Published online Mar 15, 2018. doi: 10.4251/wjgo.v10.i3.71
Peer-review started: December 8, 2017
First decision: December 22, 2017
Revised: January 9, 2018
Accepted: March 6, 2018
Article in press: March 6, 2018
Published online: March 15, 2018
Fusobacterium nucleatum (F. nucleatum) is a Gram-negative obligate anaerobe bacterium in the oral cavity and plays a role in several oral diseases, including periodontitis and gingivitis. Recently, several studies have reported that the level of F. nucleatum is significantly elevated in human colorectal adenomas and carcinomas compared to that in adjacent normal tissue. Several researchers have also demonstrated that F. nucleatum is obviously associated with colorectal cancer and promotes the development of colorectal neoplasms. In this review, we have summarized the recent reports on F. nucleatum and its role in colorectal cancer and have highlighted the methods of detecting F. nucleatum in colorectal cancer, the underlying mechanisms of pathogenesis, immunity status, and colorectal cancer prevention strategies that target F. nucleatum.
Core tip: Fusobacterium nucleatum (F. nucleatum) promotes the progress of colorectal adenomas involving in multiple potential mechanisms. F. nucleatum positivity in colorectal cancer (CRC) is different in different research groups. Some potential biomarkers may be regarded as a criterion for judging CRC prognosis. Some chemoprevention and immunotherapy strategies on F. nucleatum-positive colorectal cancer need to be further explored in the future.
- Citation: Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: A review. World J Gastrointest Oncol 2018; 10(3): 71-81
- URL: https://www.wjgnet.com/1948-5204/full/v10/i3/71.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v10.i3.71
Colorectal cancer (CRC) is the third most prevalent malignant neoplasm and the fourth most frequent cause of cancer death in the world, and the five-year survival rate is nearly 65%[1]. For a long time, the mortality rate of CRC has declined in areas where medical resources are abundant, while the mortality rate has risen in areas with poor medical conditions[2]. CRC is a complex disease that is influenced by both genetic and environmental factors such as dietary habits and lifestyle. Recently, increasing evidence has indicated an association between the intestinal microbiota and CRC[3-5].
More than 100 trillion (1014) microorganisms reside in the intestinal tract and play an extremely important role in human health. These microbes maintain intestinal homeostasis by regulating various biological activities such as mucosal barrier, immune and metabolic functions[6,7]. Once the intestinal balance is damaged, it may cause numerous intestinal diseases including inflammatory bowel diseases (IBD) and colorectal neoplasms[8-10]. There is accumulating evidence to suggest that the gut microbiota is associated with colorectal neoplasms[11-18]. Several studies have validated that the levels of Bacteroides, Prevotella, Escherichia coli, Bacteroides fragilis (ETBF), Streptococcus gallolyticus, Enterococcus faecalis, and Streptococcus bovis are significantly higher in CRC tissue compared to those in adjacent normal tissue[4,11-16,18]. ETBF has been confirmed to selectively stimulate STATA3 in the colon, induce inflammation infiltrates of T helper type 17 and promote the development of CRC[19]. Enterococcus faecalis has been reported to facilitate tumorigenesis through activating the DNA damage pathways[20]. Furthermore, the abundance of both Fusobacterium nucleatum (F. nucleatum) and C. difficile was found to be significantly higher in CRCs compared to the healthy control group[21]. Additional studies have also confirmed that F. nucleatum associates with some Gram-negative bacteria, including Streptococcus, Campylobacter spp. and Leptotrichia, and synergistically promotes the occurrence of CRC[22,23].
F. nucleatum, a common Gram-negative anaerobic bacterium, is one of the most prevalent species in the oral cavity, and several studies have demonstrated that F. nucleatum is associated with oral inflammation diseases, such as periodontitis and gingivitis[24-26]. It has also been associated with pancreatic cancer, oral cancer, and premature and term stillbirths[27-30]. In addition, F. nucleatum is closely connected with liver abscess[9,31], appendicitis and infections of the head and neck, including mastoiditis, tonsillitis and maxillary sinusitis[32-35]. Increasing evidence has indicated that the levels of F. nucleatum are significantly elevated in tumor tissues and stool specimens of CRC patients relative to those in normal controls[36-42]. Researchers have reported that F. nucleatum may contribute to the development of CRC and that it is considered to be a potential risk factor for CRC progression[17,43]. Investigators have demonstrated that a higher abundance of F. nucleatum in CRC is associated with a shorter survival time[44]. Several researchers have also shown that a high-abundance of F. nucleatum induces a series of specific tumor molecular events, including CpG island methylator phenotype (CIMP), microsatellite instability (MSI), and genetic mutations in BRAF, CHD7, CHD8 and TP53[44,45]. However, F. nucleatum was previously regarded as a passenger bacterium in human intestinal tract[46,47]. Recently, it has been considered to be a potential initiator of CRC susceptibility[37,45]. Kostic et al[48] have confirmed that F. nucleatum promotes colorectal tumorigenesis in Apcmin/+ mice. Rubinstein et al[43] have reported that F. nucleatum stimulates tumor cell growth in CRC by activating β-catenin signaling and inducing oncogenic gene expression via the FadA adhesion virulence factor. Together, these studies show that F. nucleatum plays an important role in the initiation of CRC and promoting tumor cell growth in CRC, supporting that F. nucleatum is a cause of CRC rather than a consequence. In this review, we have summarized the recent reports on F. nucleatum and its role in CRC and have highlighted the methods of detecting F. nucleatum in CRC, the underlying mechanisms of pathogenesis, immunity status, and colorectal prevention strategies that target F. nucleatum.
F. nucleatum invades human epithelial cells, activates β-catenin signaling, induces oncogenic gene expression and promotes growth of CRC cells through the FadA adhesion virulence factor.
To detect F. nucleatum in CRC, investigators have used several different methods, including fluorescent quantitative polymerase chain reaction (FQ-PCR), fluorescence in situ hybridization (FISH), quantitative real-time polymerase chain reaction (qPCR), and droplet digital polymerase chain reaction (ddPCR). Furthermore, sample collection methods also vary among studies, some of which are derived from formalin-fixed paraffin-embedded (FFPE) CRC tissues, CRC frozen tissues, genomic DNA, and feces collected from CRC patients.
As shown in Table 1, the detection method and the detection rate of F. nucleatum in CRC differ among studies. In one Chinese study, the F. nucleatum abundance was measured in frozen tissues from 101 CRC patients by FQ-PCR, and FISH analysis was conducted on 22 CRC FFPE tissues with the highest abundance of F. nucleatum to confirm the FQ-PCR results, and the positive rate of F. nucleatum was detected to be 87.13% (88/101)[40]. Analyzing 598 CRC patients in 2 American nationwide prospective cohort studies, researchers detected the abundance of F. nucleatum in FFPE tissue samples obtained from CRC patients by qPCR and found that the positive percentage of F. nucleatum accounted for 13% (76/598) of the CRC samples. This detection rate was significantly lower than that reported in the Chinese study (87.13%)[38]. In one Japanese study, the experimental specimens were obtained from CRC FFPE tissues from 511 Japanese patients, and the abundance of F. nucleatum was detected by qPCR. F. nucleatum was detected in 8.6% (44/511) of the CRC tissue samples, which was similar, albeit slightly lower, to that reported in the USA (13%)[49]. In another study, the richness of F. nucleatum was evaluated by qPCR, and the samples were prepared from genomic DNA extracted from 149 primary CRC tissue samples; F. nucleatum was detected in 74% (111/149) of the CRC tissue samples[45]. In a recent study, the samples consisted of FFPE tissues from 511 CRC patients, and F. nucleatum was detected in 56% (286/511) of the CRC patients by qPCR[39]. In another study, F. nucleatum was detected in the stool samples collected from CRC patients, and the sensitivity and specificity were found to be 72.1% (75/104) and 91.0%, respectively, while the high-abundance of F. nucleatum in patients exhibited a false positive rate of 7.0%[42]. In another study, the levels of F. nucleatum were measured in fecal specimens from Japanese CRC patients by ddPCR, and F. nucleatum was found to be present in 54% (85/158) of the specimens[50]. Furthermore, some researchers used a qPCR assay to detect F. nucleatum in FFPE tissue from CRC patients and revealed that F. nucleatum was present in 2.5% (4/157) of rectal cancers and 11% (19/178) of cecum cancers, with a significant linear trend along all subsites[51]. The percentage of F. nucleatum-enriched CRC gradually increases from rectum to cecum[51], suggesting that the rate at which F. nucleatum is present may also differ among intestinal sites.
| Author (publish date) | Total cases (n) | Positive cases (n) | Positive percentage | Detection method | Detection samples |
| Li et al[40] (3/2016) | 101 | 88 | 87.13% | FISH and FQ-PCR | Frozen tissue and FFPE tissue |
| Mima et al[38] (8/2015) | 598 | 76 | 13% | qPCR | FFPE tissue |
| Nosho et al[49] (1/2016) | 511 | 44 | 8.6% | qPCR | FFPE tissue |
| Tahara et al[45] (1/2014) | 149 | 111 | 74% | qPCR | Genomic DNA |
| Ito et al[39] (2/2015) | 511 | 286 | 56% | qPCR | FFPE tissue |
| Suehiro et al[50] (3/2017) | 158 | 85 | 54% | ddPCR | Feces |
Common specimens for detecting F. nucleatum in CRC include frozen tissues, FFPE tissues, genomic DNA and feces. The use of both frozen tissue and FFPE tissue specimens are limited by surgery or colonoscopy. Specimens derived from the feces of CRC patients are easy to obtain, but they often result in high false positive detection rates. As mentioned above, qPCR, ddPCR, FQ-PCR and FISH are applied to detect the levels of F. nucleatum. While the qPCR assay is the most popular technique to measure the abundance of F. nucleatum in CRC tissues, it is difficult to detect F. nucleatum in the feces[52]; in addition, a higher false positive rate is seen in the high abundance group of F. nucleatum[42]. It has been reported that ddPCR improved the sensitivity of F. nucleatum detection in the feces compared to qPCR, and ddPCR was demonstrated to be 1000 times more sensitive than qPCR[53]. In addition, ddPCR resulted in a higher detection rate of low concentrations of microorganisms compared with qPCR[54]. FQ-PCR is a convenient and rapid method for detecting pathogens and displays a higher sensitivity and specificity than qPCR[55]. In addition, it is difficult to contaminate FQ-PCR during experimental operation compared with qPCR[55].
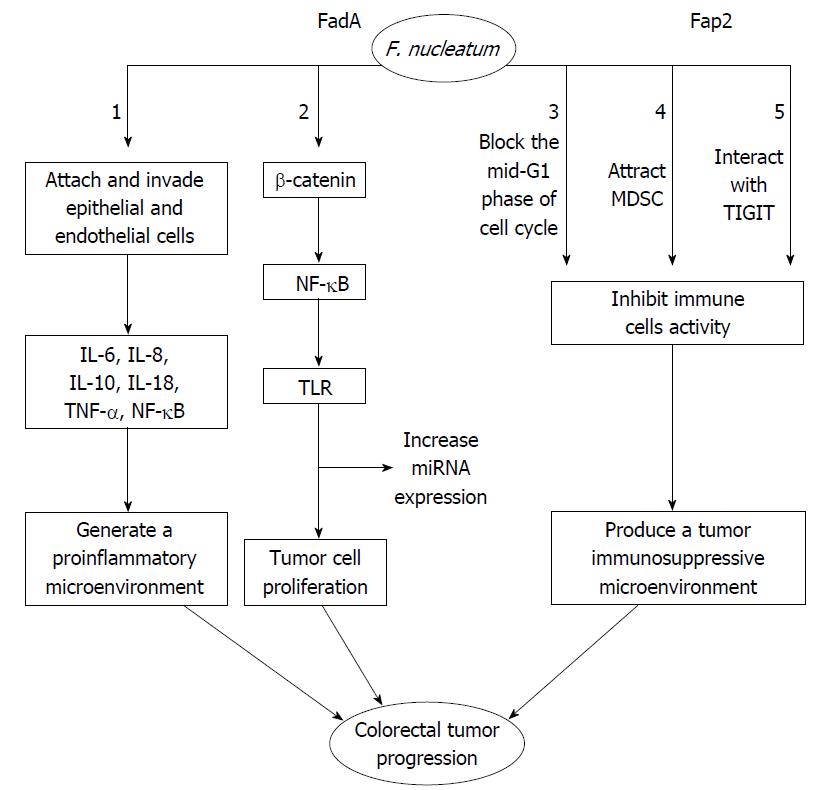
A previous study has shown that lymph node metastases are present in 52 out of 88 (59.1%) cases with a high-abundance of F. nucleatum and in 0 out of 13 (0%) subjects with a low-abundance of F. nucleatum, which indicates that a high abundance of F. nucleatum is associated with CRC progression and metastasis[40]. It has been suggested that high levels of F. nucleatum may be associated with poor outcomes of CRC. Some researchers have also reported that the load of F. nucleatum DNA in CRC tissue is correlated with higher colorectal cancer-specific mortality[44] and that F. nucleatum DNA may serve as a potential poor prognostic biomarker[44]. Fusobacterium was shown to be enriched in the mucosa-adherent microbiota and have the ability to adhere to and invade human epithelial and endothelial cells[27,52,56]. Recently, several researchers have suggested that F. nucleatum is a pathogenic bacterium rather than a bacterium that promotes colorectal carcinogenesis[43,57]. Several studies have shown that its virulence factors are closely linked with colorectal lesions. It has been demonstrated that F. nucleatum invades human epithelial cells, activates β-catenin signaling, induces oncogenic gene expression and promotes growth of CRC cells via the FadA adhesion virulence factor[43]. A second virulence factor, an autotransporter protein, Fap2, has been shown to potentiate the progress of CRC via inhibiting immune cell activity[58].
As shown in Figure 1, F. nucleatum attaches and invades human epithelial and endothelial cells[27,56]. This attachment and invasion depends on the F. nucleatum FadA adhesion protein[59,60]. The FadA protein exists in two main forms. The first form is the intact pre-FadA consisting of 129 amino that is anchored to the membrane, and the second form is the secreted mature FadA (mFadA) consisting of 111 amino acids that are secreted outside of F. nucleatum[61]. When mFadA combines with pre-FadA, the pre-FadA-mFadA is internalized, and FadAc is activated[61]. The internalization of the pre-FadA and mFadA complex ensures that F. nucleatum binds to and invades host epithelial cells[61]. The host endothelial receptor for FadA is the vascular endothelial cadherin (CDH5), which is a member of the cadherin family[59]. The CDH5 receptor is required for F. nucleatum to adhere to and invade endothelial cells[59]. F. nucleatum invasion induces the production of cytokines such as interleukin-8 (IL-8), which is regulated by the p38 MAPK signaling pathway but independent of Toll-like receptor (TLR), NOD-1, NOD-2 and Nuclear Factor-kappaB (NF-κB) signaling[62]. F. nucleatum promotes the expression of several inflammatory genes such as NF-κB and cytokines, including IL-6, IL-8 and IL-18[43]. F. nucleatum also promotes the release of inflammatory cytokines particularly IL-8, IL-10 and tumor necrosis factor-α (TNF-α) in a proinflammatory microenvironment that accelerates colorectal tumor progression[37,62,63]. Another receptor of FadA is the cell-adhesion molecule E-cadherin expressed on non-CRC and CRC cells[43]. E-cadherin is a strong tumor suppressor that inhibits tumor growth and development[64].
FadA binding to wnt7b E-cadherin on CRC cells promotes F. nucleatum adhesion and invasion of host epithelial cells, activates β-catenin signaling that leads to increased expression of Wnt genes, oncogenes, transcription factors, and inflammatory genes, and promotes tumor cells proliferation[43]. FadAc, but not mFadA, binds specifically to the E-cadherin-5, the cytoplasmic or the transmembrane domains of E-cadherin, and results in E-cadherin phosphorylation and internalization[43,65]. As a result, a series of events, which include a decrease in β-catenin phosphorylation, an accumulation of β-catenin in the cytoplasm, and translocation toward the nucleus, leads to the activation of β-catenin-regulated transcription (CRT)[43]. CRT increases the expression of wnt signaling genes such as wnt7a, wnt7b and wnt9a, the oncogenes myc and cyclin D1, transcription factors such as the lymphoid enhancer factor (LEF-1), NF-κB such as NF-κB2, T cell factor such as TCF1, TCF3 and TCF4, and proinflammatory cytokines including IL-6, IL-8 and IL-18[43]. On the other hand, F. nucleatum infected cells increase the expression of microRNA-21 (miR21) by activating TLR4 signaling to MYD88, which leads to the activation of NF-κB[41]. Subsequently, hyperactive NF-κB attaches to the promoter of miR21 and induces the oncogenic cascade in CRC[41]. Moreover, F. nucleatum reduces CD3+ T-cell density in CRC tissue[38]. A previous study has shown that FDC364, sonic extract of F. nucleatum, inhibits human T-cell responses to antigens and mitogens[66]. By blocking the mid-G1 phase of cell cycle, the F. nucleatum inhibitory protein suppresses human T-cell activity[67]. This effect may promote an immunosuppressive microenvironment that allows tumor cell growth[67]. By releasing short-chain fatty acids (acetate, propionate, and butyrate) and short-peptides (formylmethionyl-leucyl-phenylalanine), F. nucleatum also selectively attracts myeloid-derived suppressor cells (MDSCs)[48,68]. MDSCs, a group of heterogeneous cells, show strong T-cell suppressive activity in the immune response[69]. MDSCs and their effectors are key components of the neoplasm and promote tumor progression[48,70]. F. nucleatum-associated tumors increase the myeloid-lineage infiltrating cells, including CD11b+, tumor-associated macrophages (TAMs), M2-like TAMs, tumor-associated neutrophils, conventional myeloid dendritic cells (DCs) and CD103+ regulatory DCs[48]. These cells play an important role in dampening antitumor immunity and promoting tumor progression[69,71-73]. Collectively, these studies have shown that F. nucleatum produces a tumor immunosuppressive microenvironment and promotes CRC progression. Fap2, a galactose-sensitive adhesion protein, plays an important role in coaggregation and cell adhesion[74]. In F. nucleatum, the virulence factor Fap2 protein suppresses immune cell activities through interacting with TIGIT[58]. The interaction between Fap2 and TIGIT protects tumors containing F. nucleatum from host immune cell attack[58]. TIGIT is an inhibitory receptor in humans that is expressed on T cells and natural killer (NK) cells[75]. The Fap2 has also been reported to induce human lymphocyte cell death[57]. In addition, Fap2 mediates F. nucleatum enrichment by interacting with Gal-GalNAc overexpressed in colorectal tumors[76]. Gal-GalNAc is a host polysaccharide overexpressed in CRC[76]. In summary, F. nucleatum produces a tumor immunosuppressive microenvironment that promotes CRC progression.
Some researchers have demonstrated that F. nucleatum modulates the tumor immune microenvironment while promoting CRC development[48]. Recently, it has been confirmed that biomarkers such as immune antibodies, miRNA, TAMs, and tumor-infiltrating T-cell subsets play a significant role in F. nucleatum-associated CRC[44,48,77,78].
Several studies have shown that F. nucleatum infection causes high levels of serum F. nucleatum-IgA antibodies in CRC patients[77]. Researchers have confirmed that serum anti-F. nucleatum-IgA combined with CA19-9 and CEA has a higher sensitivity than CA19-9 and CEA alone in screening early CRC[77]. This study suggests that serum F. nucleatum-IgA antibodies may be regarded as a potential diagnosing biomarker for early CRC[77]. In addition, some researchers have found that the levels of the fadA gene in colon tissue from CRC patients are > 10-100 times higher in comparison with normal subjects[43]. This study also reveals a gradual increase in fadA gene copies in normal individuals compared to CRC patients[43]. The fadA gene has become a potential ideal diagnostic marker to identify individuals with CRC risk[43]. The miR-21 gene has been demonstrated to promote tumor cell growth and migration via inhibiting sec23a protein expression[79]. The data also indicated that F. nucleatum induces a high level of miR-21 expression in advanced CRC[41]. The amount of miR-21 in CRC tissues has been shown to be associated with poor clinical outcomes[41]. Studies have reported that non-coding RNAs (lncRNAs) play a crucial role in the diagnosis and prognosis of CRC[80]. One study has found that low levels of NR_034119 and NR_029373 are associated with a short survival rate of CRC[80]. These researchers suggested that several lncRNAs (NR_034119, NR_029373, NR_026817, and BANCR) are potential diagnostic biomarkers for CRC and that NR_034119 and NR_029373 are potential prognostic indicators for CRC[80]. Another study reported that the level of lncRNA PANDAR was higher in CRC cells and tissues relative to adjacent normal tissues[81], and high levels of PANDAR expression were associated with short overall survival[81]. The authors suggested that the amount of PANDAR expression may be a prognostic indicator for CRC.
A previous study reported that F. nucleatum-positive tumors increased TAM infiltration[48]. TAMs play an important role in innate immunity, and subpopulations of regulatory T-lymphocytes (Tregs) are a component of the acquired immunity. A recent study has found that intense infiltration of TAMs in colorectal tumor tissue is associated with shorter disease-free survival and overall survival of CRC[78]. Infiltration of TAMs CD68+/iNOS− in colorectal tumor stroma is confirmed to be related to the poor prognosis of CRC[78]. Some researchers have reported that tumor-infiltrating T-cell subpopulations distinctly regulate the prognosis of CRC[82]. For instance, in tumor-infiltrating T-cell subsets, CD45RO+-cell density, but not that of FOXP3+-cell, is significantly associated with a long survival of CRC patients[82]. CD45RO+-cell is considered to be a potential good prognostic biomarker for CRC[82]. The FOXP3+ transcription factor, which plays an important role in regulating the immune system, is regarded as an immunosuppressive factor. Some scholars have reported that infiltration of FOXP3+ in colorectal tumor stroma is associated with a poor prognosis in CRC[78]. However, several researchers also suggest that FOXP3+-cells are generally associated with a good prognosis of CRC[83]. An article recently published in Nature Medicine has shown that distinct tumor-infiltrating FOXP3+-T cell subpopulations have an opposite approach to determining CRC prognosis. The development of inflammatory FOXP3± (lo) non-Treg cells was shown to be associated with tumor invasion by intestinal bacteria, particularly F. nucleatum[84]. In this study, CRC patients with a high infiltration of FOXP3± (lo) T cells exhibit a significantly better prognosis, compared to those with a FOXP3± (hi) Treg cell infiltration[84]. When FOXP3± (hi) Treg cells are depleted from CRC tissues, antitumor immunity is augmented[84]. The elimination of FOXP3± (hi) Treg cells has been suggested to play a crucial role in suppressing CRC formation[84]. Recent research has also found that prudent diets such as whole grain and dietary fiber reduce the risk of F nucleatum-positive CRC[85].
In conclusion, anti-F. nucleatum-IgA, the fadA gene, and lncRNAs may be considered as potential diagnostic biomarkers during the early stage of F. nucleatum-positive CRC. The CD45RO+-cell and FOXP3± (lo) T cell biomarkers are associated with a favorable prognosis in F. nucleatum-positive CRC, while the miR-21, LncRNA PANDAR, and TAMs CD68+/iNOS− biomarkers are associated with a poor clinical prognosis of F. nucleatum-positive CRC.
Currently, cancer prevention strategies have been mainly focused on chemoprevention and immunotherapy. Chemoprevention, which involves the use of aspirin, cyclo-oxygenase-2 (COX-2) inhibitors, and selective EP2 antagonists, plays an important role in F. nucleatum-associated CRC. Immunotherapies, such as antibody treatment, immune-checkpoint blockade therapy and adoptive cell transfer therapies, may aid in the prevention of F. nucleatum-positive CRC.
Chemoprevention, including the use of aspirin, COX-2 inhibitors, and selective EP2 antagonists, plays a significant role in the mechanisms of F. nucleatum-positive CRC. For instance, some researchers have reported that regular aspirin use lowers CRC incidence and mortality and reduces the risk of distant metastasis of CRC[85,86]. Regular doses of aspirin were also associated with a lower risk of CRC and low levels of CD45RO (PTPRC)+T cells, CD3+T cells or CD8+ T cells[87]. Aspirin induces neutrophils apoptosis[88] and triggers a lipoxin-driven immune-regulatory effect[89]. Aspirin directly inhibits T-cell activation and proliferation and suppresses cytokine production involved in the T cell-mediated adaptive immune response[90]. Tumor-infiltrating immune cells have been associated with a good prognosis in CRC[91,92]. The amount of F. nucleatum is inversely proportional to CD3+ T-cell density in colorectal carcinoma tissue[38]. These data indicate that aspirin may support the host immune system and prevent the development of F. nucleatum-associated CRC.
In addition, FadA in F. nucleatum specifically binds to E-cadherin and activates Wnt signaling[43]. F. nucleatum increases expression of inflammatory genes and Wnt genes[43]. A recent study has reported that EP2 enhances the expression of NF-κB-targeted proinflammatory genes induced by TNF-α in neutrophils[93]. The levels of cytokines such as TNF-α and IL-6, COX-2, chemokine CXCL1, and Wnt are significantly higher in tumor lesions of EP2-abundant mice than those in EP2- deficient mice[93]. This study revealed that EP2 promotes colon tumorigenesis by means of expanding inflammation and shaping a tumor microenvironment[93]. PF-04418948, a selective EP2 antagonist, significantly inhibits the formation of colon tumors[93]. This suggests that selective EP2 antagonists may be promising drugs for the chemoprevention of F. nucleatum-associated CRC.
Furthermore, COX expression in BrafV600E cells may prevent CD103+ DC activation and accumulation in tumors[94]. By suppressing local T-cell effector, COX-2 also promotes immune evasion and resistance to antigen-specific cancer immunity[95]. COX-2 is also considered an inhibitor of antigen-specific tumor immunotherapy[95]. This is powerful evidence that supports that COX inhibitors reduce the risk of CRC by inhibiting inflammatory pathways, and COX inhibitors may be important for immune-based therapy in CRC patients. In conclusion, aspirin, EP2 antagonists, and COX-2 inhibitors may be important tools for preventing F. nucleatum-associated CRC.
Immunotherapies, including antibody treatment, immune-checkpoint blockade therapy and adoptive cell transfer therapies, may be effective strategies for preventing F. nucleatum-positive CRC. For example, the interaction between Fap2 and TIGIT receptor protects tumors against immune cell attack and, accordingly, inhibits antitumor immunity and supports tumor cells growth[58]. Fap2 also induces lymphocyte cell death[57]. Fap2 mediates F. nucleatum enrichment via its interaction with Gal-GalNAc that is overexpressed in CRC, which may exacerbate the inhibition of antitumor immunity[76]. Therefore, anti-Fap2 antibody development may favor antitumor immune response and be a potential immunotherapy in F. nucleatum-positive CRC. F. nucleatum inhibits T-cell activity and stimulates lymphocyte cell death, which protects tumors from immune cell attack. F. nucleatum may have immunosuppressive function in the tumor immune microenvironment.
Recently, the approach to cancer immunotherapy involves immune-checkpoint blockade, such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and programmed death protein 1 (PD-1). CTLA-4 and PD-1 have been reported to be involved in T cell-mediated antitumor immunity[96,97]. It was speculated that blockade of CTLA-4 and PD-1 may shape the antitumor immunity response and be an effective immunotherapy for F. nucleatum-associated CRC. Other CRC treatment strategies involving F. nucleatum, such as miR-21 blockade may play a significant role in F. nucleatum-positive CRC, as F. nucleatum increases expression of miR-21 by activating TLR4 signaling to NF-κB[41]. It has been demonstrated that miR-21 promotes tumor cells proliferation and migration by down-regulating the expression of the sec23a protein[79]. The inhibition of miR-21 suppresses the metastasis of colorectal tumor cells by regulating programmed cell death 4[98]. In a miR-21 knockout mouse model, expression of proinflammatory and procarcinogenic cytokines was decreased, suggesting that miR-21 deficiency promotes the apoptosis of tumor cells by suppressing STATA3 and Bcl-2 activation[99]. It has been suggested that the miR-21 blockade may be a potential treatment strategy for F. nucleatum-associated CRC. Some adoptive cell transfer therapies, such as NK cells[100], cytokine-induced killer cells[101], and tumor-infiltrating lymphocytes[102], are also being used to strengthen antitumor immunity in clinical practice. These adoptive cell transfer therapies may also be considered as an immunotherapy approach in CRC associated with F. nucleatum.
In sum, CRC prevention strategies that target F. nucleatum are mainly focused on chemoprevention, which includes the use of aspirin, COX-2 inhibitors and selective EP2 antagonists, and immunotherapy, which includes anti-Fap2 antibody treatment, CTLA-4, PD-1, miR-21 blockade therapies and adoptive cell transfer therapies.
In summary, the gut microbiota, especially F. nucleatum, has been extensively associated with CRC. F. nucleatum promotes the progression of CRC via multiple potential mechanisms. The positive detection rate of F. nucleatum in CRC samples varies among different studies. FadA combined with anti-F. nucleatum-IgA may improve the diagnosis of CRC. Several potential biomarkers, such as miR-21, LncRNA PANDAR, TAMs CD68+/iNOS−, FOXP3± (lo) T cells and CD45RO+ cells, may be considered as criteria for determining CRC prognosis. Furthermore, chemoprevention and immunotherapy strategies should be further explored in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: El-Tawil AM, Roncucci L S- Editor: Cui LJ L- Editor: A E- Editor: Li RF
| 1. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11065] [Cited by in F6Publishing: 11879] [Article Influence: 1697.0] [Reference Citation Analysis (3)] |
| 2. | Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 786] [Cited by in F6Publishing: 827] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 3. | Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer Discov. 2013;3:384-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 4. | Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 606] [Cited by in F6Publishing: 566] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 5. | Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 6. | Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 335] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 7. | Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452-10459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 659] [Cited by in F6Publishing: 567] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 8. | Arthur JC, Jobin C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes. 2013;4:253-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 9. | Ahmed Z, Bansal SK, Dhillon S. Pyogenic liver abscess caused by Fusobacterium in a 21-year-old immunocompetent male. World J Gastroenterol. 2015;21:3731-3735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 21] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, Brigidi P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20:908-922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 153] [Cited by in F6Publishing: 142] [Article Influence: 14.2] [Reference Citation Analysis (1)] |
| 11. | Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1374] [Cited by in F6Publishing: 1481] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 12. | Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10:e0119462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 165] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 13. | Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011;53:870-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 243] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 14. | Zhou Y, He H, Xu H, Li Y, Li Z, Du Y, He J, Zhou Y, Wang H, Nie Y. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget. 2016;7:80794-80802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Tsai CE, Chiu CT, Rayner CK, Wu KL, Chiu YC, Hu ML, Chuah SK, Tai WC, Liang CM, Wang HM. Associated factors in Streptococcus bovis bacteremia and colorectal cancer. Kaohsiung J Med Sci. 2016;32:196-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Krishnan S, Eslick GD. Streptococcus bovis infection and colorectal neoplasia: a meta-analysis. Colorectal Dis. 2014;16:672-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381-1390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 253] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 18. | Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 491] [Cited by in F6Publishing: 475] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 19. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1113] [Cited by in F6Publishing: 1191] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 20. | Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68:9909-9917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 21. | Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Júnior U, Nakano V, Avila-Campos MJ. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol. 2015;46:1135-1140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 22. | Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 219] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 23. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 612] [Cited by in F6Publishing: 625] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 24. | Yang NY, Zhang Q, Li JL, Yang SH, Shi Q. Progression of periodontal inflammation in adolescents is associated with increased number of Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis, and Fusobacterium nucleatum. Int J Paediatr Dent. 2014;24:226-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PLoS One. 2013;8:e71227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 26. | Fujii R, Saito Y, Tokura Y, Nakagawa KI, Okuda K, Ishihara K. Characterization of bacterial flora in persistent apical periodontitis lesions. Oral Microbiol Immunol. 2009;24:502-505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 27. | Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun. 2004;72:2272-2279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 283] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 28. | Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209-7220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 29. | Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, Nussbaum G, Elkin M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613-22623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 234] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 30. | Krejs GJ. Pancreatic cancer: epidemiology and risk factors. Dig Dis. 2010;28:355-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 31. | Yoneda M, Kato S, Mawatari H, Kirikoshi H, Imajo K, Fujita K, Endo H, Takahashi H, Inamori M, Kobayashi N. Liver abscess caused by periodontal bacterial infection with Fusobacterium necrophorum. Hepatol Res. 2011;41:194-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Yarden-Bilavsky H, Raveh E, Livni G, Scheuerman O, Amir J, Bilavsky E. Fusobacterium necrophorum mastoiditis in children - emerging pathogen in an old disease. Int J Pediatr Otorhinolaryngol. 2013;77:92-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | Jensen A, Hagelskjaer Kristensen L, Prag J. Detection of Fusobacterium necrophorum subsp. funduliforme in tonsillitis in young adults by real-time PCR. Clin Microbiol Infect. 2007;13:695-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Finegold SM, Flynn MJ, Rose FV, Jousimies-Somer H, Jakielaszek C, McTeague M, Wexler HM, Berkowitz E, Wynne B. Bacteriologic findings associated with chronic bacterial maxillary sinusitis in adults. Clin Infect Dis. 2002;35:428-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 90] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 35. | Salö M, Marungruang N, Roth B, Sundberg T, Stenström P, Arnbjörnsson E, Fåk F, Ohlsson B. Evaluation of the microbiome in children’s appendicitis. Int J Colorectal Dis. 2017;32:19-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 36. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1164] [Cited by in F6Publishing: 1324] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 37. | McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 360] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 38. | Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653-661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 420] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 39. | Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258-1268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 40. | Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227-3233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 120] [Cited by in F6Publishing: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 41. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 455] [Cited by in F6Publishing: 568] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 42. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 43. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1511] [Cited by in F6Publishing: 1353] [Article Influence: 123.0] [Reference Citation Analysis (0)] |
| 44. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 530] [Cited by in F6Publishing: 601] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 45. | Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311-1318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 320] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 46. | Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 484] [Cited by in F6Publishing: 571] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 47. | Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes. 2011;2:294-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 48. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1659] [Cited by in F6Publishing: 1581] [Article Influence: 143.7] [Reference Citation Analysis (0)] |
| 49. | Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22:557-566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 232] [Cited by in F6Publishing: 238] [Article Influence: 29.8] [Reference Citation Analysis (3)] |
| 50. | Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54:86-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 51. | Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016;7:e200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 52. | Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 565] [Cited by in F6Publishing: 651] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 53. | Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604-8610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1737] [Cited by in F6Publishing: 1774] [Article Influence: 136.5] [Reference Citation Analysis (0)] |
| 54. | Singh G, Sithebe A, Enitan AM, Kumari S, Bux F, Stenström TA. Comparison of droplet digital PCR and quantitative PCR for the detection of Salmonella and its application for river sediments. J Water Health. 2017;15:505-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Jiao H, Weng WC, Wang FJ, Cheng G, Wang W, Xie J. [Faster detection of Vibrio parahaemolyticus in foods by FQ-PCR technique]. Wei Sheng Yan Jiu. 2005;34:457-460. [PubMed] [Cited in This Article: ] |
| 56. | Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140-3146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 317] [Cited by in F6Publishing: 318] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 57. | Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78:4773-4778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 58. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 580] [Cited by in F6Publishing: 776] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 59. | Fardini Y, Wang X, Témoin S, Nithianantham S, Lee D, Shoham M, Han YW. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 173] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 60. | Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330-5340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 161] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 61. | Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000-25009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 62. | Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550-559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 63. | Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79:2597-2607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 64. | Bryant DM, Stow JL. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 2004;14:427-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 290] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 65. | Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622-634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1117] [Cited by in F6Publishing: 1130] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 66. | Shenker BJ, DiRienzo JM. Suppression of human peripheral blood lymphocytes by Fusobacterium nucleatum. J Immunol. 1984;132:2357-2362. [PubMed] [Cited in This Article: ] |
| 67. | Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. 1995;63:4830-4836. [PubMed] [Cited in This Article: ] |
| 68. | Bashir A, Miskeen AY, Hazari YM, Asrafuzzaman S, Fazili KM. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol. 2016;37:2805-2810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 69. | Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2369] [Cited by in F6Publishing: 2633] [Article Influence: 219.4] [Reference Citation Analysis (0)] |
| 70. | Sun HL, Zhou X, Xue YF, Wang K, Shen YF, Mao JJ, Guo HF, Miao ZN. Increased frequency and clinical significance of myeloid-derived suppressor cells in human colorectal carcinoma. World J Gastroenterol. 2012;18:3303-3309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 71] [Reference Citation Analysis (0)] |
| 71. | Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1025] [Cited by in F6Publishing: 1067] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 72. | Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1881] [Cited by in F6Publishing: 2010] [Article Influence: 167.5] [Reference Citation Analysis (0)] |
| 73. | Gulubova MV, Ananiev JR, Vlaykova TI, Yovchev Y, Tsoneva V, Manolova IM. Role of dendritic cells in progression and clinical outcome of colon cancer. Int J Colorectal Dis. 2012;27:159-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 74. | Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, Bachrach G. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83:1104-1113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 146] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 75. | Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci USA. 2009;106:17858-17863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 482] [Cited by in F6Publishing: 591] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 76. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 326] [Cited by in F6Publishing: 439] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 77. | Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:33440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 78. | Waniczek D, Lorenc Z, Śnietura M, Wesecki M, Kopec A, Muc-Wierzgoń M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch Immunol Ther Exp (Warsz). 2017;65:445-454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 79. | Li C, Zhao L, Chen Y, He T, Chen X, Mao J, Li C, Lyu J, Meng QH. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer. 2016;16:605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 80. | Wang R, Du L, Yang X, Jiang X, Duan W, Yan S, Xie Y, Zhu Y, Wang Q, Wang L. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142:2291-2301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 81. | Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 82. | Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, Giovannucci E, Dranoff G, Fuchs CS, Ogino S. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 380] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 83. | deLeeuw RJ, Kost SE, Kakal JA, Nelson BH. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18:3022-3029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 334] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 84. | Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 471] [Cited by in F6Publishing: 572] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 85. | Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017;3:921-927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 198] [Article Influence: 28.3] [Reference Citation Analysis (1)] |
| 86. | Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, Meade TW. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741-1750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 976] [Cited by in F6Publishing: 947] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 87. | Cao Y, Nishihara R, Qian ZR, Song M, Mima K, Inamura K, Nowak JA, Drew DA, Lochhead P, Nosho K. Regular Aspirin Use Associates With Lower Risk of Colorectal Cancers With Low Numbers of Tumor-Infiltrating Lymphocytes. Gastroenterology. 2016;151:879-892.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 88. | Negrotto S, Malaver E, Alvarez ME, Pacienza N, D’Atri LP, Pozner RG, Gómez RM, Schattner M. Aspirin and salicylate suppress polymorphonuclear apoptosis delay mediated by proinflammatory stimuli. J Pharmacol Exp Ther. 2006;319:972-979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 89. | El Kebir D, József L, Khreiss T, Pan W, Petasis NA, Serhan CN, Filep JG. Aspirin-triggered lipoxins override the apoptosis-delaying action of serum amyloid A in human neutrophils: a novel mechanism for resolution of inflammation. J Immunol. 2007;179:616-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 90. | Hussain M, Javeed A, Ashraf M, Zhao Y, Mukhtar MM, Rehman MU. Aspirin and immune system. Int Immunopharmacol. 2012;12:10-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 91. | Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960-1964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4318] [Cited by in F6Publishing: 4590] [Article Influence: 255.0] [Reference Citation Analysis (0)] |
| 92. | Mei Z, Liu Y, Liu C, Cui A, Liang Z, Wang G, Peng H, Cui L, Li C. Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br J Cancer. 2014;110:1595-1605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 222] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 93. | Ma X, Aoki T, Tsuruyama T, Narumiya S. Definition of Prostaglandin E2-EP2 Signals in the Colon Tumor Microenvironment That Amplify Inflammation and Tumor Growth. Cancer Res. 2015;75:2822-2832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 94. | Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257-1270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 769] [Cited by in F6Publishing: 740] [Article Influence: 82.2] [Reference Citation Analysis (0)] |
| 95. | Göbel C, Breitenbuecher F, Kalkavan H, Hähnel PS, Kasper S, Hoffarth S, Merches K, Schild H, Lang KS, Schuler M. Functional expression cloning identifies COX-2 as a suppressor of antigen-specific cancer immunity. Cell Death Dis. 2014;5:e1568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 96. | Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1834] [Cited by in F6Publishing: 2167] [Article Influence: 240.8] [Reference Citation Analysis (0)] |
| 97. | Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4180] [Cited by in F6Publishing: 4769] [Article Influence: 529.9] [Reference Citation Analysis (0)] |
| 98. | Nedaeinia R, Sharifi M, Avan A, Kazemi M, Nabinejad A, Ferns GA, Ghayour-Mobarhan M, Salehi R. Inhibition of microRNA-21 via locked nucleic acid-anti-miR suppressed metastatic features of colorectal cancer cells through modulation of programmed cell death 4. Tumour Biol. 2017;39:1010428317692261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 99. | Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470-1481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 100] [Article Influence: 12.5] [Reference Citation Analysis (1)] |
| 100. | Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16:112-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 363] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 101. | Pan K, Guan XX, Li YQ, Zhao JJ, Li JJ, Qiu HJ, Weng DS, Wang QJ, Liu Q, Huang LX. Clinical activity of adjuvant cytokine-induced killer cell immunotherapy in patients with post-mastectomy triple-negative breast cancer. Clin Cancer Res. 2014;20:3003-3011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 102. | Pilon-Thomas S, Kuhn L, Ellwanger S, Janssen W, Royster E, Marzban S, Kudchadkar R, Zager J, Gibney G, Sondak VK. Efficacy of adoptive cell transfer of tumor-infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. J Immunother. 2012;35:615-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |