Published online Dec 14, 2017. doi: 10.3748/wjg.v23.i46.8128
Peer-review started: February 24, 2017
First decision: April 28, 2017
Revised: September 15, 2017
Accepted: November 2, 2017
Article in press: November 2, 2017
Published online: December 14, 2017
To investigate the effect of epigallocatechin gallate (EGCG) on structural changes of gut microbiota in colorectal carcinogenesis.
An azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced colitis mouse model was established. Forty-two female FVB/N mice were randomly divided into the following three groups: group 1 (10 mice, negative control) was treated with vehicle, group 2 (16 mice, positive control) was treated with AOM plus vehicle, and group 3 (16 mice, EG) was treated with AOM plus EGCG. For aberrant crypt foci (ACF) evaluation, the colons were rapidly took out after sacrifice, rinsed with saline, opened longitudinally, laid flat on a polystyrene board, and fixed with 10% buffered formaldehyde solution before being stained with 0.2% methylene blue in saline. For tumor evaluation, the colon was macroscopically inspected and photographed, then the total number of tumors was enumerated and tumor size measured. For histological examination, the fixed tissues were paraffin-embedded and sectioned at 5 mm thickness. Microbial genomic DNA was extracted from fecal and intestinal content samples using a commercial kit. The V4 hypervariable regions of 16S rRNA were PCR-amplified with the barcoded fusion primers. Using the best hit classification option, the sequences from each sample were aligned to the RDP 16S rRNA training set to classify the taxonomic abundance in QIIME. Statistical analyses were then performed.
Treatment of mice with 1% EGCG caused a significant decrease in the mean number of ACF per mouse, when compared with the model mice treated with AOM/DSS (5.38 ± 4.24 vs 13.13 ± 3.02, P < 0.01). Compared with the positive control group, 1% EGCG treatment dependently decreased tumor load per mouse by 85% (33.96 ± 6.10 vs 2.96 ± 2.86, respectively, P < 0.01). All revealed that EGCG could inhibit colon carcinogenesis by decreasing the number of precancerous lesions as well as solid tumors, with reduced tumor load and delayed histological progression of CRC. During the cancerization, the diversity of gut microbiota increased, potential carcinogenic bacteria such as Bacteroides were enriched, and the abundance of butyrate-producing bacteria (Clostridiaceae, Ruminococcus, etc.) decreased continuously. In contrast, the structure of gut microbiota was relatively stable during the intervention of EGCG on colon carcinogenesis. Enrichment of probiotics (Bifidobacterium, Lactobacillu, etc.) might be a potential mechanism for EGCG’s effects on tumor suppression. Via bioinformatics analysis, principal coordinate analysis and cluster analysis of the tumor formation process, we found that the diversity of gut microbiota increased in the tumor model group while that in the EGCG interfered group (EG) remained relatively stable.
Gut microbiota imbalance might be a potential mechanism for the prevention of malignant transformation by EGCG, which is significant for diagnosis, treatment, prognosis evaluation, and prevention of colorectal cancer.
Core tip: Our study revealed the protective effect of epigallocatechin gallate (EGCG) on colorectal carcinogenesis and structural changes of intestinal flora in an animal model of colorectal cancer. EGCG was detected for its roles through azoxymethane/dextran sulfate sodium induced tumor (aberrant crypt foci) formation. The microbial population was compared among groups at different developmental stages by pyrosequencing of V4 regions of 16S rRNA genes. Results suggested that intestinal flora imbalance might be a potential mechanism for the prevention of malignant transformation by the green tea extract EGCG, which is significant for the diagnosis, treatment, prognosis evaluation, and prevention of colorectal cancer.
- Citation: Wang X, Ye T, Chen WJ, Lv Y, Hao Z, Chen J, Zhao JY, Wang HP, Cai YK. Structural shift of gut microbiota during chemo-preventive effects of epigallocatechin gallate on colorectal carcinogenesis in mice. World J Gastroenterol 2017; 23(46): 8128-8139
- URL: https://www.wjgnet.com/1007-9327/full/v23/i46/8128.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i46.8128
There are approximately 1.2 million people diagnosed with colorectal cancer (colon cancer) every year, and it is the third most common cancer in the human population. There is a fairly high incidence of colon cancer in developed countries[1]. The human body contains about 1000 kinds of bacteria, approximately 1014 in total, most of which are distributed in the large intestine. The total number of microbial genes is at least 150 times that of human genes[2], and both together are termed the human metagenome. Now it is considered that the gut microbiota plays an important role in the construction of the biological barrier, which helps with nutrient absorption, energy metabolism, and immune regulation[3]. Meanwhile, more and more evidence indicates that imbalance of gut microbiota plays an important role in gastrointestinal, metabolic, liver, and autoimmune diseases[4-6]. Previous studies found that similar to some tumor related microorganisms (such as Helicobacter pylori, human papilloma virus, and hepatitis B virus, etc.), many bacteria (such as toxigenic fragile bacteroides, Enterococcus faecalis, and Streptococcus spp.) may be toxic enough to cause colorectal cancer[7]. In addition, based on the study of fecal bacterial culture in patient and control populations, it was found that there were differences between the two groups[8]. With the development of molecular biology techniques, especially the next generation high throughput sequencing, detection of the complex structure of gut microbiota becomes feasible. A large number of reports have confirmed the role of bacterial flora in the pathogenesis of colorectal cancer, outlining the structure of gut microbiota in patients with colorectal cancer[9-12]. Previous studies in our group also found that there was a reduction in the abundance of colonic mucosal flora in patients with colorectal cancer, and the increase of Bactaeroides may be related to the occurrence of colorectal cancer[13]. The mechanisms by which bacteria cause colon cancer may include: inducing chronic inflammation of the intestine, producing carcinogenic metabolites, forming carcinogenic biofilms, etc[14]. Although the relationship between gut microbiota and the occurrence of colorectal cancer and its accurate mechanism remain unclear, it can be speculated that the regulation with gut microbiota as a target is a potential breakthrough point for the prevention and treatment of colorectal cancer[15]. Based on this theoretical basis, therapeutic means for the intervention of gut microbiota such as supplementation with probiotics and prebiotics, fecal bacteria transplantation, and even weight loss surgery have become research focuses[16-17].
The biological availability of epigallocatechin gallate (EGCG) is low, and its elimination half-time in blood is only 2.0-3.5 h. Most EGCG is fermented by bacteria in the large intestine, and discharged with stool. The large intestine is the main metabolic site for EGCG and is also the most active portion of the body for the interaction with bacteria. Importantly, when compared to smaller bacteria density in the proximal small intestine, the incidence of colorectal cancer is significantly higher[18]. Therefore, the interaction of EGCG with gut microbiota is very likely to affect the occurrence and development of colorectal cancer.
In this study, gut microbiota was considered a target for disease prevention and treatment, which will further broaden our understanding of the effects of drugs and diet on the body’s health by regulating gut microbiota in humans and animals.
Six-week-old female FVB/N mice (18-22 g) were purchased from Laboratory Animal Center of Shanghai East China Normal University (SCXK2011-0031) and were quarantined for 7 d before the experiment. All animals were housed in individual plastic cages (with 4 or 5 mice/cage) and maintained under controlled conditions of humidity (44% ± 5%), light (12 h light/dark cycles), and temperature (22 ± 2 °C). They had free access to drinking water and a pelleted basal diet. Azoxymethane (AOM, A5486-25 mg), a colonic carcinogen, EGCG (E4268-100MG), and dextran sodium sulfate (DSS, 42867-5G) for the induction of colitis were purchased from Sigma-Aldrich (St. Louis, MO, United States).
All animal experiments were performed in compliance with the Institutional Animal Care and Use Committee of the East China Normal University. Forty-two female FVB/N mice were randomly divided into the following three groups (Figure 1): group 1 (10 mice, negative control) was treated with vehicle; group 2 (16 mice, positive control) was treated with AOM plus vehicle; group 3 (16 mice, EG) was treated with AOM plus EGCG. AOM (30 mg/kg, total dose) was administered intraperitoneally (IP), at single doses of 10 mg/kg body weight, on the first days of week 1, week 4, and week 7. One day after each injection, the mice received 2.5% (v/v) DSS in drinking water for 3 consecutive days. One percent (v/v) of EGCG was given by gavage in group 3 throughout the experiment.
Half of all animals were sacrificed by asphyxiation with CO2 8 wk after the first AOM injection. A significant number of aberrant crypt foci (ACF) were observed at this time. The remaining animals were sacrificed at week 13 of the experiment. The gut tissue samples of all animals were collected for further investigation. Stool samples were collected at regular intervals and snap frozen in liquid nitrogen during the course of the study. The stool samples were then transferred to -80 °C until DNA extraction was performed.
For ACF evaluation, the colons were rapidly took out after sacrifice, rinsed with saline, opened longitudinally, laid flat on a polystyrene board, and fixed with 10% buffered formaldehyde solution before being stained with 0.2% methylene blue in saline. After measurement of the length (from the ileocecal junction to the anal verge), the colon specimens were cut into two parts: the distal part (5 cm from the anus) and the proximal part (the remainder of the colon). Specimens were examined using a light microscope at × 20 magnification. Only foci with four or more crypts were evaluated since they indicate early neoplastic occurrence. ACF were distinguished from their surrounding normal crypts by greater size, larger and elongated luminal opening, thicker lining, and compression of the surrounding epithelium. The total number of ACF throughout the colon was scored.
For tumor evaluation, the colon was macroscopically inspected and photographed, then the total number of tumors was enumerated and tumor size measured. Tumor load was the accumulation of diameter (average of three diameter measurements). For histological examination, colon tumors were separately excised and fixed in 10% neutral phosphate-buffered formalin.
For histological examination, the fixed tissues were paraffin-embedded and sectioned at 5 mm thickness. Sections were H&E stained and then microscopically examined by two independent researchers in a blind fashion. Microscopic observations were graded according to the morphological criteria.
Microbial genomic DNA was extracted from fecal and intestinal content samples using the TIANGEN DNA stool mini kit (TIANGEN, cat#DP328) following the manufacturer’s guidelines. The V4 hypervariable regions of 16S rRNA were PCR-amplified with the barcoded fusion primers. The PCR condition was as follows: initial denaturation at 94 °C for 5 min; denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s, repeated for 25 cycles; and final extension at 72 °C for 7 min. PCR products were extracted using a QIAGEN quick Gel Extraction Kit (QIAGEN). Then a sequencing library for each sample was constructed with the Illumina TruSeq DNA Sample Preparation Kit. For each sample, Barcoded V4 PCR amplicons were sequenced with Illumina Miseq platform. 16S rRNA amplification and sequencing services were provided by Personal Biotechnology Co.Ltd. (Shanghai, China). Sequence reads were eliminated if they contained ambiguous bases, if average paired score was lower than 25, if homopolymer run exceeded 6, if there were mismatches in the primers, or if sequence length was shorter than 100 bp. Sequences that overlapped the region between R1 and R2, longer than 10 bp without any mismatches, were assembled according to their overlap sequence. This step was ensured to remove chimeras. The sequence reads which could not be assembled were discarded. Barcode and sequencing primers were trimmed from sequence reads. Trimmed and assembled sequences were uploaded to QIIME for further analysis (This part of the study was assisted by Personal bioBiological Technology Company).
Using the best hit classification option, the sequences from each sample were aligned to the RDP 16S rRNA training set to classify the taxonomic abundance in QIIME[19]. Delineation of operational taxonomic units (OTUs) was conducted with UCLUST function in QIIME at a 97% cutoff[20]. Richness estimators (Ace, chao) and diversity estimators (Shannon index) were calculated with mothur software package[21-23]. We also conducted the UniFrac distance metrics analysis using OTUs from each sample, and performed the principal co-ordinates analysis and NMDS results in terms of the matrix of distance (This part of the study was assisted by Personal bioBiological Technology Company).
Statistical analyses were performed using SPSS version 20.0 and Graph prism 5.0. Results are expressed as mean ± SE of the mean for normally distributed data. Inter-group comparisons for body weights were performed using one-way ANOVA with correction for multiple comparisons by Tukey’s post hoc test; P < 0.05 was considered statistically different. Inter-group comparisons of ACF counts and tumor measurements were assessed with the 2 × 2 factorial designs; P < 0.05 was regarded as statistically significant for each main effect and interaction.
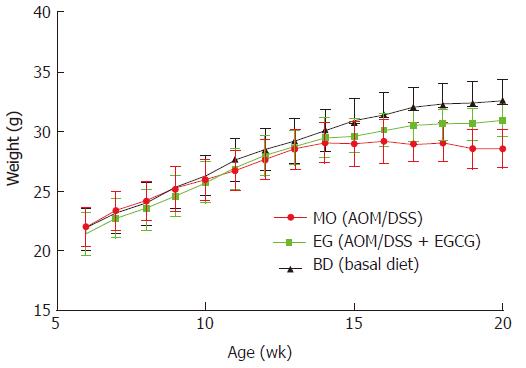
As shown in Figure 2, all mice had a steady body weight gain and the administration of AOM/DSS and 1% EGCG did not affect the growth of the mice during the first eight weeks in all groups measured at different time points. Also, we observed a significant body weight loss or toxicity in mice administered AOM/DSS only; however, no significant body weight loss was observed in mice treated with 1% EGCG after AOM/DSS. Table 1 compares the differences in the mean weights of the three groups at the time of sacrificing. In groups 1 and 2, all mice survived to the end of the experiment and two mice died in group 3, from asphyxia caused by gavage and intestinal obstruction. During the entire period of the experiment, there were no signs of toxicity or otherwise adverse conditions suggesting adverse effects caused by administration of EGCG.
In the current study, we used the well-established protocol of the mouse colorectal carcinogenesis model system and chose ACF as the end point because it is considered to be pre-neoplastic lesions and is regarded as a useful biomarker. Half of mice in each group were harvested for ACF evaluation at 8 wk. Table 2 shows the length of the large intestine as well as the number of ACF in both distal (5 cm from the anus) and proximal (the remainder of the colon) parts among the three groups. Mice fed a basal diet (n = 5) showed no evidence of ACF formation in the colonic mucosa. All AOM/DSS treated mice (n = 8, r = 100% incidence) developed ACF, whereas those treated with 1% EGCG (n = 8, r = 75% incidence) were found to have significantly fewer ACF. Almost all ACF were observed in the distal colon. In group 3, treatment of mice with 1% EGCG caused a significant decrease in the mean number of ACF per mouse, when compared with the model mice treated with AOM/DSS (5.38 ± 4.24 vs 13.13 ± 3.02, P < 0.01). These results indicate that 1% EGCG significantly inhibited ACF formation induced by AOM/DSS in FVB mice.
Starting from 10 wk after treatment, mice in the model group showed apparent diarrhea and rectal bleeding. The presence and development of inflammation manifested clearly. In the EG group, suppression of experimental colitis by EGCG was not only evident during AOM/DSS treatment, but also obvious after the cessation of DSS administration (i.e., week 10), suggesting that EGCG significantly promoted recovery from colitis. Unfortunately, a mouse treated by AOM/DSS died in the 12th week of colonic obstruction and cachexia. As a result, we decided to euthanize the remainder of mice in the 13th week in order to keep the mouse life cycle in concordance.
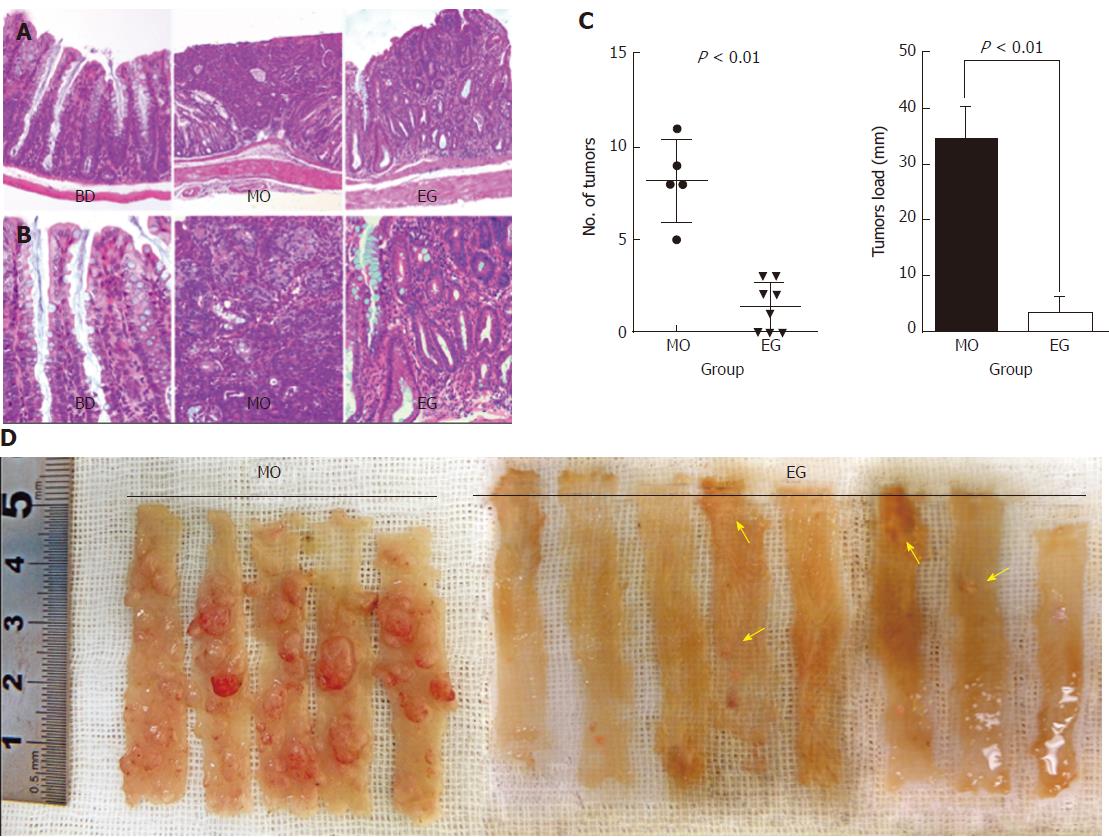
At necropsy, macroscopically, nodular, polypoid, or caterpillar-like tumors were observed in the entire colons of mice. Neither benign adenomas nor metastatic invasion of the colonic tumors to the liver, peritoneum, or regional lymph nodes were observed. Tumors mostly occurred in the distal colon. Figure 3C summarizes the total number of tumors per mouse and compares the mean number as well as tumor load between the MO and EG groups. Mice fed a basal diet showed no evidence of tumor formation at 13 wk. For AOM/DSS treated mice, colonic adenocarcinomas developed in all mice (5/5, 100%) 13 wk after the first injection of AOM, while the incidence in group 3 was 62.5% (5/8). The number of malignant colonic tumors per mice was 8.2 ± 2.2 and 1.4 ± 1.3 for the AOM/DSS and EGCG treatment groups, respectively (P < 0.01). In addition, compared with the positive control group, 1% EGCG treatment decreased tumor load per mouse by 85% (33.96 ± 6.10 vs 2.96 ± 2.86, respectively, P < 0.01) as showed in Figure 3C. Likewise, Figure 3D is a representative macroscopic morphologic depiction of the distal colon for the model and EG groups. Obvious tumorigenesis was observed in the model group. However, in the EGCG treatment group, the tumor number and size were significantly fewer and relatively small, respectively. This result suggests that the inhibition of colorectal tumor development by EGCG was due to not only a reduction in the number of but also the size of tumors. Figure 3 (A and B) shows representative H&E staining of histological sections of the three groups. In colonic tissue from the model animals, multifocal adenomatous lesions were observed without invasion into the submucosa; there was mild inflammation with cryptitis, and mild degree of loss of goblet cells, fibrosis, and apoptotic changes. For the EGCG treatment group, the mucosa revealed tightly packed glands with a normal number of goblet cells while crypt architecture remained normal. Compared to the model, the histological sections of the EGCG treatment group are more similar to those of the control group.
Table 3 shows the range of valid and trimmed sequences of different groups of mice. In this study, OTUs were defined as sharing 97% sequence identity using furthest neighbor method (http://www.mothur.org/wiki/Cluster). The total number of OTUs at 97% similarity level was 41923, with an average of 2096 OTUs per sample. The value of Good’s coverage for each group was over 93%, indicating that the 16S rRNA sequences identified in the groups represent the majority of bacteria present in the study samples.
| Group | Valid sequences (mean) | Trimmed sequences (mean) |
| BD Group | 75116-49662 (615022) | 65936-45406 (54162) |
| MO Group | 82804-25360 (56551 ) | 73924-22895 (50842 ) |
| EG Group | 79506-40705 (65124) | 74876-36795 (59693 ) |
Whereas we did not observe the plateau of the refraction curve with the current sequencing, the Shannon diversity estimates of all samples had already reached stable values at this sequencing depth, which suggests that, although identification of new phylotypes would be expected from additional sequencing, the range of diversity within the samples had been captured.
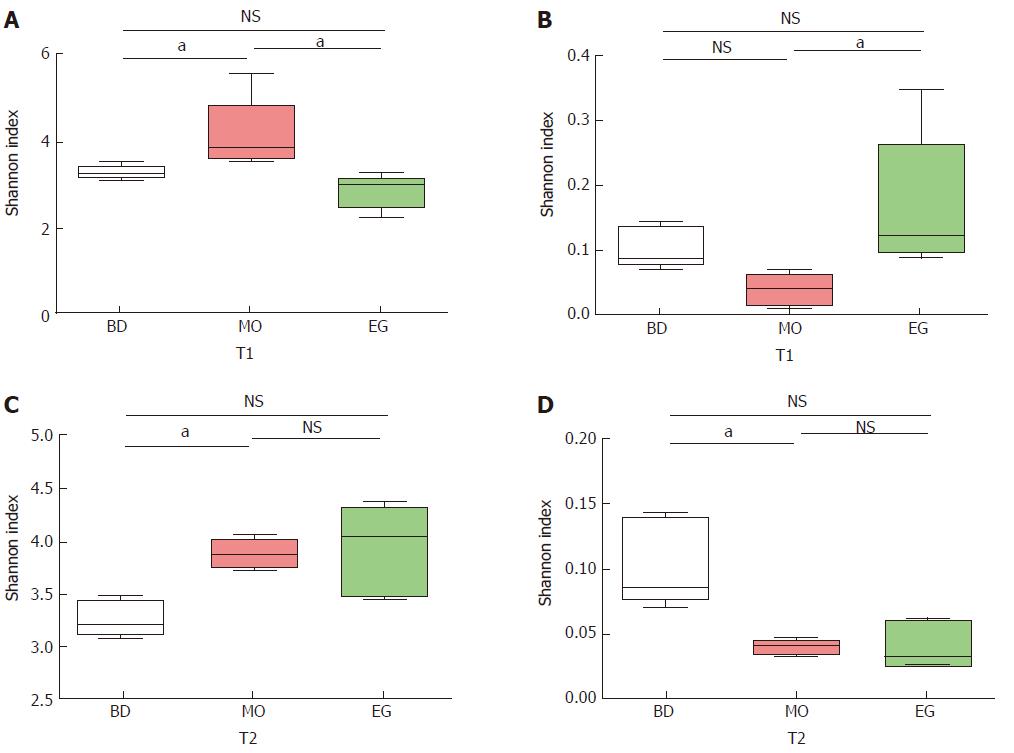
As Figure 4 shows, we examined the estimators of community richness (Chao and Ace indexes) and diversity and evenness (Shannon and Simpson indexes) in MO and EG samples during the 8th week. Statistically significant differences were seen in the Shannon and Simpson indexes between the MO group and control group (4.10 vs 3.23, P = 0.022), while no significance was seen between the EG group and control group (2.81 vs 3.23, P = 0.230) (Figure 4A). The Simpson indexes between the MO and control groups (0.04 vs 0.10, P = 0.022) had no significant differences, while significant differences were seen between the MO and EG groups (0.04 vs 0.17, P = 0.009) (Figure 4B).
During the 11st week (T2), the Shannon index in the MO group became higher than that in the BD group (3.88 vs 3.23, P = 0.003), but no significant difference between the BD and EG groups was noted (3.23 vs 3.94, P = 0.058, Figure 4C). The same result can be noted for Simpson index (Figure 4D).
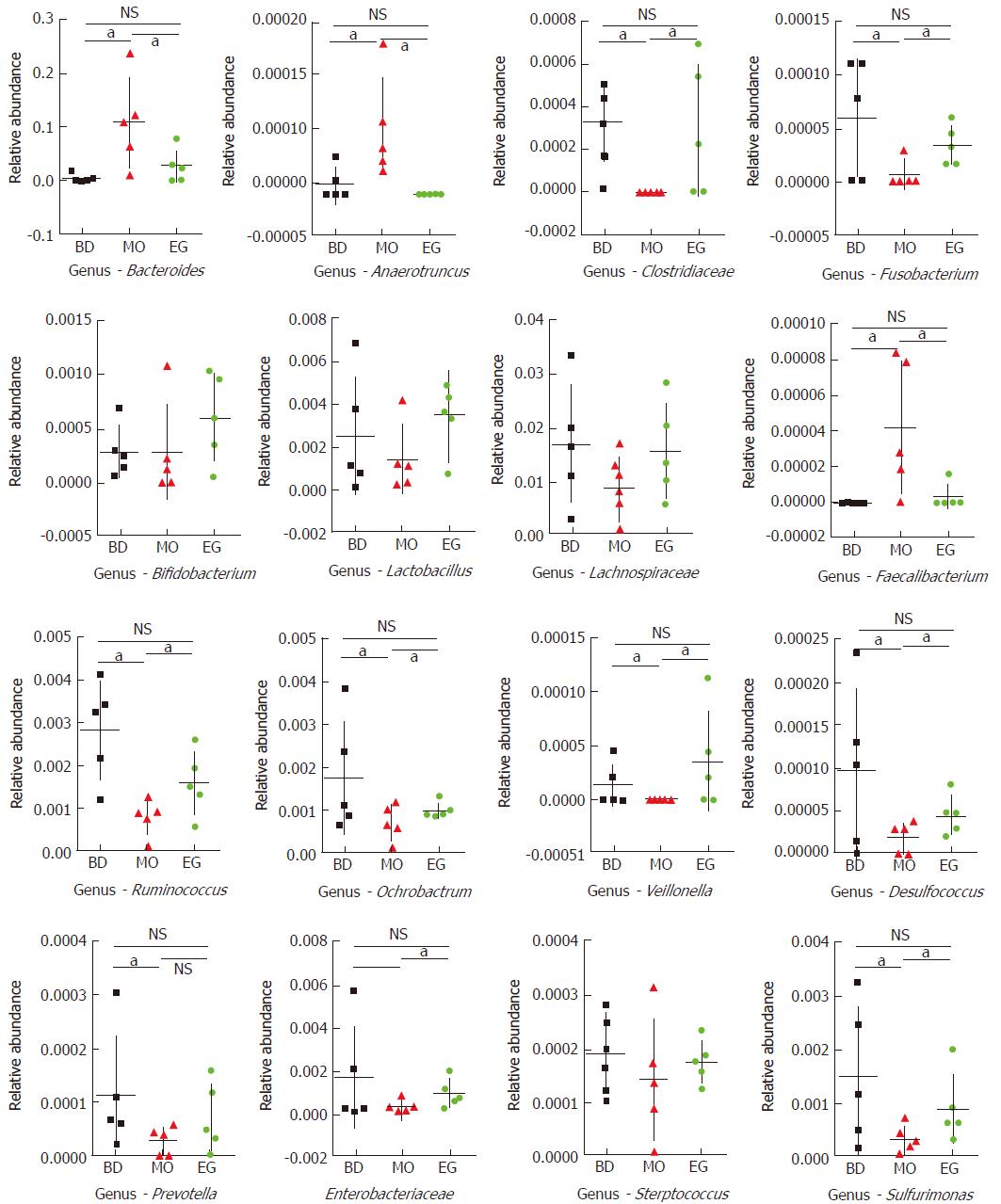
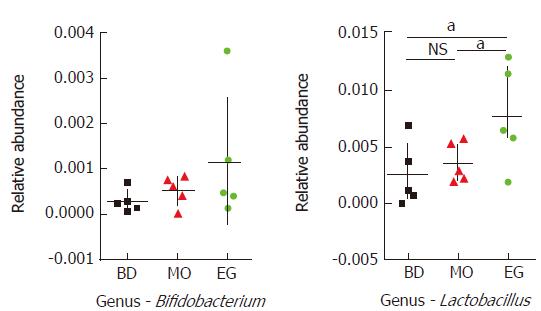
In this study, 339 genera were annotated; we only compared the abundant differences of 16 species among groups. As Figure 5 shows at the 8th week, the abundance of Bacteroides as well as Anaerotruncus in the MO group was significantly higher than that in the BD group and EG group, but Clostridiaceae which can produce butyrate, and Ruminococcus in the MO group were significantly reduced. Butyrate is recognized to alleviate intestinal inflammation. The enrichment of Fusobacterium was previously reported in colorectal cancer tissue but was not reproduced in our study. As the recognized probiotics, Bifidobacterium and Lactobacillus were increased in the EG group.
The abundance of other important butyrate-producing strains such as Lachnospiraceae and Faecalibacterium were not significantly decreased in the MO group. In addition, we found that Ochrobactrum, Veillonella, and Desulfococcus had significantly lower abundance in the MO group. In addition, common intestinal opportunistic pathogens, such as Enterobacteriaceae, Streptococcus, and Prevotella, were not found to be enriched in the MO group.
Specifically, at week 11, the abundance of Lactobacilli in the EG group was significantly higher than that in the BD group and the MO group (P1 = 0.028, P2 = 0.049). The difference of Bifidobacterium was not significant among the three groups (Figure 6).
Sporadic reports suggested that EGCG may inhibit proliferation of intestinal pathogenic bacteria (such as Clostridium perfringens, Clostridium difficile, and Bacaeroides) in vitro, but EGCG is less inhibitory to or even stimulatory to probiotic bacteria (such as Bacillus bifidus and Lactobacillus). Animal and human experiments also confirmed that EGCG may affect the growth and floral structure of specific intestinal specific bacteria, and participates in the regulation of the body’s energy metabolism[24,25]. The role of gut microbiota as an intermediate link for EGCG’s impact on colorectal cancer occurrence and development is still unclear.
EGCG acts as a means of chemical prevention of colorectal cancer, and gut microbiota is very likely to be one of its targets. Its impact on overall floral structure or specific bacteria may be related to the inhibition of colorectal cancer.
In this study, we used a mouse model of colorectal cancer (chemically induced) and intragastrically administered the mice with EGCG to inhibit colorectal cancer. At the same time, to further explore the mechanism of EGCG in the prevention of colorectal cancer, we collected the fresh feces of mice during the interference of tumor formation. Via bacterial 16s cDNA high throughput sequencing in combination with bioinformatics, we analyzed changes in the structure of gut microbiota in mice, and flora differences among the groups, to uncover the role of bacterial flora in the EGCG-mediated inhibition of carcinogenesis.
The development of colorectal cancer is generally known to take at least 3 years (progressing from normal mucosa, to adenoma, and finally to adenocarcinoma) and to include three phases, i.e., initial mutation, cancer promotion, and progression. The main purpose for the study of chemical prevention is preventive treatment of disease. ACF are a commonly acknowledged precancerous lesion of humans and rodents and is convenient for in vitro recognition after methylene blue staining. ACF often show proliferative lesions with poor differentiation, and enhanced gland cell division and proliferation. At the molecular level, mutations of the APC and ras genes are common. At the end of the eighth week in this experiment, the results showed that EGCG could effectively reduce the number of precancerous lesions of colorectal cancer. ACF in the distal intestine of the interfering group were reduced by 59%, indicating that EGCG may play a preventive role in the early stages (mutation and cancer promoting stages) of colorectal cancer. In the past, Xiao et al[26] interfered the AOM induced tumor formation process in rats with a high fat diet via green tea polyphenols (65% of the components were EGCG). That study also showed reduced ACF numbers, in particular the larger highly atypical hyperplastic ACF types. Similar results were confirmed in mice[27]. To determine the effect of EGCG on colorectal cancer formation, in another study, tumor formation in APCmin/+ was interfered by 0.08% and 0.16% of EGCG. Tumor numbers were decreased by 37% and 47%, respectively. In our study, however, at the end of the 13th week after intervention, tumor number in the interfered group was reduced by 82.9% as compared to the control group, and tumor load was reduced by 91.3%, both indicating that the inhibitory effect of EGCG on tumor formation was more obvious than in the past. The possible reason is that the concentration of EGCG in our study was 1%, much higher than those used in previous studies. On the other hand, this also suggested that EGCG plays a preventive role in colorectal cancer occurrence in a dose-dependent manner.
We conducted further studies to uncover the effect of green tea extract EGCG on the gut microbiota of mice, using macro-genomics in combination with bioinformatics analysis. Considering the complexity of gut microbiota, this study was designed with the following research objectives in mind: (1) to preliminarily understand the compositions of the gut microbiota in mice at the 8th week ( precancerous stage) and the 11th week (progressive stage) of colorectal cancer; (2) to preliminarily understand the change in gut microbiota during the process of colorectal cancer progression in mice as compared to healthy mice, and to clarify the effects of EGCG intervention on this change; and (3) through statistical analysis and bioinformatics, to try to find specific bacteria that can distinguish the structure of each group of bacteria, i.e., the specific bacterial target of EGCG intervention.
With the aid of the 454 sequencing platform for the detection of 16S rDNA V3 region or V3/V4 region[10], we analyzed the structure of gut microbiota in mice by amplification of the V4 region in this study, which was good for identification. The sequencing results were under strict quality control. The distribution map, dilution curve, and abundance cure of the sequences acquired all confirmed that the sequencing results could well reflect the structure of the gut microbiota in mice.
It is worthwhile to note that in 2011, Kosticet al[11], Castellarin et al[12], and Marchesi et al[28] simultaneously found that there were gut microbiota disorders at varying degrees in colorectal cancer patients, using sequencing techniques, and the Fusobacterium was enriched in cancer tissues as compared to the healthy tissues. Meanwhile, it was also found that the abundance of Fusobacterium was directly proportional to the rate of lymph node metastasis. It is noticeable from the above reports that despite the increasing concern about the relationship between colorectal cancer and bacteria and that exciting results were obtained by relevant studies, the studies focused mainly on the structure of gut microbiota or specific bacteria during colorectal cancer progression (especially during the middle and late stages), which was only a snapshot of the big picture. Little is known about the dynamic changes of the gut microbiota in the process of tumor formation, especially for the status of gut microbiota during the early stage of disease progression, which indeed is a key time point for chemical prevention.
At the same time, it will be informative for the elucidation of flora changes as a direct cause of colorectal cancer formation, or flora changes as a direct result of colorectal cancer occurrence and/or progression. In this study, we examined and compared the composition of gut microbiota in mice with precancerous lesions and in mice during tumor progression. This was helpful for further understanding the roles of different species of bacteria in carcinogenesis, cancer promotion, and tumor suppression, and therefore the potential EGCG target bacteria could be explored. In fact, during the precancerous stage, there were anomalies seen in gut microbiota structures in the mice of the MO group, mainly manifesting as enrichment of Bacaeroides and Anaerotruncus and reduced abundance of Clostridiaceae, Ruminococcus, Ochrobactrum, Veillonella alcalescens, Desulfococcus, etc. The enrichment of Bacteroides in the MO group was consistent with our previous study on mucosal flora in patients with colorectal cancer[15]. The relationship between Anaerotruncus and colon cancer occurrence is still unclear. Previous studies have suggested that Bacaeroides and its lower species could promote the occurrence and development of colorectal cancer by inducing chronic inflammation, producing tumor-promoting metabolites, or changing the expression of host mucosal glycosylation. Among them, toxigenic fragile bacteroides is representative and was considered a pathogenic bacterium of colorectal cancer, initially because it was found to be able to produce the toxins of toxigenic fragile bacteroides (bacteroides fragilis toxin, BFT). BFT can rapidly change the structure and function of intestinal mucosal epithelial cells (decomposition of tumor suppression protein and E-cadherin), enhancing the intranuclear expression of Wnt/B adhesion protein signaling pathway, and therefore inducing continuous proliferation of colon adenocarcinoma cells and the expression of oncogene MYC. Besides, BFT may induce inflammatory responses by inducing expression of cytokines in intestinal mucosal epithelia cells through the NF-κB signaling pathway. Notably, increased expression of NF-κB signaling pathway could also promote the carcinogenesis of mucosal epithelium. In animal experiments, continuous colonization of toxigenic fragile bacteroides into APCmin/+ mice could greatly promote the incidence of adenomas[29]. Sears et al[30] suggested that this type of bacterium might induce mucosal immune defense changes to promote tumor development through direct secretion of endotoxin or indirect changes of gut microbiota, and might supplant probiotics to further expand inflammatory responses, promoting occurrence of multiple pathogenic bacteria and ultimately promoting tumor progression.
Furthermore, it was found in this study that in the precancerous stage, the Clostridiaceae and bacteria of the Ruminococcus maintained their low abundances in the tumor model group, but not in the intervention group. The Clostridiaceae includes multiple important butyrate producing bacteria, such as Eubacterium rectale and Roseburia intestinalis. This type of bacterium was reported to be reduced in the intestinal tract of patients with colorectal cancer, obese patients, and patients with a high fat diet. Similarly, Ruminococcus is also butyrate producing bacteria, and they can effectively decompose and ferment the cellulose and polysaccharide in foods into short chain fatty acids, such as butyrate. Butyrate in the intestinal tract may provide the intestinal mucosa with energy, promoting sodium and potassium absorption. More importantly, it can reduce intestinal inflammation, protect the intestinal mucosal barrier, and even inhibit the growth of colon cancer cells[31,32]. A study by Jahns et al[33] on human colorectal cancer tissues showed that butyrate could inhibit COX2 gene expression in nodal tissues and the activities of related proteins; in vitro studies also found that butyrate might activate caspase 3 and caspase 9 to induce apoptosis of colorectal cancer cells, upregulate pro-apoptotic gene BAK expression, and activate mitochondrial pathways to induce cell apoptosis and the like.
The gut microbiota of mice in the same tumor model group was different. The structure of the gut microbiota of mice interfered with green tea extract EGCG basically remained relatively stable, and bioinformatics analysis showed that the structure of the flora in these mice was similar to that of healthy mice, with no obvious fluctuations in the number of bacteria. Instead, probiotics such as Lactobacilus and Bifidobacterium were increased in the EG group, and the relative abundance of the former was significantly increased after long-term EGCG intervention. Probiotics are defined by WHO/FAO as “live bacteria that can promote body health if taken in a proper amount”. In recent years, some studies have showed that probiotics may have certain antagonistic effects on tumors. For example, Lactobacillus may significantly reduce the incidence of mouse sarcoma, colorectal cancer, and bladder cancer. Its main mechanisms may include optimization of the combined gut microbiota to inhibit the growth of carcinogenic bacteria; production of anti-tumor active substances; and stimulation and enhancement of the host immune system to prevent chronic inflammation.
It could be inferred accordingly that EGCG plays a role in cancer inhibition by maintaining the stability of gut microbiota, inhibiting the proliferation of potential pathogenic bacteria, and promoting the enrichment of probiotics. However, gut microbiota is influenced by many factors, such as diet and region, for its role in health promotion. Further study is certainly required to uncover the causal relationship(s) between gut microbiota and colorectal cancer, the interaction between bacteria and the host, as well as the interaction between EGCG and bacteria.
Studies have shown that gut microbiota plays a certain role in the occurrence and development of cancer. Inhibition of colorectal carcinogenesis by green tea, especially epigallocatechin gallate (EGCG), has been extensively investigated, but studies on microbial flora are much fewer. The interaction of EGCG with gut microbiota is very likely to affect the occurrence and development of colorectal cancer and the research of EGCG is needed. To further confirm the protective effect of EGCG on colorectal carcinogenesis and observe structural changes of gut microbiota involved during this process, an animal model of colorectal cancer was established.
A large number of reports have confirmed the role of bacterial flora in the pathogenesis of colorectal cancer, outlining the structure of gut microbiota in patients with colorectal cancer. Previous studies found that there was a reduction in the abundance of colonic mucosal flora in patients with colorectal cancer, and the increase of Bactaeroides may be related to the occurrence of colorectal cancer. The biological availability of EGCG in the current study is low. Most EGCG is fermented by bacteria in the large intestine, and discharged with stool. The large intestine is the main metabolic site for EGCG and is also the most active portion of the body for the interaction with bacteria.
The authors used a mouse model of colorectal cancer and intragastrically administered the mice with EGCG to inhibit colorectal cancer. Via bacterial 16s cDNA high throughput sequencing in combination with bioinformatics, we analyzed changes in the structure of gut microbiota of mice, and flora differences among the groups, to uncover the role of bacterial flora in the EGCG-mediated inhibition of carcinogenesis. We conducted further studies to uncover the effect of green tea extract EGCG on the gut microbiota of mice, using macro-genomics in combination with bioinformatics analysis.
EGCG plays a role in cancer inhibition by maintaining the stability of gut microbiota, inhibiting the proliferation of potential pathogenic bacteria, and promoting the enrichment of probiotics. Gut microbiota imbalance might be a potential mechanism for the prevention of malignant transformation by the green tea extract EGCG, which is significant for the diagnosis, treatment, prognosis evaluation, and prevention of colorectal cancer.
EGCG is extracted from the green tea. Most EGCG is fermented by bacteria in the large intestine, and discharged with stool. The large intestine is the main metabolic site for EGCG and is also the most active portion of the body for the interaction with bacteria.
In general, this manuscript provides the useful information about the relationship between epigallocatechin gallate and colorectal cancer. However, there are also some problems and flaws in presentation. Authors did not describe the mechanism of how EGCG suppressed colorectal cancer. In Discussion part, there is little consideration on the obtained data in this experiment, and authors should increase the consideration of the obtained data in this experiment.
The authors would like to thank Personal bioBiological Technology Company (Shanghai, China) for the contributions to this research as well as Dr. Ma Xueyun from the East China Normal University for technical support of animal experiments.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Hosomi R S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Ma YJ
| 1. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1965] [Cited by in F6Publishing: 2135] [Article Influence: 213.5] [Reference Citation Analysis (1)] |
| 2. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7052] [Cited by in F6Publishing: 7123] [Article Influence: 508.8] [Reference Citation Analysis (3)] |
| 3. | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2779] [Cited by in F6Publishing: 2856] [Article Influence: 238.0] [Reference Citation Analysis (0)] |
| 4. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1230] [Cited by in F6Publishing: 1330] [Article Influence: 133.0] [Reference Citation Analysis (2)] |
| 5. | Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34:39-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 367] [Cited by in F6Publishing: 382] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 6. | Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 315] [Cited by in F6Publishing: 292] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 7. | Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317-328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 503] [Cited by in F6Publishing: 540] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 8. | O’Keefe SJ, Chung D, Mahmoud N, Sepulveda AR, Manafe M, Arch J, Adada H, van der Merwe T. Why do African Americans get more colon cancer than Native Africans? J Nutr. 2007;137:175S-182S. [PubMed] [Cited in This Article: ] |
| 9. | Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907-1911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 566] [Cited by in F6Publishing: 638] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 10. | Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 834] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 11. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1199] [Cited by in F6Publishing: 1323] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 12. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1164] [Cited by in F6Publishing: 1324] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 13. | Huipeng W, Lifeng G, Chuang G, Jiaying Z, Yuankun C. The differences in colonic mucosal microbiota between normal individual and colon cancer patients by polymerase chain reaction-denaturing gradient gel electrophoresis. J Clin Gastroenterol. 2014;48:138-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. 2014;111:18321-18326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 474] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 15. | Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 16. | Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 643] [Cited by in F6Publishing: 671] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 17. | Borody TJ, Brandt LJ, Paramsothy S. Therapeutic faecal microbiota transplantation: current status and future developments. Curr Opin Gastroenterol. 2014;30:97-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 18. | Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 19. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23810] [Cited by in F6Publishing: 22368] [Article Influence: 1597.7] [Reference Citation Analysis (0)] |
| 20. | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194-2200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9953] [Cited by in F6Publishing: 9057] [Article Influence: 696.7] [Reference Citation Analysis (0)] |
| 21. | Pitta DW, Pinchak E, Dowd SE, Osterstock J, Gontcharova V, Youn E, Dorton K, Yoon I, Min BR, Fulford JD. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol. 2010;59:511-522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 22. | Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306-317. [PubMed] [Cited in This Article: ] |
| 23. | Mahaffee WF, Kloepper JW. Temporal Changes in the Bacterial Communities of Soil, Rhizosphere, and Endorhiza Associated with Field-Grown Cucumber (Cucumis sativus L.). Microb Ecol. 1997;34:210-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Unno T, Sakuma M, Mitsuhashi S. Effect of dietary supplementation of (-)-epigallocatechin gallate on gut microbiota and biomarkers of colonic fermentation in rats. J Nutr Sci Vitaminol (Tokyo). 2014;60:213-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Axling U, Olsson C, Xu J, Fernandez C, Larsson S, Ström K, Ahrné S, Holm C, Molin G, Berger K. Green tea powder and Lactobacillus plantarum affect gut microbiota, lipid metabolism and inflammation in high-fat fed C57BL/6J mice. Nutr Metab (Lond). 2012;9:105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 26. | Xiao H, Hao X, Simi B, Ju J, Jiang H, Reddy BS, Yang CS. Green tea polyphenols inhibit colorectal aberrant crypt foci (ACF) formation and prevent oncogenic changes in dysplastic ACF in azoxymethane-treated F344 rats. Carcinogenesis. 2008;29:113-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Shimizu M, Shirakami Y, Sakai H, Adachi S, Hata K, Hirose Y, Tsurumi H, Tanaka T, Moriwaki H. (-)-Epigallocatechin gallate suppresses azoxymethane-induced colonic premalignant lesions in male C57BL/KsJ-db/db mice. Cancer Prev Res (Phila). 2008;1:298-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 28. | Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 396] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 29. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1113] [Cited by in F6Publishing: 1191] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 30. | Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 31. | Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1657] [Cited by in F6Publishing: 1670] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 32. | Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res. 2014;20:799-803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 33. | Jahns F, Wilhelm A, Jablonowski N, Mothes H, Radeva M, Wölfert A, Greulich KO, Glei M. Butyrate suppresses mRNA increase of osteopontin and cyclooxygenase-2 in human colon tumor tissue. Carcinogenesis. 2011;32:913-920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |