ could extremely affect the cytokine production, which in turn disturbs the all over immune response either to viral infection or in HCC production. In this review article, we summarized the documented population-based variability in cytokine gene polymorphisms, that might implicate in hepatitis infection and participate in the development of HCC.
HCC is a global health dilemma. It is the third prevalent cancer among humans with a high rate of mortality[1,2]. It is the primary liver cancer as it represents 83% of liver cancers[3-5]. It mostly occurs in patients with chronic liver diseases like cirrhosis, which is primarily a set of the hepatotropic viruses as HBV or HCV. Therefore, the highest rate of HCC incidence is high in areas with high endemic level of hepatitis viruses as in African and Asian countries[6], on contrary to Europe and United State of America in which there is a low incidence rate of hepatitis infections[1,7-9]. World health organization suggested that HCC is assumed to be in a growing till 2030 which re-ranks HCC to be the second prevalent cancer among humans[10].
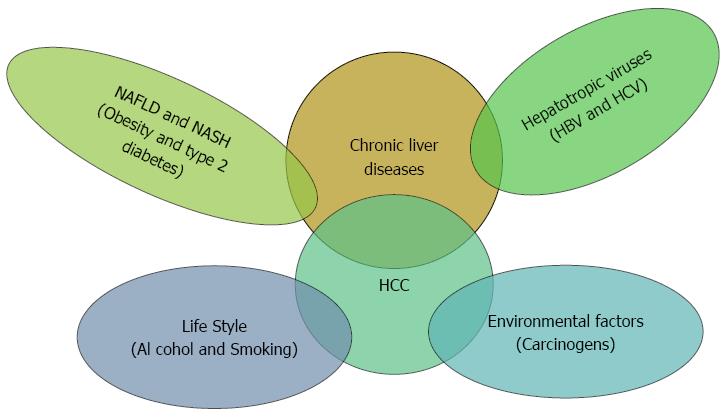
HCC is a multistep and multifactorial process (Figure 1) as there are many factors involved like environmental factors, alcohol abuse, carcinogens as aflatoxins, obesity-related hepatitis, and hepatotropic chronic viral infections[11-14]. However, there is no individual cause of HCC[15] indicating to the implication of other unknown factors such as the host genetic elements. Therefore, it is highly important to give a closer look at the molecular level of HCC development to attain a better understanding of the underlying intricate mechanisms of the hepatocarcinogenesis.
Hepatitis is a critical player in the initiation, promotion, and progression of HCC. HBV and HCV were reported to be the commonest risk factors of HCC[16-19]. Nearly 70%-80% of HCC cases are attributed to hepatitis viruses[2]. Generally, HCC is more prevalent among HBV patients than HCV ones as 75%-85% of HCC patients have chronic HBV infection[1,7,20]. Both HBV and HCV patients develop liver cirrhosis in which 20% of cirrhotic patients develop HCC. On the other hand, only a minor fraction of hepatitis viruses’ carriers develops HCC in their lifetime, indicating to the presence of other cofactors involved in liver carcinogenesis[5,21]. The exact mechanism of hepatitis-related HCC is still controversial and unknown at the mean time as there is a plethora of intracellular and extracellular factors involved in the process of carcinogenesis. To date, one of the most implicated factors in carcinogenesis is the inflammation, which stimulates angiogenesis, DNA damage, and malignant tumor cell growth[22,23]. Interestingly, inducible nitric oxide synthase was reported to be the culprit of disease progression and HCC development among HCV patients through induction of nitric oxide production[24-26]. Inflammation is highly dependent on the potential of the immune response mediated by the cytokines, in which strong immune response induces chronic hepatitis and weak response can be an oncogenic cofactor in HBV or HCV carriers as the persistent infection in the hepatic microenvironment promote the progression of HCC[7,20,27,28].
Cytokines, what are they? Cytokines are immunomodulatory glycoproteins, which orchestrate the immune response and modulate the activities of the immune cells. Cytokines are produced by a wide range of cell types whether immune and non-immune cells. Cytokines in the terms of ligands bind to specific receptors promoting the signal transduction within the target cell. The transducer signals activate key genes responsible for mitotic cell division, growth and differentiation, migration, or even apoptosis. In addition, they play a critical role in modulation of the innate and adaptive immune systems during viral infections and direct inhibition of viral replication. Cytokines are classified according to many criteria as the producing cells, the function, the receptors and the target cells. They are classified into monokines (monocyte lineage) or lymphokines (lymphocytes) according to the producing cells[29-31]. They are also classified according to the function into T helper (Th)1, Th2, and Th17 cytokines. Th1 cells secrete proinflammatory cytokines as IL-2, IFN-γ and TNF-α. Otherwise, Th2 cells secrete IL-4, IL-5, IL-10 and IL-13. Th17 cells produce IL-17, IL-17F and IL-22, all of which regulate inflammatory responses of tissue cells. Overproduction of Th2 cytokines typically promotes B-cell hyperactivity and humoral immune responses, whereas T cell hyperactivity and inflammation frequently associate with an excess of Th1 and Th17 cytokines. According to the criterion of classification, cytokines are classified into either pro-inflammatory as IL-1, IL-6 and TNF-α or anti-inflammatory as IL-4 and IL-10 in addition to other cytokines which have pleiotropic roles in the immune response such as TGF-β1[32,33].
In the hepatic microenvironment, cytokines play an important role in the defense against hepatitis viruses and immunopathogensis of hepatitis infections. They are prominently involved in the liver damage during HCV infection, while they have been reported in previous studies to be determining factor of the HBV infection’s outcome. Proinflammatory cytokines as IL-1β and TNF-α were reported to be elevated among hepatitis patients. HCV-specific T cells mediate ongoing viral clearance and liver injury by either directly killing infected hepatocytes or releasing pro-inflammatory cytokines, such as IFN-γ and TNF-α. Increased mRNA levels of IFN-γ and TNF-α are found in the livers of chronic HCV-infected patients. Importantly, the expression of these pro-inflammatory cytokines correlates with both the extent of hepatic inflammation and the development of fibrosis[34-38].
The resolution of hepatitis virus infections through inhibition of viral replication is associated with enhanced CD8+ T-cells response which is mediated by Th1 cytokines. Thus, the alterations in Th1 cytokines are involved in the liver injury and fibrosis resulted from chronic inflammation, which leads to enhanced remodeling of the hepatic tissue due to subsequent regeneration, subsequently proto-oncogenes activation leading to HCC development[39-43]. HCC development is a complex and a multidimensional process so there are many theories about the involvement of cytokines in the HCC. Nevertheless, Cytokines were reported to suppress the tumor formation through controlling the inflammatory response against infection[44]. Cytokines are traditionally attracted of inflammatory cytokines, which may be the first step in inflammation-based carcinogenesis, in addition cytokines may trigger the hepatocarcinogenesis itself through growth signaling, angiogenesis and invasive metastasis[45-47]. Previous studies reported higher levels of different cytokines in HCC patients as IL-1β, IL-6, IL-10, IL-15, IL-18, TNF-α, IFN-α, IFN-γ, and TGF-β1[48-55], which may represent a mounting proof of the involvement of cytokines in liver carcinogenesis. Therefore, looking in depth at the molecular levels of hepatocarcinogenesis may give a clear view of what actually happens during hepatitis-related HCC.
DNA sequence between two randomly opted human genomes is 99.9% identical. The other 0.1% represents polymorphic regions that differ among individuals and occurs at a frequency of at least 1% in population so it is different from the mutations. Gene polymorphisms are characterized by the presence of multiple alleles for a given gene due to alterations in the nucleotide sequence. The polymorphisms take the following main forms: SNPs, variable number of tandem repeats (VNTRs) and microsatellites also known as Simple Sequence Repeats or short tandem repeats (STRs)[44,45]. The genes of cytokines and their receptors have been shown to be highly polymorphic. So what can such variations affect the cytokines and their function? The answer comes to surface when gene polymorphisms were found to influence the expression of cytokines controlling the immune response which open the gate to understand the intricate mechanisms of diseases at the molecular level. In recent years, several studies have demonstrated that cytokine gene polymorphisms correlated with susceptibility to, or protection from, disease development[56,57].
The polymorphisms of cytokine and cytokine receptor genes occur either in the coding or the non-coding regions. The genetic polymorphisms within the coding region that lead to amino acid substitution are not common, on the other hand, the genetic polymorphism are so common in the non-coding region as the promoter, intron and the untranslated regions. There is a growing evidence of that cytokine production capacity is genetically determined by the cytokine gene polymorphisms either in coding or non-coding regions. Given that the functional consequences of polymorphisms may contribute to the establishment of a given disease phenotype, reports have focused on discovering associations between specific gene polymorphisms and disease status with the main target of identifying susceptibility groups[29,58,59]. Many reports focused on the genetic polymorphisms of cytokines as host genetic factors in hepatitis-related HCC as they may clear the complexity of HCC occurrence due to HBV or HCV as etiological factors (Table 1). In addition to disease development, polymorphisms in the cytokine genes have been reported to be significantly associated with response to therapies and spontaneous HCV/HBV clearance[5,60-62].
| Cytokine Gene | Polymorphism | Risk factor (Ref.) | Protective factor (Ref.) | No association with HCC (Ref.) |
| IL-1β | -511T/C | Hirankarn et al[73] 2006 | ||
| Wang et al[75] 2003 | ||||
| Tanaka et al[76] 2003 | ||||
| -31C/T | Wang et al[75] 2003 | |||
| IL-1RN | Intron 286 bp | Chen et al[65] 2005 | ||
| Allele 2 VNTR | Saxena et al[74] 2013 | Zhang et al[71] 2004 | Wang et al[75] 2003 | |
| TNF-α | -1031T/C | Wei et al[87] 2011 | ||
| -863C/A | Wei et al[87] 2011 | |||
| -857C/T | Wei et al[87] 2011 | |||
| -308G/A | Akkiz et al[78] 2009 | Wilson et al[82] 1992 | ||
| Shin et al[2] 2015 | Yang et al[81] 2009 | |||
| Jeng et al[79] 2009 | ||||
| Talaat et al[80] 2012 | ||||
| Wei et al[87] 2011 | ||||
| -238G/A | Wei et al[87] 2011 | |||
| IL-6 | -597G/A | Tang et al[1] 2014 | ||
| Saxena et al[103] 2014 | ||||
| Chang et al[104] 2015 | ||||
| -572G/C | Park et al[105] 2003 | Tang et al[1] 2014 | ||
| Saxena et al[103] 2014 | ||||
| Chang et al[104] 2015 | ||||
| -174G/C | Ognjanovic et al[11] 2009 | Falleti et al[101] 2010 | Nieters et al[100] 2005 | |
| Terry et al[102] 2000 | ||||
| Giannitrapani et al[27] 2011 | ||||
| IL-10 | -1082G/A | Migita et al[119] 2005 | Bouzgarrou et al[121] 2009 | |
| Tseng et al[120] 2006 | ||||
| Saxena et al[103] 2014 | ||||
| -819T/C | Migita et al[119] 2005 | Bouzgarrou et al[121] 2009 | ||
| Tseng et al[120] 2006 | ||||
| Saxena et al[103] 2014 | ||||
| TGF-β1 | -509C/T | Grainger et al[148] 1999 | Wei et al[153] 2012 | Wang et al[131] 2005 |
| Ma et al[5] 2015 | Qi et al[140] 2009 | Shi et al[151] 2012 | ||
| +869T/C | Ben-Ari et al[145] 2003 | |||
| Ribeiro et al[146] 2007 | ||||
| Migita et al[119] 2005 | ||||
| Xin et al[147] 2012 | ||||
| IL-18 | -607A/C | Karra et al[175] 2015 | Zhang et al[177] 2005 | |
| Bouzgarrou et al[167] 2008 | Hirankarn et al[73] 2006 | |||
| -137C/G | Zhang et al[177] 2005 | |||
| Hirankarn et al[73] 2006 | ||||
| 148G/C | Kim et al[178] 2009 | |||
| IL-16 | rs11556218T/G | Li et al[20] 2011 | ||
| rs4072111C/T | Li et al[20] 2011 | |||
| rs4778889 | Romani et al[196] 2014 | |||
| IL-13 | rs20541G/A | Deng et al[201] 2015 | ||
| IFN-γ | +874T/A | Nieters et al[100] 2005 | Dai et al[212] 2006 | Barrett et al[213] 2003 |
| Bouzgarrou et al[121] 2009 | Abbas et al[214] 2005 | |||
| IL-2 | +114 T/G | Peng et al[13] 2014 | ||
| Wang et al[222] 2015 | ||||
| IL-27 | -954A/G | Peng et al[14] 2013 | ||
| 2905T/G | ||||
| IL-4 | 2590C/T | Lu et al[227] 2014 | ||
| 233C/T | Wu et al[228] 2015 | |||
| IL-12A/IL-12B | rs3212227A/C | Saxena et al[4] 2014b | ||
| rs2243115T/G | Nieters et al[100] 2005 | |||
| Liu et al[7] 2011 | ||||
| rs568408G/A | Liu et al[7] 2011 | |||
| IL-21 | rs13143866 | Wu et al[228] 2015 | ||
| rs2221903 | ||||
| rs907715 | Wu et al[228] 2015 | |||
| IL-17A/IL-17F | rs4711998 | Xi et al[229] 2015 | ||
| rs2275913 | ||||
| rs763780 |
IL-1 gene family is located on chromosome 2q13 spanning a region of 430 kb, consists of IL-1α and IL-1β in addition to IL-1 receptor antagonist (IL-1RN). IL-1α and IL-1β are potent pro-inflammatory which produced by macrophages and their inflammatory mediated response is inhibited by IL-1RN and the acute phase protein[63,64]. The genes of this family have been reported to be highly polymorphic in which the polymorphisms modulate IL-1β production. The most studied SNPs of IL-1 gene family are IL-1β -511T/C, -31C/T and +3593C/T, in addition to the 86bp of VNTR in intron 2 of IL-1RN gene which has 5 different alleles (1/1, 1/4 and 1/3 and 1/2 and 2/2, allele1 with 6 repeats, allele2 with 4 repeats, allele3 with 5 repeats)[65]. IL-1β-31C/T is located in TATA box which reported to affect the DNA-protein interactions as it is associated with increased binding to transcription initiation factor. It was reported to be in strong linkage disequilibrium (LD) with IL-1β -511T/C. Those genetic polymorphisms were suggested to be involved in the hepatocarcinogenesis[66-68]. Chen et al[68] reported that the synergistic interaction between IL-1β -511T allele and -31C allele leads to increase of IL-1β production. Both alleles were reported to be associated with high level of IL-1β in the plasma of chronic HBV patients[69-71].
IL-1RN-allele2 was also reported to be linked to increased IL-1β production[72]. In the study of Chen et al[65] allele2 was found to be a risk factor for HBV-related HCC in the presence of IL-1β (-511CC and -31TT) genotypes. Nevertheless, both genotypes did not be risk factors for HCC in the absence of allele2 suggesting the synergistic effect of them on the production of IL-1β. The IL-1β -511C allele was found to be a risk factor while IL-1RN was not a risk factor for HBV-related HCC[73]. On the other hand, several studies found no association between IL-1β -511C allele and HBV-related HCC. Saxena et al[74] stressed on the importance of IL-1RN allele 1 and 2 as a risk factor for HCC. On contrary, Zhang et al[71] reported that allele 2 isn’t a risk factor for HBV-related HCC. In the study by Wang et al[75] IL-1β -31TT genotype and IL-1β -511/-31CT haplotype were found to be associated with an increased risk for HCV-related HCC and the IL-1RN VNTR polymorphism was not a risk factor. This is also demonstrated by Tanaka et al[76] as they found that IL-1β -511TT genotype was a possible risk factor in HCV-related HCC. Thus, some studies promote the role of IL-1β in HCC incidence as it is associated with high levels of IL-1β which reported to be determined genetically. In addition, IL-1β-31C/T and IL-1β-511T/C are suggested to be modulating factors of IL-1β production, therefore they may be candidate genetic factors in the liver hepatocarcinogenesis and they need to be further investigated to verify them as cofactors in hepatitis-related HCC.
TNF-α is a proinflammatory cytokine that affects the growth, differentiation, and survival of various cells. It is mainly produced by macrophages, in addition to other array of cells, including neutrophils, keratinocytes, mast cells, endothelial cells, neurons, NK cells, fibroblasts, T and B lymphocytes and tumor cells[77]. TNF-α has been reported to promote chronic inflammation-related carcinogenesis. TNF-α gene is located on chromosome 6p21.3 of the major histocompatibility complex class III region. The most studied polymorphisms in TNF-α gene lies in the promoter region TNF-α (-1031T/C, -863C/A, -857C/T, -308G/A and -238G/A). These polymorphisms have been reported to modulate the production of TNF-α[2,78].
TNF-α-308G/A is considered a critical risk factor for liver carcinogenesis[79,80], but TNF-α-238G/A was found to have a passive role in HCC risk[81]. TNF-α-308GG genotype has been reported to be associated with a decrease of HCC incidence[82]. On the other hand, other studies reported higher risk of HCC in patients carrying TNF-α-308A allele[78]. Many studies reported the correlation between TNF-α promoter alleles as TNF-α-308A and TNF-α-238A and the different capacity levels of TNF-α production[83,84]. Additionally, it was reported that the TNF-α promoter alleles as TNF-α-308A and TNF-α-238A which were correlated with high TNF-α levels and the resolution of HBV infection, suggesting the protective roles of them[85,86]. In the meta-analysis study of Wei et al[87], they found that TNF-α (-308G/A, -238G/A, and -863C/A) polymorphisms are associated with the risk for HCC. On the other hand, -857C/T and -1031T/C polymorphisms weren’t associated with risk to HCC. In a meta analysis of Shin et al[2] no single TNF-α polymorphisms (-1031 T>C, -863 C>A, -857 C>T, -308 G>A, and -238 G>A) are associated with HCC cases with HBV infection.
Haplotype studies also revealed some surprising results concerning the risk of HCC. The homozygous haplotype (CGG/CGG) of the TNF-α (-863A/C, -308G/A and -238G/A) was reported to be protective against HCC[88], while, TCGG haplotype of the TNF-α (-1031T/C, -863A/C, -308G/A and -238G/A) was associated with HCC[89]. Additionally, GGCCT haplotype of the TNF-α (-238G/A, -308G/A, -857C/T, -863A/C and -1031T/C) was reported to be associated HBV clearance[90]. Therefore, the haplotypes containing GG represent protection factors against HCC, while AG of -308G/A and -238G/A implies high risk for hepatitis-related HCC[91].
IL-6 is a pluripotent cytokine which is a central player in regulating several cellular processes as proliferation and differentiation in addition to its pivotal role in the acute phase response. It is also the control balance valve between proinflammatory and anti-inflammatory responses[92]. IL-6 is reputed to be the lost ring between the inflammation response and liver carcinogenesis[93] as IL-6 was reported to inhibit apoptosis and induce metastasis[1,41,94]. It has also been reported to be the main player at the first stage of the immune response, whereas IL-6 promotes the secretion of acute phase proteins and the proliferation of lymphocytes[95]. Moreover, IL-6 was found to be associated with liver fibrosis and cirrhosis[96]. Therefore, IL-6 is a chief player in the viral infection and the counteracting inflammatory response. IL-6 gene, located on chromosome 7p21 with 5kbp length, has five exons interspersed with four introns[97]. The most studied genetic polymorphisms in IL-6 gene lie in the promoter region (-597G/A, -572G/C, -174G/C, and -373A/T). These SNPs, which are in a strong linkage disequilibrium, has a heavy impact on the production capacity of IL-6 at the transcriptional level[1,62,98] and they have been reported to be associated with chronic hepatitis[27,93,99].
IL-6 -174G/C has been extensively studied; however, the results are still controversial concerning its function in hepatitis-related HCC. Nieters et al[100] study reported no association between IL-6 -174G/C and the risk to HCC while a study by Ognjanovic et al[11] found that IL-6 -174GG showed a great inclination to HCC risk. IL-6 -174CC has been reported to coincide with low IL-6 levels, which is associated with protection against HCC in cirrhotic patients[101]. Another study reported that IL-6-174GG correlated with high levels of IL-6 while IL-6-174CC is correlated with low levels of Il-6 among HCC patients, which suggest the risk of HCC in G allele carriers[27,102]. IL-6 (-572G/C and -597G/A) were found to be not associated with hepatitis-related HCC[103]. In the meta-analysis of Chang et al[104] (2015), IL-6 (-572G/C, -597G/A and -174G/C) might not be significantly correlated to the progressive HBV to HCC. On the other hand, IL-6 -572G/C was found to be associated with the risk of HBV-related HCC in a study on Korean population[105].
There is a great disparity in the correlation between IL-6 gene polymorphism and hepatitis-related HCC according to the surveyed literature.
IL-10, an immunosuppressor cytokine produced by macrophages, down regulates Th1 cytokines such as IL-1, IL-6 and TNFα[106,107] and it plays a role in the compromised host immune response during chronic viral infections[34,108]. Its production has been reported to be controlled at the genetic level[109]. IL-10 (-1082G/A, -819T/C and -592A/C) are the most studied SNPs of the IL-10 gene as they have been reported to have a deep impact on the production of IL-10 leading to different production capacities among individuals. That effect can be attributed to that IL-10 -1082A/G and -592A/C and their haplotypes mediate the expression of IL-10 by differences in the nuclear binding activity[110,111]. The reports suggested that -1082G allele and GCC haplotype were correlated with high expression of IL-10, however, ATA haplotype was correlated with lower expression levels of IL-10. On the contrary, other studies reported that IL-10GCC and AAGCC haplotype of IL-10 (-3575, -2763, -1082, -819, and -592) were correlated with lower expression levels of IL-10[112,113]. Therefore, there are conflicting results which may be attributed to ethnic variations and those studies were conducted in different populations. Several studies have reported the correlation between IL-10 gene promoter polymorphism and progression of HBV[114], HCV[115,116], chronic liver diseases[117,118] and hepatitis-related HCC[103,119,120]. On the other hand, other reports suggest no association between IL-10 gene promoter polymorphism and hepatitis[121].
TGF-β1, produced by many cell populations as T-cells and platelets, hepatic stellate cells and hepatocytes in the liver, is a pluripotent multifunctional cytokine involved in a plethora of cellular processes[122,123]. It regulates cell growth, differentiation and induces the fibrosis, though up regulation of collagen and extracellular matrix[124,125]. Moreover, it has the ability to inhibit B-cells and T-cells in addition to its anti-inflammatory action by down regulation NK cells and Th1[86,126]. It normally inhibits cell growth, but in tumor cells, it promotes the progression of the tumor by suppressing the immune response[127-129]. TGF-β1 was reported to be a versatile player in the liver, whereas it is involved in many physiological processes[130] and many studies focused on the correlation between TGF-β1 and chronic liver diseases as liver fibrosis[131-133]. TGF-β1 gene, containing 7 exons separated by 6 introns, lies on chromosome 19q13.1-13.3. The production capacity of TGF-β1 has been reported to be genetically controlled[127,134,135]. Many genetic polymorphisms have been reported either in the coding or non-coding regions. The most studied polymorphisms were in both regions, whereas TGF-β1-988C/A, -800G/A, -509C/T and in an insertion/deletion at position 72 in the 5’UTR region of intron 4, in addition to +869T/C (Codon 10), +915G/C (Codon 25) in exon 1 and +788C/T (codon 263) in exon 5[136].
In the coding regions, the variations cause non-synonymous change in the amino acid sequence of the signal peptide. While codon 10 variation causes the exchange of leucine with proline, codon 25 causes the exchange of arginine with proline, and codon 263 variation causes the exchange of threonine with isoleucine[137,138] which lead to the formation of hydrophobic signal sequence and subsequently cause alternation in the export efficiency of the TGF-β1 pre-pro-protein through the endoplasmic reticulum. The correlations between chronic liver diseases and TGF-β1 gene polymorphisms have been studied extensively[119,131,139-141]. TGF-β1 (+869T/C and +915G/C) were reported to be effective SNPs[142,143] and TGF-β1 +869T allele was reported to be correlated with high levels of TGF-β1[137,138]. There are some other reports that found no association between +869T/C and TGF-β1 levels among hepatitis patients[119,139,144]. However, some studies reported the negative role of TGF-β1 +869T/C in hepatitis infection[145,146], other studies reported the association between +869CC genotype and the reduced risk of HBV-related HCC[119,147].
TGF-β1-800G/A hasn’t been studied with hepatitis-related HCC but it was reported that TGF-β1-800A allele may reduce the binding affinity of cAMP-responsive element binding protein and subsequently lower TGF-β1 levels. On the other hand, TGF-β1-509C/T has been studied with hepatitis-related HCC. It is reported that -509T allele correlates with high levels of TGF-β1 and high transcriptional activity more than -509C allele[148,149]. Moreover, Grainger et al[148] reported that TGF-β1-509TT genotype correlates with higher TGF-β1 levels than the genotypes -509CT or -509CC and this was attributed to that -509C/T lies within the consensus transcription factor binding site Yin Yang 1 regulatory region[150]. While the genotype TGF-β1-509TT was reported in Ma et al[5] meta analysis to be associated with the risk for HCV-related HCC, the TGF-β1-509CC was reported to be associated with the risk for HBV-related HCC[140]. On the other hand, Wang et al[131] reported that TGF-β1-509C allele was associated with the progression of Liver cirrhosis and TGF-β1-509T allele wasn’t associated with the risk to HCC[151]. TGF-β1-509T allele was reported to be correlated with high levels of TGF-β1[152], while Qi et al[140] reported that TGF-β1-509C allele was correlated with high levels of TGF-β1 in HBV-related HCC. Controversially, some studies reported that the genotypes -509TT and -509CT were associated with reduced risk to HCC[85,140,153].
IL-18, mainly produced by the activated macrophages and kupffer cells[64], is a proinflammatory cytokine which has been reported to be involved in liver injury[41,154]. It is also reported that IL-18 mediates cellular immune response against viral infections[155,156]. It was reported to be up regulated during hepatitis infection and liver cirrhosis as well[157,158]. Therefore, it may be able to inhibit HBV and HCV replication. The antiviral activity of IL-18 was attributed to its ability to induce NK cells and T-cells to produce IFN-γ and IFN-α/β in the hepatic microenvironment so, IL-18 is frequently called IFN-γ-inducing factor[41,158,159]. IL-18 activates cellular immune response via Fas ligand and perforin-mediated T-cell and NK-cell cytotoxicity. Additional ly, it acts synergistically with IL-12 to induce Th1 polarization which up regulates the production of IFN-γ, IL-1, IL-6 and TNF-α in addition to its efforts to decrease IL-10 production[160-162] and it was found to promote chemokines production as IL-8. IL-18 was reported to mediate anti-tumor immune response and it may regress liver cancer, though cytotoxic T-cells and NK cells[163,164]. High levels of IL-18 were reported to be correlated with HCC[165-167].
IL-18 gene is located on chromosome 11q22.2-q22.3. The most studied SNPs in IL-18 gene are -607A/C and -137C/G which were reported to be in a strong LD[168]. These SNPs was reported to affect the expression levels of IL-18[169,170], as -137C allele disrupt H4TF-binding site and -607C/A disrupt cAMP responsive element binding site. Additionally, the haplotype AC was reported to be associated with reduced levels of IL-18[171,172]. IL-18 gene promoter polymorphisms were reported to be associated with HBV and HCV infections in addition to HCC[173-175]. In a recent study, -607AA genotype and -607A allele were reported to be protective among HBV patients[175]. Moreover, -607CC genotype and -607C allele were reported to be associated with increased risk to the progression of the infection to cirrhosis and HCC development[167], while -607AA genotype was associated with reduced risk to HCC progression[175,176]. On the contrary, it was found that IL-18 -607AA genotype is associated with risk to disease progression while IL-18 -137C allele was reported to be a protective allele[41,177]. On the other side, IL-18 -148G/C has also been studied in which IL-18 -148C allele was found to be correlated with low levels of IL-18[178], which gives rise to the risk of HCC. These controversial assert that more studies needed to assess the precise role of IL-18 gene promoter polymorphism in the progression of HBV and HCV infections.
IL-16 is a chemotactic proinflammatory cytokine produced mainly by the activated CD8+ T-cells, mast cells and B-cells in addition to dendritic cells (DCs) and neural cells in addition to epithelial cells during inflammation[179,180]. It is involved in many cellular functions which is formerly known as a lymphocyte chemoattractant factor[181]. It activates many immune cells as DCs, Monocytes, macrophages and T-cells, through binding to CD4[182,183]. It induces the production of Th1 cytokines as IL-1β, IL-6 and TNF-α by monocytes[184,185]. IL-16 also triggers a cellular mediated immune response against viral infections[180,186]. IL-16 is also able to upregulate CD25 and IL-4 during the course of viral infections[184,185]. On the other side, IL-16 was found to activate many cellular pathways as p56lck tyrosine kinase, the stress-activated protein kinase, and the p38 mitogen-activated protein kinase[187,188]. Moreover, IL-16 is reported to play a role in carcinogenesis through activation of signal transducer and activator of transcription[189]. All of these reports assert on the role in HBV and HCV infection in the liver carcinogenesis.
IL-16 gene is located on chromosome 15q26.3 with seven exons interrupted by six introns and it is transcribed into two versions lymphatic and neural IL-16[190,191]. SNPs of the IL-16 gene coding region was reported to affect IL-16 production and function as IL-16 (rs11556218T/G) in exon 6 which causes the exchange of asparagine with lysine, and IL-16 (rs4072111C/T) which leads to exchange of proline with serine in addition to IL-16 (rs4778889T/C) at the position of -295 in the gene promoter which reported to affect the transcriptional level of IL-16[192-195]. The studies on hepatitis-related HCC and IL-16 gene polymorphisms are so far scanty so there an urgent need for further studies. In a previous study, IL-16 (rs11556218T/G), IL-16 (rs4072111C/T) and IL-16 (rs4778889T/C) were studied in HBV patients and the data showed that genotypes IL-16 (rs11556218TG and GG) and allele IL-16 (rs11556218G) are associated with high risk of HBV-related HCC. In addition, they found that IL-16 (rs4072111TT) was also associated with high risk for HCC among HBV patients[20]. In another study, IL-16 (rs4778889CC) genotype was reported to be associated with the progression of HBV infection[196].
IL-13 is a pleiotropic Th2-derived cytokine which is involved in inducing apoptosis, B-cells and mast cell proliferation, and anti-inflammatory responses through inhibition of Th1 cytokines and chemokines as IL-8. The crucial roles of IL-13 have been studied in tumors as it was reported that IL-13 was over expressed in tumor tissues and targeting IL-13 in cancer may have a potent role in cancer immunotherapy[197,198].
IL-13 gene is located on chromosome 5q31 with four exons[199-201]. IL-13 gene polymorphisms were reported to affect the IL-13 levels as IL-13 -1112C/T, -1512A/C and -1024C/T, and in the coding region IL-13 +2044G/A (in which arginine is replaced by glycine) and IL-13 +130G/A. Moreover, it was found that -1024C/T may affect the binding of STAT transcription factors which profoundly affect the expression levels of IL-13 in the activated T-cells[202,203]. IL-13 gene polymorphisms have been scarcely studied in hepatitis-related HCC but in a recent study by Deng et al[201] two SNPs rs1800925C/T and rs20541G/A have been studied in HBV and HCC patients. They found a significant correlation between rs2054G/A and HBV-related HCC. Moreover, they found that GA genotype was significantly associated with reduced risk for HCC, in addition to the haplotype TG which has a protective role against HCC and the haplotype CA which has an increased risk to liver carcinogenesis. Therefore, more studies are needed to assess the role of IL-13 gene polymorphisms in the hepatitis-related HCC as no conclusion can be addressed from the scrape of studied on IL-13 and HCC.
Viral infections as hepatitis viruses induce the production of IFNs through the innate immune response. There are three types of IFNs, Type I (IFN-α/β), Type II (IFN-γ) and Type III (IFN-λ, IL-28A/B and IL-29) which induce IFN-stimulated genes (ISGs) during the infection course[204]. ISGs are involved in antiviral immune response, inflammation and apoptosis[205]. IFN-γ is Th1 antiviral cytokine and it mediates inflammation and it was found to be associated with hepatitis-related HCC[206,207]. It was reported to be anti-fibrogenic cytokine, however, it is suggested to be involved in liver fibrosis[43,207]. The imbalance between Th1 and Th2 responses may be mediated by the genetic polymorphisms of both IFN-γ and IL-10 genes which affect the outcome of the hepatitis infection[123,208].
IFN-γ gene polymorphisms were studied primarily in hepatitis infections and HCC. IFN-γ +874T/A (rs2430561), +2109C/A and the microsatellite CAn repeat were the most studied polymorphisms[9,209]. IFN-γ +874T allele and +874TT genotype were reported to be in complete LD with CA12 repeat[210]. IFN-γ gene polymorphisms were found to affect the IFN-γ levels in which +874T allele correlates with high expression levels of IFN-γ[211]. On the other side, +874T allele was reported to be associated with the risk of progression of hepatitis infection, in addition, both homozygous and heterozygous genotypes IFN-γ (+874TT and +874TA) was reported to be associated with the risk to liver cirrhosis and HCC incidence among HCV patients[121,212]. On the contrary, +874A allele and +874AA genotype were reported to be associated with the risk of HBV-related HCC incidence[100]. Other reports suggest no correlation between IFN-γ +874T/A and chronic liver diseases[213,214]. The meta-analysis of Sun et al[215] (2015) indicated that there is no significant correlation between elevated risk of HBV-infected HCC and IFN-γ +874A/T polymorphism. Gene polymorphisms in Type III IFNs were mainly focused on the response to IFN therapy[216], as there is not many specific studies focus on the effect of this polymorphism on the chronic hepatitis infections and its progression to HCC. The meta-analysis of Zhou et al[217] (2015) pointed that the IFN-γ +874A/T polymorphism might correlate to HBV related HCC risk.
IL-2 is a cytokine produced by the activated CD4+ and CD8+T-cells and DCs. IL-2 plays a central role in the immune response and its regulation as it stimulates the proliferation and differentiation of NKs. Moreover, it stimulates the production of TGF-α and IFN-γ[13,217]. IL-2 has been reported to be a suspected player in the inhibition of the carcinogenesis and the immune response against tumors, therefore it was suggested to be a candidate for cancer immunotherapy[217]. IL-2 gene has five exons with five introns and it is located on chromosome 4q26-q27. IL-2 gene polymorphisms have been found to affect the levels of secreted IL-2 and those polymorphisms have been reported to be associated with a wide range of cancers[218-221]. HCC risk and IL-2 gene polymorphisms has been scarcely studied. IL-2 +114T/G has been studied by Peng et al[13,14], they documented the increased association between IL-2 +114GG genotype, IL-2 +114G allele and HBV-related HCC[222].
IL-27 is a cytokine of the IL-12 family. IL-27 is secreted after APCs activation, which lead the induction of Th1 response and IFN-γ secretion. Recent reports suggested that IL-27 induce CTLs response which impair cancer progression and induce IFN-γ inducible protein-10 (IP-10) which impair the angiogenesis process[223-225]. IL-27 gene consists of five exons and it is located on chromosome 16p11. There are few studies on the IL-27 gene polymorphism and the hepatitis-related HCC risk. Ali et al[226] reported that IL-27 (-964A/G, 2905T/G and 4730T/C) gene may not contribute to HBV infection. Peng et al[14] studied the association between HBV-related HCC and IL-27-954A/G and IL-27 2905T/G, they found that there is no association between the hepatitis-related HCC and IL-27 gene polymorphism.
IL-4 is chiefly Th2 pleiotropic cytokine which is produced by a variety of cells as mast cells, basophils and eosinophils. IL-4 gene is located on chromosome 5q31 and the gene was reported to have polymorphisms as 2590C/T (rs2243250), 2979G/T (rs2227284), 233C/T (rs2070874), and +3437C/G (rs2227282)[227,228]. Few studies have been conducted on the role of IL-4 gene polymorphisms in the HCC. In meta-analysis of Lu et al[227], IL-4 gene polymorphisms may have a role in hepatitis-related HCC as 2590C/T and 233C/T which are linked with increased risk for HBV-related HCC and IL-4 -590C/T which has been described to be associated with HBV progression to cirrhosis[4] and HBV-related HCC[228].
IL-12 is Th1 cytokine produced by APCs. It activates NKs and CTLs in addition to the induction of IFN-γ production. The critical role of IL-12 in Th1 immune response suggest that IL-12 may have a role in the immune response against cancer cells[7,13]. IL-12 gene polymorphisms are still unknown for its role in hepatitis-related HCC. While some studies reported that IL12B rs3212227A/C and IL12A rs2243115 T/G have been reported to have no role in HBV-related HCC[7,100], Saxena et al[4] reported that IL12B rs3212227A/C has a suspicious role in HCC, which may have attributed to the ethnic variations. On the other hand, Liu et al[7] reported that IL12A rs568408 G/A may be a genetic element in HBV-related HCC.
Several association studies have been performed on cytokine gene polymorphism and hepatitis related HCC. They vary considerably between different populations studied. Due to differences in sample size, the ethnicity of the study population, the disease stage, and even the genotyping method, it is difficult to conclude definite associations based on the available data. Moreover, a thorough understanding of host-virus interactions, which may or may not end up with HCC is still in progress. Several important biological and clinical pertinent questions and their correlation with documented cytokine gene polymorphisms still need to be addressed. This is of maximum importance not only for a better understanding of liver malignancy but also for global viral hepatitis immunobiology. Thus, an integrated work, including larger set of individual s in various sets of populations is required.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Ning BT, Shalaby SM, Utama A, Yin YH S- Editor: Yu J L- Editor: A E- Editor: Ma S
| 1. | Tang S, Yuan Y, He Y, Pan D, Zhang Y, Liu Y, Liu Q, Zhang Z, Liu Z. Genetic polymorphism of interleukin-6 influences susceptibility to HBV-related hepatocellular carcinoma in a male Chinese Han population. Hum Immunol. 2014;75:297-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 2. | Shin SP, Kim NK, Kim JH, Lee JH, Kim JO, Cho SH, Park H, Kim MN, Rim KS, Hwang SG. Association between hepatocellular carcinoma and tumor necrosis factor alpha polymorphisms in South Korea. World J Gastroenterol. 2015;21:13064-13072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Shi Z, Du C. Tumor necrosis factor alpha 308 G/A polymorphism and hepatocellular carcinoma risk in a Chinese population. Genet Test Mol Biomarkers. 2011;15:569-572. [PubMed] [Cited in This Article: ] |
| 4. | Saxena R, Chawla YK, Verma I, Kaur J. IL-6(-572/-597) polymorphism and expression in HBV disease chronicity in an Indian population. Am J Hum Biol. 2014;26:549-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Ma J, Liu YC, Fang Y, Cao Y, Liu ZL. TGF-β1 polymorphism 509 C& gt; T is associated with an increased risk for hepatocellular carcinoma in HCV-infected patients. Genet Mol Res. 2015;14:4461-4468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153-156. [PubMed] [Cited in This Article: ] |
| 7. | Liu L, Xu Y, Liu Z, Chen J, Zhang Y, Zhu J, Liu J, Liu S, Ji G, Shi H. IL12 polymorphisms, HBV infection and risk of hepatocellular carcinoma in a high-risk Chinese population. Int J Cancer. 2011;128:1692-1696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | van den Bosch MA, Defreyne L. Hepatocellular carcinoma. Lancet. 2012;380:469-470; author reply 470-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Kim HJ, Chung JH, Shin HP, Jeon JW, Park JJ, Cha JM, Joo KR, Lee JI. Polymorphisms of interferon gamma gene and risk of hepatocellular carcinoma in korean patients with chronic hepatitis B viral infection. Hepatogastroenterology. 2013;60:1117-1120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 10. | Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030-3044. [PubMed] [Cited in This Article: ] |
| 11. | Ognjanovic S, Yuan JM, Chaptman AK, Fan Y, Yu MC. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis. 2009;30:758-762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010;30:3-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 13. | Peng Q, Qin X, He Y, Chen Z, Deng Y, Li T, Xie L, Zhao J, Li S. Association of IL27 gene polymorphisms and HBV-related hepatocellular carcinoma risk in a Chinese population. Infect Genet Evol. 2013;16:1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Peng Q, Li H, Lao X, Deng Y, Chen Z, Qin X, Li S. Association of IL-2 polymorphisms and IL-2 serum levels with susceptibility to HBV-related hepatocellular carcinoma in a Chinese Zhuang population. Infect Genet Evol. 2014;27:375-381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 154] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 16. | Donato F, Gelatti U, Limina RM, Fattovich G. Southern Europe as an example of interaction between various environmental factors: a systematic review of the epidemiologic evidence. Oncogene. 2006;25:3756-3770. [PubMed] [Cited in This Article: ] |
| 17. | Kew MC. Interaction between hepatitis B and C viruses in hepatocellular carcinogenesis. J Viral Hepat. 2006;13:145-149. [PubMed] [Cited in This Article: ] |
| 18. | Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823-3833. [PubMed] [Cited in This Article: ] |
| 19. | Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25:3834-3847. [PubMed] [Cited in This Article: ] |
| 20. | Li S, Deng Y, Chen ZP, Huang S, Liao XC, Lin LW, Li H, Peng T, Qin X, Zhao JM. Genetic polymorphism of interleukin-16 influences susceptibility to HBV-related hepatocellular carcinoma in a Chinese population. Infect Genet Evol. 2011;11:2083-2088. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2881] [Cited by in F6Publishing: 2983] [Article Influence: 229.5] [Reference Citation Analysis (0)] |
| 22. | Phoa N, Epe B. Influence of nitric oxide on the generation and repair of oxidative DNA damage in mammalian cells. Carcinogenesis. 2002;23:469-475. [PubMed] [Cited in This Article: ] |
| 23. | Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci. 2009;1155:206-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 289] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 24. | Machida K, Cheng KT, Sung VM, Lee KJ, Levine AM, Lai MM. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J Virol. 2004;78:8835-8843. [PubMed] [Cited in This Article: ] |
| 25. | Sun MH, Han XC, Jia MK, Jiang WD, Wang M, Zhang H, Han G, Jiang Y. Expressions of inducible nitric oxide synthase and matrix metalloproteinase-9 and their effects on angiogenesis and progression of hepatocellular carcinoma. World J Gastroenterol. 2005;11:5931-5937. [PubMed] [Cited in This Article: ] |
| 26. | Atik E, Onlen Y, Savas L, Doran F. Inducible nitric oxide synthase and histopathological correlation in chronic viral hepatitis. Int J Infect Dis. 2008;12:12-15. [PubMed] [Cited in This Article: ] |
| 27. | Giannitrapani L, Soresi M, Giacalone A, Campagna ME, Marasà M, Cervello M, Marasà S, Montalto G. IL-6 -174G/C polymorphism and IL-6 serum levels in patients with liver cirrhosis and hepatocellular carcinoma. OMICS. 2011;15:183-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Deng Y, Li M, Wang J, Xie L, Li T, He Y, Lu Q, Li R, Tan A, Qin X. Susceptibility to hepatocellular carcinoma in the Chinese population--associations with interleukin-6 receptor polymorphism. Tumour Biol. 2014;35:6383-6388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, Oksenberg J, McNicholl J, Pociot F, Hardt C. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1:3-19. [PubMed] [Cited in This Article: ] |
| 30. | Fattovich G. Natural history and prognosis of hepatitis B. Semin Liver Dis. 2003;23:47-58. [PubMed] [Cited in This Article: ] |
| 31. | Ito H, Ando K, Ishikawa T, Saito K, Takemura M, Imawari M, Moriwaki H, Seishima M. Role of TNF-alpha produced by nonantigen-specific cells in a fulminant hepatitis mouse model. J Immunol. 2009;182:391-397. [PubMed] [Cited in This Article: ] |
| 32. | Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 575] [Cited by in F6Publishing: 598] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 33. | Lourenço EV, La Cava A. Cytokines in systemic lupus erythematosus. Curr Mol Med. 2009;9:242-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Koziel MJ. Cytokines in viral hepatitis. Semin Liver Dis. 1999;19:157-169. [PubMed] [Cited in This Article: ] |
| 35. | Hui CK, Lau GK. Immune system and hepatitis B virus infection. J Clin Virol. 2005;34 Suppl 1:S44-S48. [PubMed] [Cited in This Article: ] |
| 36. | Huang CF, Lin SS, Ho YC, Chen FL, Yang CC. The immune response induced by hepatitis B virus principal antigens. Cell Mol Immunol. 2006;3:97-106. [PubMed] [Cited in This Article: ] |
| 37. | Tilg H, Wilmer A, Vogel W, Herold M, Nölchen B, Judmaier G, Huber C. Serum levels of cytokines in chronic liver diseases. Gastroenterology. 1992;103:264-274. [PubMed] [Cited in This Article: ] |
| 38. | Kishihara Y, Hayashi J, Yoshimura E, Yamaji K, Nakashima K, Kashiwagi S. IL-1 beta and TNF-alpha produced by peripheral blood mononuclear cells before and during interferon therapy in patients with chronic hepatitis C. Dig Dis Sci. 1996;41:315-321. [PubMed] [Cited in This Article: ] |
| 39. | Penna A, Del Prete G, Cavalli A, Bertoletti A, D’Elios MM, Sorrentino R, D’Amato M, Boni C, Pilli M, Fiaccadori F. Predominant T-helper 1 cytokine profile of hepatitis B virus nucleocapsid-specific T cells in acute self-limited hepatitis B. Hepatology. 1997;25:1022-1027. [PubMed] [Cited in This Article: ] |
| 40. | Rico MA, Quiroga JA, Subirá D, Castañón S, Esteban JM, Pardo M, Carreño V. Hepatitis B virus-specific T-cell proliferation and cytokine secretion in chronic hepatitis B e antibody-positive patients treated with ribavirin and interferon alpha. Hepatology. 2001;33:295-300. [PubMed] [Cited in This Article: ] |
| 41. | Falasca K, Ucciferri C, Dalessandro M, Zingariello P, Mancino P, Petrarca C, Pizzigallo E, Conti P, Vecchiet J. Cytokine patterns correlate with liver damage in patients with chronic hepatitis B and C. Ann Clin Lab Sci. 2006;36:144-150. [PubMed] [Cited in This Article: ] |
| 42. | Matsuzaki K, Murata M, Yoshida K, Sekimoto G, Uemura Y, Sakaida N, Kaibori M, Kamiyama Y, Nishizawa M, Fujisawa J. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma. Hepatology. 2007;46:48-57. [PubMed] [Cited in This Article: ] |
| 43. | Gigi E, Raptopoulou-Gigi M, Kalogeridis A, Masiou S, Orphanou E, Vrettou E, Lalla TH, Sinakos E, Tsapas V. Cytokine mRNA expression in hepatitis C virus infection: TH1 predominance in patients with chronic hepatitis C and TH1-TH2 cytokine profile in subjects with self-limited disease. J Viral Hepat. 2008;15:145-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 44. | Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540-550. [PubMed] [Cited in This Article: ] |
| 45. | Vingerhoets J, Michielsen P, Vanham G, Bosmans E, Paulij W, Ramon A, Pelckmans P, Kestens L, Leroux-Roels G. HBV-specific lymphoproliferative and cytokine responses in patients with chronic hepatitis B. J Hepatol. 1998;28:8-16. [PubMed] [Cited in This Article: ] |
| 46. | Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6912] [Cited by in F6Publishing: 7713] [Article Influence: 482.1] [Reference Citation Analysis (0)] |
| 47. | Capone F, Costantini S, Guerriero E, Calemma R, Napolitano M, Scala S, Izzo F, Castello G. Serum cytokine levels in patients with hepatocellular carcinoma. Eur Cytokine Netw. 2010;21:99-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 30] [Reference Citation Analysis (0)] |
| 48. | d’Arville CN, Nouri-Aria KT, Johnson P, Williams R. Gene expression regulation for interferon-alpha in hepatocellular carcinoma. J Hepatol. 1993;17:339-346. [PubMed] [Cited in This Article: ] |
| 49. | Jaskiewicz K, Chasen MR. Differential expression of transforming growth factor alpha, adhesions molecules and integrins in primary, metastatic liver tumors and in liver cirrhosis. Anticancer Res. 1995;15:559-562. [PubMed] [Cited in This Article: ] |
| 50. | Kakumu S, Okumura A, Ishikawa T, Yano M, Enomoto A, Nishimura H, Yoshioka K, Yoshika Y. Serum levels of IL-10, IL-15 and soluble tumour necrosis factor-alpha (TNF-alpha) receptors in type C chronic liver disease. Clin Exp Immunol. 1997;109:458-463. [PubMed] [Cited in This Article: ] |
| 51. | Huang YS, Hwang SJ, Chan CY, Wu JC, Chao Y, Chang FY, Lee SD. Serum levels of cytokines in hepatitis C-related liver disease: a longitudinal study. Zhonghua Yi Xue Zazhi (Taipei). 1999;62:327-333. [PubMed] [Cited in This Article: ] |
| 52. | Tangkijvanich P, Vimolket T, Theamboonlers A, Kullavanijaya P, Suwangool P, Poovorawan Y. Serum interleukin-6 and interferon-gamma levels in patients with hepatitis B-associated chronic liver disease. Asian Pac J Allergy Immunol. 2000;18:109-114. [PubMed] [Cited in This Article: ] |
| 53. | Chia CS, Ban K, Ithnin H, Singh H, Krishnan R, Mokhtar S, Malihan N, Seow HF. Expression of interleukin-18, interferon-gamma and interleukin-10 in hepatocellular carcinoma. Immunol Lett. 2002;84:163-172. [PubMed] [Cited in This Article: ] |
| 54. | Kitaoka S, Shiota G, Kawasaki H. Serum levels of interleukin-10, interleukin-12 and soluble interleukin-2 receptor in chronic liver disease type C. Hepatogastroenterology. 2003;50:1569-1574. [PubMed] [Cited in This Article: ] |
| 55. | Ikeguchi M, Hirooka Y. Interleukin-2 gene expression is a new biological prognostic marker in hepatocellular carcinomas. Onkologie. 2005;28:255-259. [PubMed] [Cited in This Article: ] |
| 56. | Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 264] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 57. | Keen LJ. The extent and analysis of cytokine and cytokine receptor gene polymorphism. Transpl Immunol. 2002;10:143-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Dutra WO, Moreira PR, Souza PE, Gollob KJ, Gomez RS. Implications of cytokine gene polymorphisms on the orchestration of the immune response: lessons learned from oral diseases. Cytokine Growth Factor Rev. 2009;20:223-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Ollier WE. Cytokine genes and disease susceptibility. Cytokine. 2004;28:174-178. [PubMed] [Cited in This Article: ] |
| 60. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 61. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1747] [Cited by in F6Publishing: 1746] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 62. | Giannitrapani L, Soresi M, Balasus D, Licata A, Montalto G. Genetic association of interleukin-6 polymorphism (-174 G/C) with chronic liver diseases and hepatocellular carcinoma. World J Gastroenterol. 2013;19:2449-2455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 39] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 63. | Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382-384. [PubMed] [Cited in This Article: ] |
| 65. | Chen CC, Yang SY, Liu CJ, Lin CL, Liaw YF, Lin SM, Lee SD, Chen PJ, Chen CJ, Yu MW. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol. 2005;34:1310-1318. [PubMed] [Cited in This Article: ] |
| 66. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [PubMed] [Cited in This Article: ] |
| 67. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. The role of interleukin-1 polymorphisms in the pathogenesis of gastric cancer. Nature. 2001;412:99. [PubMed] [Cited in This Article: ] |
| 68. | Chen H, Wilkins LM, Aziz N, Cannings C, Wyllie DH, Bingle C, Rogus J, Beck JD, Offenbacher S, Cork MJ. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519-529. [PubMed] [Cited in This Article: ] |
| 69. | Hulkkonen J, Laippala P, Hurme M. A rare allele combination of the interleukin-1 gene complex is associated with high interleukin-1 beta plasma levels in healthy individuals. Eur Cytokine Netw. 2000;11:251-255. [PubMed] [Cited in This Article: ] |
| 70. | Wen AQ, Wang J, Feng K, Zhu PF, Wang ZG, Jiang JX. Effects of haplotypes in the interleukin 1beta promoter on lipopolysaccharide-induced interleukin 1beta expression. Shock. 2006;26:25-30. [PubMed] [Cited in This Article: ] |
| 71. | Zhang PA, Li Y, Xu P, Wu JM. Polymorphisms of interleukin-1B and interleukin-1 receptor antagonist genes in patients with chronic hepatitis B. World J Gastroenterol. 2004;10:1826-1829. [PubMed] [Cited in This Article: ] |
| 72. | Hwang IR, Kodama T, Kikuchi S, Sakai K, Peterson LE, Graham DY, Yamaoka Y. Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology. 2002;123:1793-1803. [PubMed] [Cited in This Article: ] |
| 73. | Hirankarn N, Kimkong I, Kummee P, Tangkijvanich P, Poovorawan Y. Interleukin-1beta gene polymorphism associated with hepatocellular carcinoma in hepatitis B virus infection. World J Gastroenterol. 2006;12:776-779. [PubMed] [Cited in This Article: ] |
| 74. | Saxena R, Chawla YK, Verma I, Kaur J. Interleukin-1 polymorphism and expression in hepatitis B virus-mediated disease outcome in India. J Interferon Cytokine Res. 2013;33:80-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 75. | Wang Y, Kato N, Hoshida Y, Yoshida H, Taniguchi H, Goto T, Moriyama M, Otsuka M, Shiina S, Shiratori Y. Interleukin-1beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology. 2003;37:65-71. [PubMed] [Cited in This Article: ] |
| 76. | Tanaka Y, Furuta T, Suzuki S, Orito E, Yeo AE, Hirashima N, Sugauchi F, Ueda R, Mizokami M. Impact of interleukin-1beta genetic polymorphisms on the development of hepatitis C virus-related hepatocellular carcinoma in Japan. J Infect Dis. 2003;187:1822-1825. [PubMed] [Cited in This Article: ] |
| 77. | Anderson GM, Nakada MT, DeWitte M. Tumor necrosis factor-alpha in the pathogenesis and treatment of cancer. Curr Opin Pharmacol. 2004;4:314-320. [PubMed] [Cited in This Article: ] |
| 78. | Akkiz H, Bayram S, Bekar A, Ozdil B, Akgöllü E, Sümbül AT, Demiryürek H, Doran F. G-308A TNF-alpha polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: case-control study. Cancer Epidemiol. 2009;33:261-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 79. | Jeng JE, Tsai HR, Chuang LY, Tsai JF, Lin ZY, Hsieh MY, Chen SC, Chuang WL, Wang LY, Yu ML. Independent and additive interactive effects among tumor necrosis factor-alpha polymorphisms, substance use habits, and chronic hepatitis B and hepatitis C virus infection on risk for hepatocellular carcinoma. Medicine (Baltimore). 2009;88:349-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 80. | Talaat RM, Esmail AA, Elwakil R, Gurgis AA, Nasr MI. Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer. 2012;31:29-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 81. | Yang JJ, Ko KP, Cho LY, Shin A, Gwack J, Chang SH, Shin HR, Yoo KY, Kang D, Park SK. The role of TNF genetic variants and the interaction with cigarette smoking for gastric cancer risk: a nested case-control study. BMC Cancer. 2009;9:238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 82. | Wilson AG, di Giovine FS, Blakemore AI, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet. 1992;1:353. [PubMed] [Cited in This Article: ] |
| 83. | Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195-3199. [PubMed] [Cited in This Article: ] |
| 84. | Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562-566. [PubMed] [Cited in This Article: ] |
| 85. | Kim YJ, Lee HS, Yoon JH, Kim CY, Park MH, Kim LH, Park BL, Shin HD. Association of TNF-alpha promoter polymorphisms with the clearance of hepatitis B virus infection. Hum Mol Genet. 2003;12:2541-2546. [PubMed] [Cited in This Article: ] |
| 86. | Li C, Zhi-Xin C, Li-Juan Z, Chen P, Xiao-Zhong W. The association between cytokine gene polymorphisms and the outcomes of chronic HBV infection. Hepatol Res. 2006;36:158-166. [PubMed] [Cited in This Article: ] |
| 87. | Wei Y, Liu F, Li B, Chen X, Ma Y, Yan L, Wen T, Xu M, Wang W, Yang J. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Dig Dis Sci. 2011;56:2227-2236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 88. | Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841-848. [PubMed] [Cited in This Article: ] |
| 89. | Niro GA, Fontana R, Gioffreda D, Valvano MR, Lacobellis A, Facciorusso D, Andriulli A. Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int. 2005;25:1175-1181. [PubMed] [Cited in This Article: ] |
| 90. | Du T, Guo XH, Zhu XL, Li JH, Lu LP, Gao JR, Gou CY, Li Z, Liu Y, Li H. Association of TNF-alpha promoter polymorphisms with the outcomes of hepatitis B virus infection in Chinese Han population. J Viral Hepat. 2006;13:618-624. [PubMed] [Cited in This Article: ] |
| 91. | Wang B, Wang J, Zheng Y, Zhou S, Zheng J, Wang F, Ma X, Zeng Z. A study of TNF-alpha-238 and -308 polymorphisms with different outcomes of persistent hepatitis B virus infection in China. Pathology. 2010;42:674-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 92. | Hirano T, Akira S, Taga T, Kishimoto T. Biological and clinical aspects of interleukin 6. Immunol Today. 1990;11:443-449. [PubMed] [Cited in This Article: ] |
| 93. | Hsia CY, Huo TI, Chiang SY, Lu MF, Sun CL, Wu JC, Lee PC, Chi CW, Lui WY, Lee SD. Evaluation of interleukin-6, interleukin-10 and human hepatocyte growth factor as tumor markers for hepatocellular carcinoma. Eur J Surg Oncol. 2007;33:208-212. [PubMed] [Cited in This Article: ] |
| 94. | Prieto J. Inflammation, HCC and sex: IL-6 in the centre of the triangle. J Hepatol. 2008;48:380-381. [PubMed] [Cited in This Article: ] |
| 95. | Fielding CA, McLoughlin RM, McLeod L, Colmont CS, Najdovska M, Grail D, Ernst M, Jones SA, Topley N, Jenkins BJ. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J Immunol. 2008;181:2189-2195. [PubMed] [Cited in This Article: ] |
| 96. | Panek H. [Functional shaping of the occlusal surfaces of fixed dentures]. Protet Stomatol. 1976;26:105-111. [PubMed] [Cited in This Article: ] |
| 97. | Kishimoto T. Interleukin-6: from basic science to medicine--40 years in immunology. Annu Rev Immunol. 2005;23:1-21. [PubMed] [Cited in This Article: ] |
| 98. | Hösel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773-1782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 270] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 99. | Ataseven H, Bahcecioglu IH, Kuzu N, Yalniz M, Celebi S, Erensoy A, Ustundag B. The levels of ghrelin, leptin, TNF-alpha, and IL-6 in liver cirrhosis and hepatocellular carcinoma due to HBV and HDV infection. Mediators Inflamm. 2006;2006:78380. [PubMed] [Cited in This Article: ] |
| 100. | Nieters A, Yuan JM, Sun CL, Zhang ZQ, Stoehlmacher J, Govindarajan S, Yu MC. Effect of cytokine genotypes on the hepatitis B virus-hepatocellular carcinoma association. Cancer. 2005;103:740-748. [PubMed] [Cited in This Article: ] |
| 101. | Falleti E, Fabris C, Vandelli C, Colletta C, Cussigh A, Smirne C, Fontanini E, Cmet S, Minisini R, Bitetto D. Genetic polymorphisms of interleukin-6 modulate fibrosis progression in mild chronic hepatitis C. Hum Immunol. 2010;71:999-1004. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 102. | Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138-18144. [PubMed] [Cited in This Article: ] |
| 103. | Saxena R, Chawla YK, Verma I, Kaur J. Association of interleukin-10 with hepatitis B virus (HBV) mediated disease progression in Indian population. Indian J Med Res. 2014;139:737-745. [PubMed] [Cited in This Article: ] |
| 104. | Chang L, Lan T, Wu L, Li C, Yuan Y, Liu Z. The association between three IL-6 polymorphisms and HBV-related liver diseases: a meta-analysis. Int J Clin Exp Med. 2015;8:17036-17045. [PubMed] [Cited in This Article: ] |
| 105. | Park BL, Lee HS, Kim YJ, Kim JY, Jung JH, Kim LH, Shin HD. Association between interleukin 6 promoter variants and chronic hepatitis B progression. Exp Mol Med. 2003;35:76-82. [PubMed] [Cited in This Article: ] |
| 106. | Ralph P, Nakoinz I, Sampson-Johannes A, Fong S, Lowe D, Min HY, Lin L. IL-10, T lymphocyte inhibitor of human blood cell production of IL-1 and tumor necrosis factor. J Immunol. 1992;148:808-814. [PubMed] [Cited in This Article: ] |
| 107. | Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001;9:86-92. [PubMed] [Cited in This Article: ] |
| 108. | Cacciarelli TV, Martinez OM, Gish RG, Villanueva JC, Krams SM. Immunoregulatory cytokines in chronic hepatitis C virus infection: pre- and posttreatment with interferon alfa. Hepatology. 1996;24:6-9. [PubMed] [Cited in This Article: ] |
| 109. | Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1-8. [PubMed] [Cited in This Article: ] |
| 110. | Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, O’Brien TR, Vlahov D, Buchbinder S, Giorgi J. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci USA. 2000;97:14467-14472. [PubMed] [Cited in This Article: ] |
| 111. | Rees LE, Wood NA, Gillespie KM, Lai KN, Gaston K, Mathieson PW. The interleukin-10-1082 G/A polymorphism: allele frequency in different populations and functional significance. Cell Mol Life Sci. 2002;59:560-569. [PubMed] [Cited in This Article: ] |
| 112. | Keijsers V, Verweij CL, Westendorp RGJ, Breedveld FC, Huizinga TWJ. IL-10 polymorphisms in relation to production and rheumatoid arthritis. Arthritis Rheum. 1997;40 Suppl 9: S179. [Cited in This Article: ] |
| 113. | Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915-3922. [PubMed] [Cited in This Article: ] |
| 114. | Miyazoe S, Hamasaki K, Nakata K, Kajiya Y, Kitajima K, Nakao K, Daikoku M, Yatsuhashi H, Koga M, Yano M. Influence of interleukin-10 gene promoter polymorphisms on disease progression in patients chronically infected with hepatitis B virus. Am J Gastroenterol. 2002;97:2086-2092. [PubMed] [Cited in This Article: ] |
| 115. | Vidigal PG, Germer JJ, Zein NN. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. J Hepatol. 2002;36:271-277. [PubMed] [Cited in This Article: ] |
| 116. | Knapp S, Hennig BJ, Frodsham AJ, Zhang L, Hellier S, Wright M, Goldin R, Hill AV, Thomas HC, Thursz MR. Interleukin-10 promoter polymorphisms and the outcome of hepatitis C virus infection. Immunogenetics. 2003;55:362-369. [PubMed] [Cited in This Article: ] |
| 117. | Hamada H, Yatsuhashi H, Yano K, Arisawa K, Nakao K, Yano M. Interleukin-10 promoter polymorphisms and liver fibrosis progression in patients with chronic hepatitis C in Japan. J Hepatol. 2003;39:457-458. [PubMed] [Cited in This Article: ] |
| 118. | Paladino N, Fainboim H, Theiler G, Schroder T, Muñoz AE, Flores AC, Galdame O, Fainboim L. Gender susceptibility to chronic hepatitis C virus infection associated with interleukin 10 promoter polymorphism. J Virol. 2006;80:9144-9150. [PubMed] [Cited in This Article: ] |
| 119. | Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505-510. [PubMed] [Cited in This Article: ] |
| 120. | Tseng LH, Lin MT, Shau WY, Lin WC, Chang FY, Chien KL, Hansen JA, Chen DS, Chen PJ. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens. 2006;67:127-133. [PubMed] [Cited in This Article: ] |
| 121. | Bouzgarrou N, Hassen E, Farhat K, Bahri O, Gabbouj S, Maamouri N, Ben Mami N, Saffar H, Trabelsi A, Triki H. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Hum Immunol. 2009;70:230-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 122. | Chin D, Boyle GM, Parsons PG, Coman WB. What is transforming growth factor-beta (TGF-beta)? Br J Plast Surg. 2004;57:215-221. [PubMed] [Cited in This Article: ] |
| 123. | Funkenstein B, Olekh E, Jakowlew SB. Identification of a novel transforming growth factor-beta (TGF-beta6) gene in fish: regulation in skeletal muscle by nutritional state. BMC Mol Biol. 2010;11:37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 124. | Taylor LM, Khachigian LM. Induction of platelet-derived growth factor B-chain expression by transforming growth factor-beta involves transactivation by Smads. J Biol Chem. 2000;275:16709-16716. [PubMed] [Cited in This Article: ] |
| 125. | Bernabeu C, Lopez-Novoa JM, Quintanilla M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim Biophys Acta. 2009;1792:954-973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 195] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 126. | Regateiro FS, Howie D, Cobbold SP, Waldmann H. TGF-β in transplantation tolerance. Curr Opin Immunol. 2011;23:660-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 127. | Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350-1358. [PubMed] [Cited in This Article: ] |
| 128. | Leivonen SK, Kähäri VM. Transforming growth factor-beta signaling in cancer invasion and metastasis. Int J Cancer. 2007;121:2119-2124. [PubMed] [Cited in This Article: ] |
| 129. | Oh S, Kim E, Kang D, Kim M, Kim JH, Song JJ. Transforming growth factor-β gene silencing using adenovirus expressing TGF-β1 or TGF-β2 shRNA. Cancer Gene Ther. 2013;20:94-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Kanzler S, Lohse AW, Keil A, Henninger J, Dienes HP, Schirmacher P, Rose-John S, zum Büschenfelde KH, Blessing M. TGF-beta1 in liver fibrosis: an inducible transgenic mouse model to study liver fibrogenesis. Am J Physiol. 1999;276:G1059-G1068. [PubMed] [Cited in This Article: ] |
| 131. | Wang H, Mengsteab S, Tag CG, Gao CF, Hellerbrand C, Lammert F, Gressner AM, Weiskirchen R. Transforming growth factor-beta1 gene polymorphisms are associated with progression of liver fibrosis in Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2005;11:1929-1936. [PubMed] [Cited in This Article: ] |
| 132. | Guo JC, Bao JF, Chen QW, Li XO, Shi JP, Lou GQ, Shi WZ, Xun YH. [Level of serum and liver tissue TGF-beta1 in patients with liver fibrosis due to chronic hepatitis B]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Zazhi. 2008;22:354-357. [PubMed] [Cited in This Article: ] |
| 133. | Wang H, Zhao YP, Gao CF, Ji Q, Gressner AM, Yang ZX, Weiskirchen R. Transforming growth factor beta 1 gene variants increase transcription and are associated with liver cirrhosis in Chinese. Cytokine. 2008;43:20-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 134. | Fujii D, Brissenden JE, Derynck R, Francke U. Transforming growth factor beta gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet. 1986;12:281-288. [PubMed] [Cited in This Article: ] |
| 135. | Derynck R, Rhee L, Chen EY, Van Tilburg A. Intron-exon structure of the human transforming growth factor-beta precursor gene. Nucleic Acids Res. 1987;15:3188-3189. [PubMed] [Cited in This Article: ] |
| 136. | Watanabe Y, Kinoshita A, Yamada T, Ohta T, Kishino T, Matsumoto N, Ishikawa M, Niikawa N, Yoshiura K. A catalog of 106 single-nucleotide polymorphisms (SNPs) and 11 other types of variations in genes for transforming growth factor-beta1 (TGF-beta1) and its signaling pathway. J Hum Genet. 2002;47:478-483. [PubMed] [Cited in This Article: ] |
| 137. | Yamada Y, Miyauchi A, Goto J, Takagi Y, Okuizumi H, Kanematsu M, Hase M, Takai H, Harada A, Ikeda K. Association of a polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to osteoporosis in postmenopausal Japanese women. J Bone Miner Res. 1998;13:1569-1576. [PubMed] [Cited in This Article: ] |
| 138. | Gewaltig J, Mangasser-Stephan K, Gartung C, Biesterfeld S, Gressner AM. Association of polymorphisms of the transforming growth factor-beta1 gene with the rate of progression of HCV-induced liver fibrosis. Clin Chim Acta. 2002;316:83-94. [PubMed] [Cited in This Article: ] |
| 139. | Suzuki S, Tanaka Y, Orito E, Sugauchi F, Hasegawa I, Sakurai M, Fujiwara K, Ohno T, Ueda R, Mizokami M. Transforming growth factor-beta-1 genetic polymorphism in Japanese patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2003;18:1139-1143. [PubMed] [Cited in This Article: ] |
| 140. | Qi P, Chen YM, Wang H, Fang M, Ji Q, Zhao YP, Sun XJ, Liu Y, Gao CF. -509C& gt; T polymorphism in the TGF-beta1 gene promoter, impact on the hepatocellular carcinoma risk in Chinese patients with chronic hepatitis B virus infection. Cancer Immunol Immunother. 2009;58:1433-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 141. | Talaat RM, Dondeti MF, El-Shenawy SZ, Khamiss OA. Transforming Growth Factor- β 1 Gene Polymorphism (T29C) in Egyptian Patients with Hepatitis B Virus Infection: A Preliminary Study. Hepat Res Treat. 2013;2013:293274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 142. | Perrey C, Turner SJ, Pravica V, Howell WM, Hutchinson IV. ARMS-PCR methodologies to determine IL-10, TNF-alpha, TNF-beta and TGF-beta 1 gene polymorphisms. Transpl Immunol. 1999;7:127-128. [PubMed] [Cited in This Article: ] |
| 143. | Macedo LC, Isolani AP, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphisms with the humoral immune response to recombinant vaccine against HBV in infants. J Med Virol. 2010;82:929-933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 144. | Powell EE, Edwards-Smith CJ, Hay JL, Clouston AD, Crawford DH, Shorthouse C, Purdie DM, Jonsson JR. Host genetic factors influence disease progression in chronic hepatitis C. Hepatology. 2000;31:828-833. [PubMed] [Cited in This Article: ] |
| 145. | Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144-150. [PubMed] [Cited in This Article: ] |
| 146. | Ribeiro CS, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphism with hepatitis B infection evolution in adult patients. Mem Inst Oswaldo Cruz. 2007;102:435-440. [PubMed] [Cited in This Article: ] |
| 147. | Xin Z, Zhang W, Xu A, Zhang L, Yan T, Li Z, Wu X, Zhu X, Ma J, Li K. Polymorphisms in the potential functional regions of the TGF-β 1 and TGF-β receptor genes and disease susceptibility in HBV-related hepatocellular carcinoma patients. Mol Carcinog. 2012;51 Suppl 1:E123-E131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 148. | Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, Carter ND, Spector TD. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8:93-97. [PubMed] [Cited in This Article: ] |
| 149. | Luedecking EK, DeKosky ST, Mehdi H, Ganguli M, Kamboh MI. Analysis of genetic polymorphisms in the transforming growth factor-beta1 gene and the risk of Alzheimer’s disease. Hum Genet. 2000;106:565-569. [PubMed] [Cited in This Article: ] |
| 150. | Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFbeta1 allele association with asthma severity. Hum Genet. 2001;109:623-627. [PubMed] [Cited in This Article: ] |
| 151. | Shi HZ, Ren P, Lu QJ, Niedrgethmnn M, Wu GY. Association between EGF, TGF-β1 and TNF-α gene polymorphisms and hepatocellular carcinoma. Asian Pac J Cancer Prev. 2012;13:6217-6220. [PubMed] [Cited in This Article: ] |
| 152. | Radwan MI, Pasha HF, Mohamed RH, Hussien HI, El-Khshab MN. Influence of transforming growth factor-β1 and tumor necrosis factor-α genes polymorphisms on the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis C patients. Cytokine. 2012;60:271-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 153. | Wei ZH, Lu JX, Pu J, Wang ZX, Ya HY, Tang RG, Long XK. Association of the TGF-β1 gene promoter polymorphisms with hepatocellular carcinoma. Cancer Res Clin. 2012;24:447-450. [DOI] [Cited in This Article: ] |
| 154. | Finotto S, Siebler J, Hausding M, Schipp M, Wirtz S, Klein S, Protschka M, Doganci A, Lehr HA, Trautwein C. Severe hepatic injury in interleukin 18 (IL-18) transgenic mice: a key role for IL-18 in regulating hepatocyte apoptosis in vivo. Gut. 2004;53:392-400. [PubMed] [Cited in This Article: ] |
| 155. | Higashi N, Gesser B, Kawana S, Thestrup-Pedersen K. Expression of IL-18 mRNA and secretion of IL-18 are reduced in monocytes from patients with atopic dermatitis. J Allergy Clin Immunol. 2001;108:607-614. [PubMed] [Cited in This Article: ] |
| 156. | Mariño E, Cardier JE. Differential effect of IL-18 on endothelial cell apoptosis mediated by TNF-alpha and Fas (CD95). Cytokine. 2003;22:142-148. [PubMed] [Cited in This Article: ] |
| 157. | Vecchiet J, Falasca K, Cacciatore P, Zingariello P, Dalessandro M, Marinopiccoli M, D’Amico E, Palazzi C, Petrarca C, Conti P. Association between plasma interleukin-18 levels and liver injury in chronic hepatitis C virus infection and non-alcoholic fatty liver disease. Ann Clin Lab Sci. 2005;35:415-422. [PubMed] [Cited in This Article: ] |
| 158. | Neuman MG, Benhamou JP, Marcellin P, Valla D, Malkiewicz IM, Katz GG, Trepo C, Bourliere M, Cameron RG, Cohen L. Cytokine--chemokine and apoptotic signatures in patients with hepatitis C. Transl Res. 2007;149:126-136. [PubMed] [Cited in This Article: ] |
| 159. | Murata K, Yamamoto N, Kawakita T, Saito Y, Yamanaka Y, Sugimoto K, Shiraki K, Nakano T, Tameda Y. Up-regulation of IL-18 by interferon alpha-2b/ribavirin combination therapy induces an anti-viral effect in patients with chronic hepatitis C. Hepatogastroenterology. 2005;52:547-551. [PubMed] [Cited in This Article: ] |
| 160. | Cho D, Song H, Kim YM, Houh D, Hur DY, Park H, Yoon D, Pyun KH, Lee WJ, Kurimoto M. Endogenous interleukin-18 modulates immune escape of murine melanoma cells by regulating the expression of Fas ligand and reactive oxygen intermediates. Cancer Res. 2000;60:2703-2709. [PubMed] [Cited in This Article: ] |
| 161. | Park H, Byun D, Kim TS, Kim YI, Kang JS, Hahm ES, Kim SH, Lee WJ, Song HK, Yoon DY. Enhanced IL-18 expression in common skin tumors. Immunol Lett. 2001;79:215-219. [PubMed] [Cited in This Article: ] |
| 162. | Kanda N, Shimizu T, Tada Y, Watanabe S. IL-18 enhances IFN-gamma-induced production of CXCL9, CXCL10, and CXCL11 in human keratinocytes. Eur J Immunol. 2007;37:338-350. [PubMed] [Cited in This Article: ] |
| 163. | Osaki T, Hashimoto W, Gambotto A, Okamura H, Robbins PD, Kurimoto M, Lotze MT, Tahara H. Potent antitumor effects mediated by local expression of the mature form of the interferon-gamma inducing factor, interleukin-18 (IL-18). Gene Ther. 1999;6:808-815. [PubMed] [Cited in This Article: ] |
| 164. | Chang CY, Lee J, Kim EY, Park HJ, Kwon CH, Joh JW, Kim SJ. Intratumoral delivery of IL-18 naked DNA induces T-cell activation and Th1 response in a mouse hepatic cancer model. BMC Cancer. 2007;7:87. [PubMed] [Cited in This Article: ] |
| 165. | Nada O, Abdel-Hamid M, Ismail A, El Shabrawy L, Sidhom KF, El Badawy NM, Ghazal FA, El Daly M, El Kafrawy S, Esmat G. The role of the tumor necrosis factor (TNF)--Fas L and HCV in the development of hepatocellular carcinoma. J Clin Virol. 2005;34:140-146. [PubMed] [Cited in This Article: ] |
| 166. | Asakawa M, Kono H, Amemiya H, Matsuda M, Suzuki T, Maki A, Fujii H. Role of interleukin-18 and its receptor in hepatocellular carcinoma associated with hepatitis C virus infection. Int J Cancer. 2006;118:564-570. [PubMed] [Cited in This Article: ] |
| 167. | Bouzgarrou N, Hassen E, Schvoerer E, Stoll-Keller F, Bahri O, Gabbouj S, Cheikh I, Maamouri N, Mammi N, Saffar H. Association of interleukin-18 polymorphisms and plasma level with the outcome of chronic HCV infection. J Med Virol. 2008;80:607-614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 168. | Wei YS, Lan Y, Liu YG, Tang H, Tang RG, Wang JC. Interleukin-18 gene promoter polymorphisms and the risk of esophageal squamous cell carcinoma. Acta Oncol. 2007;46:1090-1096. [PubMed] [Cited in This Article: ] |
| 169. | Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112:146-152. [PubMed] [Cited in This Article: ] |
| 170. | Arimitsu J, Hirano T, Higa S, Kawai M, Naka T, Ogata A, Shima Y, Fujimoto M, Yamadori T, Hagiwara K. IL-18 gene polymorphisms affect IL-18 production capability by monocytes. Biochem Biophys Res Commun. 2006;342:1413-1416. [PubMed] [Cited in This Article: ] |
| 171. | Barbaux S, Poirier O, Godefroy T, Kleinert H, Blankenberg S, Cambien F, Tiret L. Differential haplotypic expression of the interleukin-18 gene. Eur J Hum Genet. 2007;15:856-863. [PubMed] [Cited in This Article: ] |
| 172. | Khripko OP, Sennikova NS, Lopatnikova JA, Khripko JI, Filipenko ML, Khrapov EA, Gelfgat EL, Yakushenko EV, Kozlov VA, Sennikov SV. Association of single nucleotide polymorphisms in the IL-18 gene with production of IL-18 protein by mononuclear cells from healthy donors. Mediators Inflamm. 2008;2008:309721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 173. | Ksiaa Cheikhrouhou L, Sfar I, Aounallah-Skhiri H, Aouadi H, Jendoubi-Ayed S, Ben Abdallah T, Ayed K, Lakhoua-Gorgi Y. Cytokine and apoptosis gene polymorphisms influence the outcome of hepatitis C virus infection. Hepatobiliary Pancreat Dis Int. 2011;10:280-288. [PubMed] [Cited in This Article: ] |
| 174. | Teixeira AC, Mendes CT, Marano LA, Deghaide NH, Secaf M, Elias J, Muglia V, Donadi EA, Martinelli AL. Alleles and genotypes of polymorphisms of IL-18, TNF-α and IFN-γ are associated with a higher risk and severity of hepatocellular carcinoma (HCC) in Brazil. Hum Immunol. 2013;74:1024-1029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 175. | Karra VK, Gumma PK, Chowdhury SJ, Ruttala R, Polipalli SK, Chakravarti A, Kar P. IL-18 polymorphisms in hepatitis B virus related liver disease. Cytokine. 2015;73:277-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 176. | Li N, Gao YF, Zhang TC, Chen P, Li X, Su F. Relationship between interleukin 18 polymorphisms and susceptibility to chronic hepatitis B virus infection. World J Hepatol. 2012;4:105-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 177. | Zhang PA, Wu JM, Li Y, Yang XS. Association of polymorphisms of interleukin-18 gene promoter region with chronic hepatitis B in Chinese Han population. World J Gastroenterol. 2005;11:1594-1598. [PubMed] [Cited in This Article: ] |
| 178. | Kim YS, Cheong JY, Cho SW, Lee KM, Hwang JC, Oh B, Kimm K, Lee JA, Park BL, Cheong HS. A functional SNP of the Interleukin-18 gene is associated with the presence of hepatocellular carcinoma in hepatitis B virus-infected patients. Dig Dis Sci. 2009;54:2722-2728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 179. | Keane J, Nicoll J, Kim S, Wu DM, Cruikshank WW, Brazer W, Natke B, Zhang Y, Center DM, Kornfeld H. Conservation of structure and function between human and murine IL-16. J Immunol. 1998;160:5945-5954. [PubMed] [Cited in This Article: ] |
| 180. | Cruikshank WW, Kornfeld H, Center DM. Interleukin-16. J Leukoc Biol. 2000;67:757-766. [PubMed] [Cited in This Article: ] |
| 181. | Center DM, Cruikshank W. Modulation of lymphocyte migration by human lymphokines. I. Identification and characterization of chemoattractant activity for lymphocytes from mitogen-stimulated mononuclear cells. J Immunol. 1982;128:2563-2568. [PubMed] [Cited in This Article: ] |
| 182. | Cruikshank WW, Center DM, Nisar N, Wu M, Natke B, Theodore AC, Kornfeld H. Molecular and functional analysis of a lymphocyte chemoattractant factor: association of biologic function with CD4 expression. Proc Natl Acad Sci USA. 1994;91:5109-5113. [PubMed] [Cited in This Article: ] |
| 183. | Center DM, Kornfeld H, Cruikshank WW. Interleukin 16 and its function as a CD4 ligand. Immunol Today. 1996;17:476-481. [PubMed] [Cited in This Article: ] |
| 184. | Jaruga B, Hong F, Sun R, Radaeva S, Gao B. Crucial role of IL-4/STAT6 in T cell-mediated hepatitis: up-regulating eotaxins and IL-5 and recruiting leukocytes. J Immunol. 2003;171:3233-3244. [PubMed] [Cited in This Article: ] |
| 185. | Durán A, Rodriguez A, Martin P, Serrano M, Flores JM, Leitges M, Diaz-Meco MT, Moscat J. Crosstalk between PKCzeta and the IL4/Stat6 pathway during T-cell-mediated hepatitis. EMBO J. 2004;23:4595-4605. [PubMed] [Cited in This Article: ] |
| 186. | Lynch EA, Heijens CA, Horst NF, Center DM, Cruikshank WW. Cutting edge: IL-16/CD4 preferentially induces Th1 cell migration: requirement of CCR5. J Immunol. 2003;171:4965-4968. [PubMed] [Cited in This Article: ] |
| 187. | Ryan TC, Cruikshank WW, Kornfeld H, Collins TL, Center DM. The CD4-associated tyrosine kinase p56lck is required for lymphocyte chemoattractant factor-induced T lymphocyte migration. J Biol Chem. 1995;270:17081-17086. [PubMed] [Cited in This Article: ] |
| 188. | Krautwald S. IL-16 activates the SAPK signaling pathway in CD4+ macrophages. J Immunol. 1998;160:5874-5879. [PubMed] [Cited in This Article: ] |
| 189. | Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1169] [Cited by in F6Publishing: 1283] [Article Influence: 91.6] [Reference Citation Analysis (1)] |
| 190. | Bannert N, Avots A, Baier M, Serfling E, Kurth R. GA-binding protein factors, in concert with the coactivator CREB binding protein/p300, control the induction of the interleukin 16 promoter in T lymphocytes. Proc Natl Acad Sci USA. 1999;96:1541-1546. [PubMed] [Cited in This Article: ] |
| 191. | Kim HS. Assignment of human interleukin 16 (IL16) to chromosome 15q26.3 by radiation hybrid mapping. Cytogenet Cell Genet. 1999;84:93. [PubMed] [Cited in This Article: ] |
| 192. | Nakayama EE, Wasi C, Ajisawa A, Iwamoto A, Shioda T. A new polymorphism in the promoter region of the human interleukin-16 (IL-16) gene. Genes Immun. 2000;1:293-294. [PubMed] [Cited in This Article: ] |
| 193. | Gao LB, Rao L, Wang YY, Liang WB, Li C, Xue H, Zhou B, Sun H, Li Y, Lv ML. The association of interleukin-16 polymorphisms with IL-16 serum levels and risk of colorectal and gastric cancer. Carcinogenesis. 2009;30:295-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 194. | Gao LB, Liang WB, Xue H, Rao L, Pan XM, Lv ML, Bai P, Fang WL, Liu J, Liao M. Genetic polymorphism of interleukin-16 and risk of nasopharyngeal carcinoma. Clin Chim Acta. 2009;409:132-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 195. | Qin X, Peng Q, Lao X, Chen Z, Lu Y, Lao X, Mo C, Sui J, Wu J, Zhai L. The association of interleukin-16 gene polymorphisms with IL-16 serum levels and risk of nasopharyngeal carcinoma in a Chinese population. Tumour Biol. 2014;35:1917-1924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 196. | Romani S, Hosseini SM, Mohebbi SR, Kazemian S, Derakhshani S, Khanyaghma M, Azimzadeh P, Sharifian A, Zali MR. Interleukin-16 gene polymorphisms are considerable host genetic factors for patients’ susceptibility to chronic hepatitis B infection. Hepat Res Treat. 2014;2014:790753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 197. | Formentini A, Braun P, Fricke H, Link KH, Henne-Bruns D, Kornmann M. Expression of interleukin-4 and interleukin-13 and their receptors in colorectal cancer. Int J Colorectal Dis. 2012;27:1369-1376. [PubMed] [Cited in This Article: ] |
| 198. | Zhu VF, Yang J, Lebrun DG, Li M. Understanding the role of cytokines in Glioblastoma Multiforme pathogenesis. Cancer Lett. 2012;316:139-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 199. | Brombacher F. The role of interleukin-13 in infectious diseases and allergy. Bioessays. 2000;22:646-656. [PubMed] [Cited in This Article: ] |
| 200. | Hölscher C. Interleukin-13: genes, receptors and signal transduction. Interleukin-13. Lands Bioscience: Georgetown 2003; 1. [Cited in This Article: ] |
| 201. | Deng Y, Xie M, Xie L, Wang J, Li T, He Y, Li R, Li S, Qin X. Association between polymorphism of the interleukin-13 gene and susceptibility to hepatocellular carcinoma in the Chinese population. PLoS One. 2015;10:e0116682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 202. | Vladich FD, Brazille SM, Stern D, Peck ML, Ghittoni R, Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest. 2005;115:747-754. [PubMed] [Cited in This Article: ] |
| 203. | Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, Yu L, Stern DA, Graves PE, Lohman IC. Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J Immunol. 2006;177:8633-8642. [PubMed] [Cited in This Article: ] |
| 204. | Stark GR. How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev. 2007;18:419-423. [PubMed] [Cited in This Article: ] |
| 205. | de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912-920. [PubMed] [Cited in This Article: ] |
| 206. | Sobue S, Nomura T, Ishikawa T, Ito S, Saso K, Ohara H, Joh T, Itoh M, Kakumu S. Th1/Th2 cytokine profiles and their relationship to clinical features in patients with chronic hepatitis C virus infection. J Gastroenterol. 2001;36:544-551. [PubMed] [Cited in This Article: ] |
| 207. | Tang JT, Fang JY, Gu WQ, Li EL. T cell immune response is correlated with fibrosis and inflammatory activity in hepatitis B cirrhotics. World J Gastroenterol. 2006;12:3015-3019. [PubMed] [Cited in This Article: ] |
| 208. | Falleti E, Fabris C, Toniutto P, Fontanini E, Cussigh A, Caldato M, Rossi E, Bitetto D, Minisini R, Smirne C. Genetic polymorphisms of inflammatory cytokines and liver fibrosis progression due to recurrent hepatitis C. J Interferon Cytokine Res. 2007;27:239-246. [PubMed] [Cited in This Article: ] |
| 209. | Henri S, Stefani F, Parzy D, Eboumbou C, Dessein A, Chevillard C. Description of three new polymorphisms in the intronic and 3’UTR regions of the human interferon gamma gene. Genes Immun. 2002;3:1-4. [PubMed] [Cited in This Article: ] |
| 210. | Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol. 2000;61:863-866. [PubMed] [Cited in This Article: ] |
| 211. | Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet. 1999;26:1-3. [PubMed] [Cited in This Article: ] |
| 212. | Dai CY, Chuang WL, Hsieh MY, Lee LP, Hou NJ, Chen SC, Lin ZY, Hsieh MY, Wang LY, Tsai JF. Polymorphism of interferon-gamma gene at position +874 and clinical characteristics of chronic hepatitis C. Transl Res. 2006;148:128-133. [PubMed] [Cited in This Article: ] |
| 213. | Barrett S, Collins M, Kenny C, Ryan E, Keane CO, Crowe J. Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol. 2003;71:212-218. [PubMed] [Cited in This Article: ] |
| 214. | Abbas Z, Moatter T, Hussainy A, Jafri W. Effect of cytokine gene polymorphism on histological activity index, viral load and response to treatment in patients with chronic hepatitis C genotype 3. World J Gastroenterol. 2005;11:6656-6661. [PubMed] [Cited in This Article: ] |
| 215. | Sun J, Liao K, Wang Y, Zhang L. Interferon-γ polymorphism and hepatocellular carcinoma susceptibility: a meta analysis. Int J Clin Exp Med. 2015;8:5667-5674. [PubMed] [Cited in This Article: ] |
| 216. | Hagihara H, Nouso K, Kobayashi Y, Iwasaki Y, Nakamura S, Kuwaki K, Toshimori J, Miyatake H, Ohnishi H, Shiraha H. Effect of pegylated interferon therapy on intrahepatic recurrence after curative treatment of hepatitis C virus-related hepatocellular carcinoma. Int J Clin Oncol. 2011;16:210-220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 217. | Zhou H, Wang L, Li X, Song J, Jiang T, Wu X, Zhou S. Interferon-γ +874A/T polymorphism and hepatocellular carcinoma risk: a meta-analysis. Med Sci Monit. 2015;21:689-693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 218. | Wei YS, Lan Y, Zhang L, Wang JC. Association of the interleukin-2 polymorphisms with interleukin-2 serum levels and risk of nasopharyngeal carcinoma. DNA Cell Biol. 2010;29:363-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 219. | López RV, Zago MA, Eluf-Neto J, Curado MP, Daudt AW, da Silva-Junior WA, Zanette DL, Levi JE, de Carvalho MB, Kowalski LP. Education, tobacco smoking, alcohol consumption, and IL-2 and IL-6 gene polymorphisms in the survival of head and neck cancer. Braz J Med Biol Res. 2011;44:1006-1012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 220. | Song H, Chen L, Cha Z, Bai J. Interleukin 2 gene polymorphisms are associated with non-Hodgkin lymphoma. DNA Cell Biol. 2012;31:1279-1284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 221. | Bleeker A, Tjiam IA, Volkers AC, Smith-Bogers J. Analysis of external and internal interlibrary loan requests: aid in collection management. Bull Med Libr Assoc. 1990;78:345-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 222. | Wang Y, Shu Y, Jiang H, Sun B, Ma Z, Tang W. Lack of association between interleukin-2 (IL-2) gene rs2069762 polymorphism and cancer risk: a meta-analysis. Int J Clin Exp Med. 2015;8:12557-12565. [PubMed] [Cited in This Article: ] |
| 223. | Zicca E, Quirino A, Marascio N, Nucara S, Fabiani F, Trapasso F, Perrotti N, Strazzulla A, Torti C, Liberto MC. Interleukin 27 polymorphisms in HCV RNA positive patients: is there an impact on response to interferon therapy? BMC Infect Dis. 2014;14 Suppl 5:S5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 224. | Lyu S, Ye L, Wang O, Huang G, Yang F, Liu Y, Dong S. IL-27 rs153109 polymorphism increases the risk of colorectal cancer in Chinese Han population. Onco Targets Ther. 2015;8:1493-1497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 225. | Zhang M, Tan X, Huang J, Ke Z, Ge Y, Xiong H, Lu W, Fang L, Cai Z, Wu S. Association of 3 Common Polymorphisms of IL-27 Gene with Susceptibility to Cancer in Chinese: Evidence From an Updated Meta-Analysis of 27 Studies. Med Sci Monit. 2015;21:2505-2513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 226. | Ali YBM, El-Masry SA, El-Akhras BA, El-Shenawy SZ, El-SayedIH . Association of IL-27 gene polymorphism and risk of HBV viral infection in Egyptian population. Egyptian Journal of Medical Human Genetics. 2014;15:53-59. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 227. | Lu Y, Wu Z, Peng Q, Ma L, Zhang X, Zhao J, Qin X, Li S. Role of IL-4 gene polymorphisms in HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2014;9:e110061. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 228. | Wu Z, Qin W, Zeng J, Huang C, Lu Y, Li S. Association Between IL-4 Polymorphisms and Risk of Liver Disease: An Updated Meta-Analysis. Medicine (Baltimore). 2015;94:e1435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 229. | Xi XE, Liu Y, Lu Y, Huang L, Qin X, Li S. Interleukin-17A and interleukin-17F gene polymorphisms and hepatitis B virus-related hepatocellular carcinoma risk in a Chinese population. Med Oncol. 2015;32:355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |