Published online Nov 28, 2020. doi: 10.3748/wjg.v26.i44.6923
Peer-review started: August 31, 2020
First decision: September 24, 2020
Revised: October 24, 2020
Accepted: November 13, 2020
Article in press: November 13, 2020
Published online: November 28, 2020
Inflammatory bowel disease (IBD) is a complex, immune-mediated gastrointestinal disorder with ill-defined etiology, multifaceted diagnostic criteria, and unpredictable treatment response. Innovations in IBD diagnostics, including developments in genomic sequencing and molecular analytics, have generated tremendous interest in leveraging these large data platforms into clinically meaningful tools. Artificial intelligence, through machine learning facilitates the interpretation of large arrays of data, and may provide insight to improving IBD outcomes. While potential applications of machine learning models are vast, further research is needed to generate standardized models that can be adapted to target IBD populations.
Core Tip: Artificial intelligence (AI) is a novel technological advancement that is rapidly growing in the field of inflammatory bowel disease. The use of AI and machine learning has been shown to aid in diagnosing and understanding severity of disease, predicting treatment response along with likelihood of disease recurrence and assisting with colorectal neoplasia screening in this patient population. Further studies are needed to understand the full impact AI may have on improving Crohn’s disease and ulcerative colitis patient outcomes.
- Citation: Kohli A, Holzwanger EA, Levy AN. Emerging use of artificial intelligence in inflammatory bowel disease. World J Gastroenterol 2020; 26(44): 6923-6928
- URL: https://www.wjgnet.com/1007-9327/full/v26/i44/6923.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i44.6923
Inflammatory bowel disease (IBD), comprised of ulcerative colitis (UC) and Crohn’s disease (CD), is a set of chronic, immunologically-mediated diseases of the gut that arise from a complex interaction of host genetics, their environment, and gut microbiome[1]. Due to its chronic relapsing nature, IBD is associated with considerable morbidity and impairment of quality of life.
Accurate diagnosis of IBD relies on a combination of clinical, endoscopic, histologic, laboratory, and radiographic data. Significant heterogeneity exists in both the quality of these critical diagnostic components and their subsequent interpretation, as they are inherently dependent on confounders, such as the technical skill and experience of the provider. Consequently, diagnostic algorithms and management of IBD can vary considerably amongst gastroenterologists.
Recently, “big data” from large clinical trials, electronic health records, medical imaging, biobanks, and multiomic (genomic, transcriptomic, metabolomic, and proteomic) databases have been increasingly employed in an effort to improve diagnostic accuracy and predictability of treatment response[2]. However, the use of big data in development of predictive models is confounded by high dimensionality of clinical and non-clinical factors[2]. To overcome this challenge, machine learning (ML) has been increasingly utilized to organize and interpret these large datasets in an effort to identify clinically meaningful patterns and translate them into improved patient outcomes[3]. This review highlights the nascent efforts to incorporate artificial intelligence (AI) and machine learning in the field of IBD.
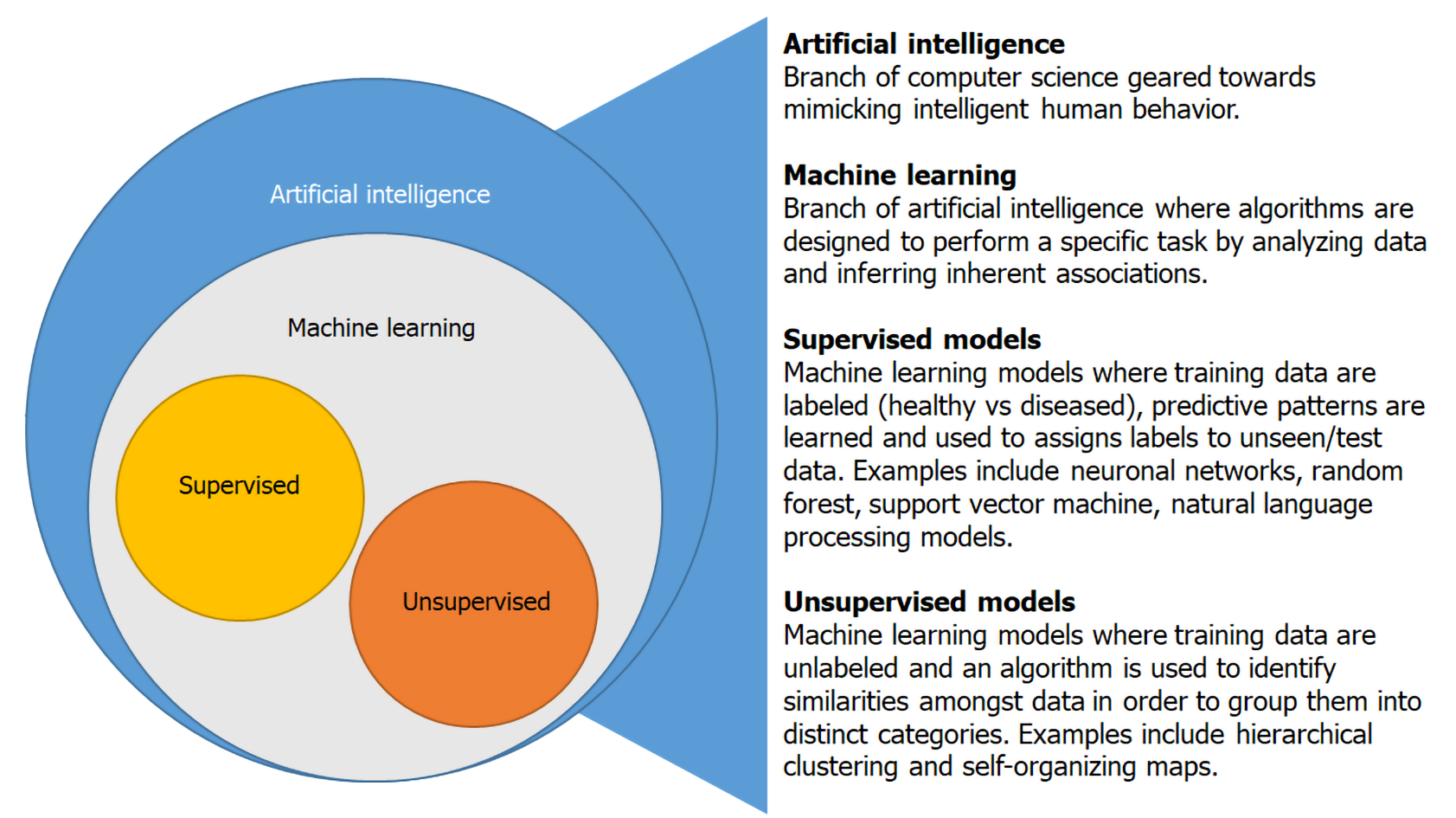
AI is an interdisciplinary branch of computer science focused on the development of machines that are programmed to perform tasks that imitate intelligent human behavior[2]. Machine learning is a subset of AI that utilizes algorithms with the goal of identifying patterns or generating predictive models as a result of “learning” from an input dataset[3]. This is achieved through supervised and unsupervised learning (Figure 1). In supervised learning, an algorithm is trained on a labeled training dataset to recognize patterns associated with specific groups (healthy vs diseased)[2]. The algorithm then uses what it has learned from the training dataset to place unseen data into specific categories. The most commonly employed supervised ML models are random forests, neuronal networks and support vector machines[3]. Unsupervised models use an unlabeled training dataset and the algorithm identifies patterns within the data guided by similar characteristics without knowledge of associated diagnosis or outcome[4]. Machine learning algorithms can provide a framework to identify previously undiscovered patterns within IBD and advance our understanding of IBD pathogenesis.
Endoscopic evaluation with mucosal biopsies remains an integral component to diagnosing IBD, yet it carries several limitations. In addition to being invasive, endoscopic assessment of disease severity remains subjective with high interobserver variability despite the use of scoring systems such as the endoscopic Mayo score. In addition, endoscopy may not be able to adequately distinguish between subtle overlapping features of the various IBD phenotypes, resulting in diagnostic dilemmas. Recent research has focused on integrating ML into the diagnostic paradigm in order to overcome some of these limitations.
Genome wide association studies have identified over 240 gene loci that have been linked to increased risk of developing IBD and can assist in distinguishing CD and UC[5,6]. Matrix factorization based ML models, using a combination of genome sequence data and biological knowledge have also been developed to distinguish patients with CD from healthy individuals (AUC = 0.816) without the need of histology[7]. In addition, ML assisted metagenomic, proteomic, and microbial prediction models have been utilized to identify predictive signatures distinguishing CD and UC, allowing for improved characterization of the IBD subtypes and risk stratification[8-10]. For example, Seeley et al[11] were able to discern between CD and UC with 76.9% accuracy using a histology based, mass spectrometry trained, support vector machine learning model by analyzing protein signatures from colonic tissue. Another study model used featured a selection algorithm combined with a support vector machine program to differentiate UC patients from healthy subjects based on the expression of 32 genes in colon tissue samples[12].
Similarly, the inherent subjectivity of endoscopic and radiographic assessment has led to a great interest in automating image interpretation. ML assisted analysis of computed tomography and magnetic resonance imaging has been shown to effectively identify structural bowel damage, such as stricturing disease in CD[13-15]. Image analyzing programs have also been adapted to improve the efficiency of previously time consuming manual review of video capsule endoscopy images[16].
AI has also been utilized to enhance interpretation of endoscopic images to assess disease severity[17-19].
For example, a convolutional neural network (CNN) was able to distinguish remission (Mayo 0-1) from moderate-to-severe disease (Mayo 2-3) with a sensitivity of 83.0% and specificity of 96.0%[18]. Another CNN model was able to detect severe CD ulcerations with high accuracy, 0.91 for grade 1 vs grade 3 ulcers[20]. Computer-aided diagnosis systems have also been shown to reliably predict persistent histologic inflammation during endocytoscopy with a sensitivity of 74%, specificity 97%, and accuracy of 91%[21].
Clearly, computer assisted imaging assessment has the potential to improve how we interpret diagnostic imaging to assess disease activity. As a result, it is expected that use of AI assisted imaging will continue to expand as the technology evolves.
Despite numerous pharmacologic advances over the past decade, clinicians are not yet able to predict treatment response in IBD. The prevailing trial and error approach has resulted in substantial variation in response rates to IBD therapy. This inefficiency, in combination with the significant pharmacoeconomic impact of failed therapies, has led to a growing interest in developing personalized approaches to IBD management.
To this end, machine learning algorithms have been developed to analyze predictive indicators of response for several IBD medications. One study was able to demonstrate that an ML algorithm could outperform conventional thiopurine metabolite testing to predict response[22]. Subsequent studies by Waljee et al[23,24] incorporated clinical trial data from the GEMINI I and GEMINI II studies with vedolizumab, and demonstrated that ML models could also be used to predict steroid free remission for UC and CD patients. In another study, a ML model used molecular and clinical data to identify biomarkers predictive of response to infliximab in refractory UC (accuracy = 70%). The authors identified tumor necrosis factor, interferon gamma, and lipopolysaccharide as potential regulators of infliximab response[25]. More recently, multi-omic factor analysis was used to identify transcriptomic and genomic biomarker panels that were predictive of ustekinumab response[26].
As a result of chronic inflammation, IBD patients are at increased risk for dysplasia and colorectal cancer. Patients with extensive colitis have up to a 19-fold increase in colorectal cancer risk when compared to the general population[27]. Despite the introduction of high-definition endoscopes and dye-based chromoendoscopy, the morbidity and mortality ascribed to IBD neoplasia has led to great interest in integrating AI-assisted detection systems into traditional colonoscopy. Multiple AI algorithms have been developed to alert the endoscopist of polyps in real-time, using visual or auditory cues during colonoscopy[28]. CNNs have been successfully used to improve adenoma detection in the general population, even for more experienced endoscopists[29]. The CNN model, when compared to expert review of machine-overlaid videos, had a sensitivity and specificity of 0.98 and 0.93, respectively[30]. Another computer-aided diagnosis system detected polyps by evaluating polyp boundaries and generating energy maps that corresponded to the presence of a polyp[31]. Machine learning may also aid in differentiation of colitis associated neoplasia, sporadic colorectal adenomas, and non-neoplastic lesions[32]. Artificial neuronal networks, when applied to complimentary deoxyribonucleic acid microarray data, have the potential to discriminate the subtle differences between polyp subtypes[32]. This may have longstanding effects on decreasing colorectal malignancy and may also guide surveillance in this population.
While AI has the ability to identify high risk subgroups and inform treatment decisions, there remain several obstacles to its routine implementation into clinical practice. Analysis of big datasets have generated several interesting disease observations but these have not necessarily translated into clinically meaningful benefits. The cross-sectional nature of ML datasets, lack of validated AI models, and paucity of biologic explanations for proposed associations makes it difficult to establish causation or adhere to AI algorithm generated decision recommendations. Additionally, the datasets that machine learning systems are dependent upon can be incomplete or of poor quality which can result in systematic errors or bias[2].
The highly sensitive nature of clinical data makes it logistically difficult to share freely amongst organizations, an obstacle potentially overcome by universal electronic medical records. There is also a need for adherence to unified data formats, as well as development of secure cloud storage facilities for easy extraction of large volumes of data[33]. Furthermore, varying degrees of clinical data are in the form of written notes, making data collection for input into mathematical ML models difficult. This can be overcome by development of natural language processing software to extract data from plain text[34]. Other challenges include the high dimensionality of clinical data, over-fitting of ML models, data security issues, and reliability of models to be generalized to the target population[2].
Overcoming the obstacles to machine learning in IBD will require collaborative efforts between clinicians, statisticians, and bioinformaticians to develop algorithms capable of generating clinically meaningful outputs. Prospective randomized trials are needed to confirm the efficacy and safety of AI assisted decision making before it can truly be translated to the bedside.
In summary, AI is a rapidly growing discipline that has the potential to revolutionize the field of inflammatory bowel disease. Machine learning approaches offer the ability to effectively synthesize and incorporate large amounts of data to improve diagnostic accuracy, uncover new disease associations, identify at risk individuals, and guide therapeutic decision making. While challenges to the routine use of AI in IBD remain, continued exploration of possible applications are expected to accelerate the drive toward precision medicine.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rodrigues AT S-Editor: Zhang L L-Editor: A P-Editor: Ma YJ
| 1. | Kuchroo VK, Ohashi PS, Sartor RB, Vinuesa CG. Dysregulation of immune homeostasis in autoimmune diseases. Nat Med. 2012;18:42-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 2. | Stafford IS, Kellermann M, Mossotto E, Beattie RM, MacArthur BD, Ennis S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit Med. 2020;3:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 3. | Romagnoni A, Jégou S, Van Steen K, Wainrib G, Hugot JP; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Rep. 2019;9:10351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 4. | Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci Rep. 2017;7:2427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 5. | Matalka II, Al-Omari FA, Salama RM, Mohtaseb AH. A novel approach for quantitative assessment of mucosal damage in inflammatory bowel disease. Diagn Pathol. 2013;8:156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, Andersen V, Andrews JM, Annese V, Brand S, Brant SR, Cho JH, Daly MJ, Dubinsky M, Duerr RH, Ferguson LR, Franke A, Gearry RB, Goyette P, Hakonarson H, Halfvarson J, Hov JR, Huang H, Kennedy NA, Kupcinskas L, Lawrance IC, Lee JC, Satsangi J, Schreiber S, Théâtre E, van der Meulen-de Jong AE, Weersma RK, Wilson DC; International Inflammatory Bowel Disease Genetics Consortium; Parkes M; Vermeire S; Rioux JD; Mansfield J; Silverberg MS; Radford-Smith G; McGovern DP; Barrett JC; Lees CW. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 493] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 7. | Jeong CS, Kim D. Inferring Crohn's disease association from exome sequences by integrating biological knowledge. BMC Med Genomics. 2016;9 Suppl 1:35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput Biol. 2016;12:e1004977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 288] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 9. | Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1756] [Cited by in F6Publishing: 1877] [Article Influence: 156.4] [Reference Citation Analysis (0)] |
| 10. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1945] [Cited by in F6Publishing: 2068] [Article Influence: 206.8] [Reference Citation Analysis (0)] |
| 11. | Seeley EH, Washington MK, Caprioli RM, M'Koma AE. Proteomic patterns of colonic mucosal tissues delineate Crohn's colitis and ulcerative colitis. Proteomics Clin Appl. 2013;7:541-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Khorasani HM, Usefi H, Peña-Castillo L. Detecting ulcerative colitis from colon samples using efficient feature selection and machine learning. Sci Rep. 2020;10:13744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 13. | Stidham RW, Enchakalody B, Waljee AK, Higgins PDR, Wang SC, Su GL, Wasnik AP, Al-Hawary M. Assessing Small Bowel Stricturing and Morphology in Crohn's Disease Using Semi-automated Image Analysis. Inflamm Bowel Dis. 2020;26:734-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 14. | Tielbeek JA, Vos FM, Stoker J. A computer-assisted model for detection of MRI signs of Crohn's disease activity: future or fiction? Abdom Imaging. 2012;37:967-973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Mahapatra D, Vos FM, Buhmann JM. Active learning based segmentation of Crohns disease from abdominal MRI. Comput Methods Programs Biomed. 2016;128:75-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Kumar R, Zhao Q, Seshamani S, Mullin G, Hager G, Dassopoulos T. Assessment of Crohn's disease lesions in wireless capsule endoscopy images. IEEE Trans Biomed Eng. 2012;59:355-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, Oshima S, Akiyama S, Motobayashi M, Nagahori M, Saito E, Matsuoka K, Watanabe M. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology. 2020;158:2150-2157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 128] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 18. | Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, Nallamothu BK, Waljee AK. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients with Ulcerative Colitis. JAMA Netw Open. 2019;2:e193963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 19. | GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 604] [Cited by in F6Publishing: 1035] [Article Influence: 207.0] [Reference Citation Analysis (0)] |
| 20. | Maeda Y, Kudo SE, Mori Y, Misawa M, Ogata N, Sasanuma S, Wakamura K, Oda M, Mori K, Ohtsuka K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. 2019;89:408-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 21. | Misawa M, Kudo SE, Mori Y, Nakamura H, Kataoka S, Maeda Y, Kudo T, Hayashi T, Wakamura K, Miyachi H, Katagiri A, Baba T, Ishida F, Inoue H, Nimura Y, Mori K. Characterization of Colorectal Lesions Using a Computer-Aided Diagnostic System for Narrow-Band Imaging Endocytoscopy. Gastroenterology 2016; 150: 1531-1532. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 22. | Waljee AK, Joyce JC, Wang S, Saxena A, Hart M, Zhu J, Higgins PD. Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin Gastroenterol Hepatol. 2010;8:143-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, Higgins PDR. Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment Pharmacol Ther. 2018;47:763-772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, Higgins PDR. Predicting Corticosteroid-Free Biologic Remission with Vedolizumab in Crohn's Disease. Inflamm Bowel Dis. 2018;24:1185-1192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 25. | Zarringhalam K, Enayetallah A, Reddy P, Ziemek D. Robust clinical outcome prediction based on Bayesian analysis of transcriptional profiles and prior causal networks. Bioinformatics. 2014;30:i69-i77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Verstockt B, Sudahakar P, Creyns B, Verstockt S, Cremer J, Wollants WJ, Organe S, Korcsmaros T, Madgwick M, Van Assche G. DOP70 An integrated multi-omics biomarker predicting endoscopic response in ustekinumab treated patients with Crohn's disease. J Crohns Colitis. 2019;13: :S072-S073. [DOI] [Cited in This Article: ] |
| 27. | Gillen CD, Walmsley RS, Prior P, Andrews HA, Allan RN. Ulcerative colitis and Crohn's disease: a comparison of the colorectal cancer risk in extensive colitis. Gut. 1994;35:1590-1592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 375] [Cited by in F6Publishing: 390] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 28. | Alagappan M, Brown JRG, Mori Y, Berzin TM. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc. 2018;10:239-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 106] [Cited by in F6Publishing: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 29. | Karnes WE, Alkayali T, Mittal M, et al. Su1642 Automated Polyp Detection Using Deep Learning: Leveling the Field. Gastrointest Endosc. 2017;85:AB376-AB377. [DOI] [Cited in This Article: ] |
| 30. | Abadir AP, Ali MF, Karnes W, Samarasena JB. Artificial Intelligence in Gastrointestinal Endoscopy. Clin Endosc. 2020;53:132-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 31. | Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, Sánchez FJ. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 32. | Selaru FM, Xu Y, Yin J, Zou T, Liu TC, Mori Y, Abraham JM, Sato F, Wang S, Twigg C, Olaru A, Shustova V, Leytin A, Hytiroglou P, Shibata D, Harpaz N, Meltzer SJ. Artificial neural networks distinguish among subtypes of neoplastic colorectal lesions. Gastroenterology. 2002;122:606-613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Mandl KD, Mandel JC, Murphy SN, Bernstam EV, Ramoni RL, Kreda DA, McCoy JM, Adida B, Kohane IS. The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc. 2012;19:597-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Hou JK, Chang M, Nguyen T, Kramer JR, Richardson P, Sansgiry S, D'Avolio LW, El-Serag HB. Automated identification of surveillance colonoscopy in inflammatory bowel disease using natural language processing. Dig Dis Sci. 2013;58:936-941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |