Published online Dec 12, 2012. doi: 10.5501/wjv.v1.i6.162
Revised: June 6, 2012
Accepted: November 7, 2012
Published online: December 12, 2012
Non-Hodgkin’s lymphoma (NHL) is among the haematological malignancies with high prevalence worldwide, causing estimated 355 900 new cases and 191 400 deaths in 2008. High prevalence of NHL is documented in economically more developed areas while low prevalence is observed in less developed areas of the globe. A wide array of environmental factors have been reported to be either directly involved or in modifying the risk of NHL development. In addition to these factors, a number of infectious agents, chiefly viruses have also been implicated in the development of NHL. This article reviews the available literature to discuss the role of hepatitis viruses in NHL development, possible mechanisms of lymphomagenesis and also identify the areas in which further research is required to better understand this disease. A brief discussion on the clinical aspects such as classification, staging, treatment approaches have also been included in this article.
- Citation: Datta S, Chatterjee S, Policegoudra RS, Gogoi HK, Singh L. Hepatitis viruses and non-Hodgkin’s lymphoma: A review. World J Virol 2012; 1(6): 162-173
- URL: https://www.wjgnet.com/2220-3249/full/v1/i6/162.htm
- DOI: https://dx.doi.org/10.5501/wjv.v1.i6.162
Cancer is a multifaceted disease, and arise mostly due to the changes in the somatic genetic material where, the interaction with the external factors play a very important role, apparently both modifying the effect of each other[1,2]. The disease, according to the GLOBOCAN 2008 data published by the IARC (International Agency for Research on Cancer), is the most important cause of death in developed countries; while second most important cause of death in developing countries and is responsible for about 7.6 million deaths in 2008, worldwide[3].
Cancer is a heterogeneous class of diseases displaying a wide range of pattern, origin site, distribution and malignancy. Of the wide range, lymphomas constitute an important group of cancers of the white blood cells that arise in lymphoid tissues and generally remain localized in lymph nodes or certain locations other than the bone marrow. Lymphomas are broadly separated into two groups, namely Hodgkin’s lymphoma (HL) and non-Hodgkin’s lymphoma (NHL). The characteristic presence of large, usually multinucleate cells called ‘Reed-Sternberg cells’ in tumour biopsy samples differentiate HL from NHL[4]. About 80% to 90% NHLs are of B-cell origin, while rest are of T-cell origin[5]. The staging and diagnostic approaches have been reviewed elsewhere[6,7].
NHL is among the haematological malignancies with high prevalence worldwide. NHL ranks 8th and 11th among the most common cancers in men and women respectively, contributing 5.1% of all cancer cases and 2.7% of all cancer deaths[8]. An estimated 355 900 new cases and 191 400 deaths have been attributed to NHL in 2008[3,9]. NHL is more frequent in developed areas, with the highest incidence rates found in Australia/New Zealand, Western, Northern and Southern Europe, and North America, while lowest rates are found in South-Central and Eastern Asia, Eastern Europe and the Caribbean[3,8]. The incidence of NHL is usually low in Africa, but in some sub-Saharan areas (particularly in East Africa) incidence of Burkitt’s lymphoma (a subtype of NHL) caused by Epstein-Barr virus (EBV) among children is remarkably high[9]. NHL incidence rates are also increasing in certain developing countries such as Thailand and Uganda, probably due to the acquired immunodeficiency syndrome epidemic.
Worldwide, the occurrence of NHL has been found to be higher in men with age-standardized rate per 100 000 (ASR, standardized to the World Standard Population) of 6.1 as compared to 4.2 for women[3,8]. The ASR of NHL incidence has been found to be 10.3 and 4.2 in males from more developed and less developed areas respectively. The ASR for NHL related mortality in males has been found to be 3.6 and 3.0 in more developed areas and less developed areas respectively. On the other hand, in females, the ASR of NHL incidence and mortality has been found to be 7.0 and 2.2 respectively from more developed areas, compared to incidence and mortality to 2.8 and 1.9 respectively from less developed areas[3,8]. Furthermore, the rise in incidences of NHL has been found to be consistent across the globe which still remains an enigma[10]. As NHL is a group of related yet diverse cancers, originating from different and complex etiologies, the issue of increase in the incidence is poorly understood.
It has already been recognized that the major factors for the development of NHL include genetic alterations/damage to the cells and/or factors that are associated with immunosuppression. Further, NHL tumours have a high rate of genetic alterations like translocations, detectable in up to 90% of NHL cases[6,10,11]. Recently, Lan and colleagues have shown that polymorphisms in the Th1/Th2 cytokine genes may contribute to lymphomagenesis[12]. In addition, compromised immune system may also increase the risk of NHL incidence by allowing cancerous cells to escape the surveillance of immune system as higher rates of NHL is observed in people with inherited or acquired immunodeficiency syndromes and in people receiving immunosuppressive therapy[13,14]. However, genetic predisposition on the higher incidence of NHL among certain families is still a debatable issue[15]. The common genetic changes in NHL, may occur due to varied reasons: rearrangements of Immunoglobulin heavy chain (IgH), Immunoglobulin κ light chain (Igk), T cell receptor β chain (TCRb), T cell receptor γ chain (TCRg); and translocations of BCL-6, C-MYC, etc.[6].
Similar to most of the other cancers, several environmental factors have been implicated in the origin of NHL. Exposures to different agricultural chemicals like certain herbicides (phenoxy-, triazine- groups), insecticides (organo-chlorine and organophosphates), and industrial chemicals like polychlorinated biphenyls and polybrominated biphenyl, dioxins, organic solvents seems to be significant risk factors for the onset of the disease[15-17]. It was found that exposed population (like farmers) of these chemicals have higher rates of NHL than non-exposed populace[18,19]. Again, long term exposure of nitrate in drinking water, UV light, X-ray, radionuclides or electromagnetic fields, etc. have also been implicated in the development of NHL[15,20]. Further, use of hair dyes, alcohol, tobacco, certain diets, certain immunological conditions may also facilitate in the development of NHL[15]. However, positive correlation among the above mentioned risk factors and the incidence of NHL is still a dubious issue[4,7,8,10,15-20].
During the last 3 decades, the role of infectious agents, mainly viruses, in oncogenesis has become increasingly significant. Approximately 15% to 20% of cancers are associated with viral infections[21]. Apart from the above mentioned genetic and environmental etiologic factors, development of NHL has also been attributed to different viruses[22,23]. For several years EBV has been considered as important cause of NHL. In the later years, human immunodeficiency virus (HIV), hepatitis viruses B, C and G (HBV, HCV, HGV/GBV-C), HTLV-1, HHV 8 and Simian Virus 40 (SV40) have also been implicated in the development of NHL[7,14,15,22]. This review will mainly focus on the hepatitis viruses, HBV, HCV and HGV/GBV-C.
Both the hepatitis viruses, HBV and HCV have been strongly associated with the hepatocellular carcinoma (HCC)[24]. Previously these two viruses were thought to be solely hepatotrophic, but lately their occult lymphotrophic characteristic has been proved in human subjects as well as in animal models[25-27]. Although, the association of HBV and HCV with NHL has been established, it remains the question if these associations simply reflect causal relationships. In this context, Marcucci and Mele, have put forward 3 possibilities to explain these associations- (1) the immunosuppressive effect of the tumor increases the risk of viral infection or reactivation; (2) some previously unknown virus with a similar mode of transmission might trigger the oncogenic signal; and (3) the actual causal relationships between hepatitis viruses and NHL[10]. The authors refuted the first two possibilities and found the third to be correlating with the available literature. The first possibility being ruled out based on the fact that NHL is observed in a number of patients in which immune deficiency is not significant, while the second possibility was refuted for lack of any evidence[10]. Available studies apparently support the third possibility of the oncogenic role of hepatitis viruses in development of NHL[22,23]. However the association of HCV and HBV with the development of NHL is much weaker as compared to the major risk for HCC development caused by HBV and HCV[24,28].
Mostly, the studies on the lymphomagenic role of hepatitis viruses have remained focused on HCV[10]. The simple evidence of an oncogenic role of HCV in NHL came from antiviral therapy studies, which shows, peginterferon and ribavirin (standard antiviral therapy against HCV) could completely or partially restrict lymphoma in HCV positive, but not in HCV-negative NHL patients[29,30]. More interestingly, in most patients, in a study, it was found that antiviral treatment results in disappearance of Ig heavy chain (IgHC) and t (14;18) translocation[31], suggesting the role of HCV in causing the genetic changes that are associated with NHL.
The association between HCV and NHL is strongest in geographic areas with highest prevalence of the viral infection[15]. In a recent meta-analysis of 15 selected studies, the pooled relative risk (RR) of all NHL among HCV-positive persons was found to be 2.5 (95% CI: 2.1-3.1) in case-control studies and 2.0 (95% CI: 1.8-2.2) in cohort studies[32]. Interestingly, the RR was significantly elevated in geographic areas with high HCV prevalence compared to areas with low HCV prevalence, which correlate well with previous studies from countries with low HCV prevalence, that could not observed any association between HCV and NHL[33,34]. It was suggested that undetectable association in countries with low HCV prevalence, was mainly due to relatively small sample size of HCV positive subjects[35].
HCV is a positive, single-stranded RNA virus of the Flaviviridae family[36]. During its replicative cycle it goes through a negative-stranded RNA, but replication does not include a DNA step, hence integration of HCV nucleic acid sequences into the host genome seems improbable, lacking a critical property of classical oncogenic retroviruses[24,35]. The HCV genome produces a single polyprotein that is proteolytically processed by viral and cellular proteases to produce structural (nucleocapsid, E1, E2) and nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Studies have demonstrated that NS5A acts as a transcriptional activator, interacts with other proteins and plays a crucial role in hepatocarcinogenesis[37]. In addition, it participates in HCV protein maturation and RNA replication, regulates gene expression in hepatocytes, stimulates cell proliferation, inhibits apoptosis and influences interferon effect[38]. It has been proposed that the E2 protein of HCV may be accountable for chronic antigen-driven polyclonal B-cell proliferation, leading to lymphomagenesis[39]. However, the oncogenic mechanism remains unclear.
In comparison to HCV, the association of HBV with NHL has been studied less thoroughly, despite the fact that the first reports were published almost simultaneously on positive association between these two viruses and NHL[40,41]. The association between HBV and NHL was studied by several authors in both HBV endemic countries (e.g., South Korea, China) and non-endemic countries (e.g., USA, Australia)[40,42-49]. As discussed by Nath and colleagues[50], results of previous retrospective case-control studies have generally supported an association (odds ratios: 1.5-3.6). However, the available data may be an underestimate of the real association between HBV and NHL, because another form of silent HBV infection, known as occult hepatitis B infection has been identified and established in the recent years[25,51].
Occult HBV infections is defined for patients who test negative for the most widely practised HBsAg detection, but carry HBV-DNA in serum or tissues or both[25,51]. Moreover, replication-competent HBV-DNA is supposed to persists in the liver or lymphocytes or in both the compartments for many years or even life long, indicating complete HBV eradication to be an infrequent event[25,52]. HBV DNA has been detected within lymphocytes but whether HBV could directly transform lymphocytes is uncertain, as some studies have not been able to detect HBV in NHL cells[42,44]. In addition, the long incubation period of HBV makes it difficult to precisely estimate the significance of HBV in NHL[10].
HBV is a small partially dsDNA prototype virus of the Hepadnaviridae family[53]. During replication, it undergoes transformation into covalently closed circular dsDNA and replicate through an RNA intermediate. It can also integrate into the host genome[54]. HBV is characterized by a genome consisting of 4 overlapping open-reading frames: the S gene, encoding envelope proteins; the core gene, encoding the core and “e” proteins; the P gene, encoding DNA polymerase; and the “x” gene, encoding a transcriptional transactivator. The HBV NS X protein, a key regulatory protein of the virus that modulates viral replication, pathogenesis, interacts with a wide range of cellular proteins including P53 has largely been held responsible for the carcinogenic properties of HBV[55].
Discovered lately, HGV/GBV-C are two viral agents that have been shown to be different strains of the same virus based on sequence similarity[56]. Overall, the worldwide prevalence of HGV/GBV-C in blood donors ranges from 0.9% to 10%. HGV/GBV-C is a parenterally transmitted virus, which in most cases occurs in the setting of co-infection of HBV and/or HCV. This coinfection has been attributed to similar modes of transmission, as HBV and HCV. The genome of HGV/ GBV-C is a positive-sense RNA having sequence and organization similar to other viruses belonging to the Flaviviridae family. The viral genome contains a continuous open reading frame (ORF) headed by a 458 nucleotide long 5' untranslated region (UTR) followed by a 315 nucleotide long 3' UTR. The ORF encodes a polyprotein of 2873 amino acids with a helicase motif, two chymotrypsin-like protease motifs and an RNA-dependent RNA polymerase motif[56].
Although initially associated with hepatitis, consideration of HGV/ GBV-C primarily as a hepatotropic virus is still under debate[57]. It has been demonstrated that in absence of coinfection with other hepatotropic viruses, liver injury or viral replicative forms are usually not detectable in the liver[58,59]. Since the majority of HGV/ GBV-C positive patients with HCC are also found to be coinfected with either HBV or HCV, it is difficult to assess the true role of HGV in the etiology of HCC[56]. Nevertheless, results from a number of studies clearly indicate a primary lymphotropic nature of GBV-C/HGV as the viral replicative forms (an indicator of active GBV-C/HGV replication) have been detected in circulating lymphocytes, bone marrow, spleen, mononuclear cells and lymph nodes in a proportion of GBV-C/HGV infected patients[57,59-63].
Considering the similarity between HCV and HGV/ GBV-C a similar relationship to the development of lymphoma has been expected[64]. In anticipation, some recent studies have shown the positive correlation between HGV/GBV-C and NHL. Renzo and colleagues reported that, in a series of unselected and untransfused patients in Italy, the prevalence of HGV infections were significantly higher in patients suffering from lymphoproliferative disease compared to healthy subjects[65]. Similar reports on the association of HGV and NHL can also be found from the countries like Germany, Canada, and Greece[64,66-69]. On the contrary, a study from Turkey, suggested neither HCV nor HGV can be linked to NHL[70]. Considering relatively late discovery studies on the exact role of HGV/GBV-C and NHL development are scarce and further studies are needed to firmly conclude about any correlation.
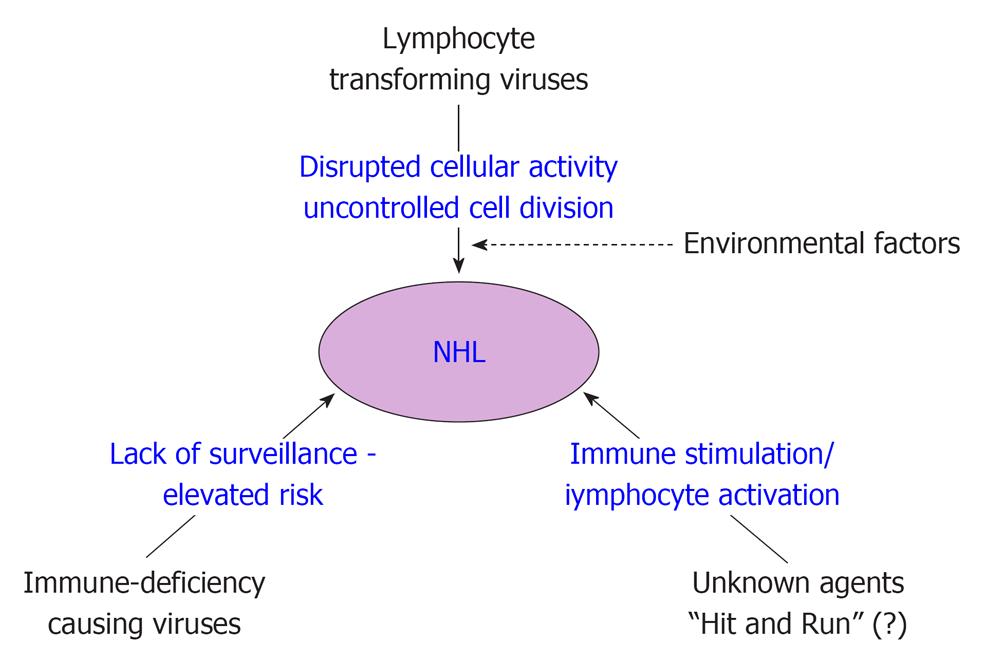
Based on the results of the association studies, Engels classified known or suspected infectious agents of NHL into three broad groups to explain the mechanism of NHL development[23]. These three mechanisms are schematically shown in Figure 1. First comes the lymphocyte infecting and transforming viruses, which disrupt normal cell functions and promote cell division. Second are those infectious agents that lead to immune deficiency (e.g., HIV) resulting in elevated risk of NHLs. Third group includes certain yet unknown infectious agents that may increase NHL risk through continual immune stimulation and lymphocytes activation. Two other hypothesis were also proposed to explain the mechanism; the ‘‘hit-and-run’’ hypothesis, which assumes that an agent significantly initiates oncogenic stimulus in the lymphocyte and disappears till NHL develops, and other ‘‘hygiene” hypothesis assumes that exposure to common infectious agents in early childhood, modulates NHL risk later in life. Although previous studies seem to support the initial three mechanisms, but the last two hypotheses (hit-and-run and hygiene hypothesis) are difficult to verify or prove since the oncogenic stimulus/agent is not detectable/lost at the time of NHL diagnosis.
Recently different workers have proposed that different individual etiological mechanisms are not mutually exclusive, but development of NHL is a multi causal event[10,27,71,72]. Recently, Marcucci and Mele hypothesised that each of the individual etiological agents provides subliminal oncogenic signals and are thus not strong enough to cause pathogenesis by them alone[10]. However, they suggested that integration of two or more oncogenic signals give rise to a supraliminal composite signal, necessary for lymphomagenesis. To explain the observed geographic discrepancies between incidence rates of NHL and prevalence rates of viral infections, it was suggested that geographic areas with a low prevalence of viral infection may have a high prevalence of a yet undefined environmental factor that may integrate with the viral oncogenic signal in a tissue specific manner[10,73].
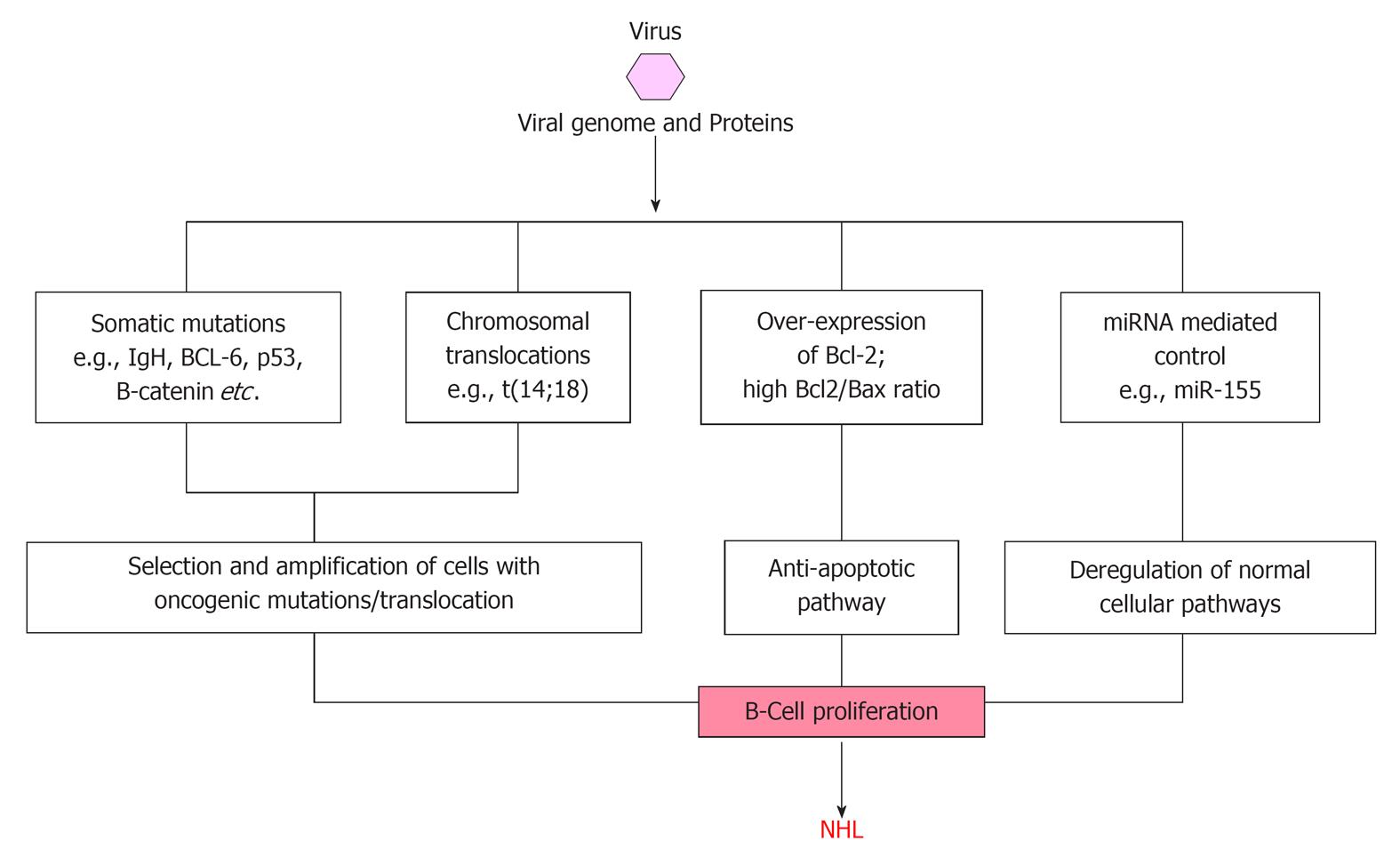
Among the three hepatitis viruses, namely HBV, HCV and HGV/GBV-C, mechanism of HCV related NHL development is most widely studied (Figure 2). HCV is considered a classical example of infectious agent causing NHL through persistent immune stimulation, associated with a range of immune-related conditions that can lead to NHL[23,74]. However, a direct oncogenic role of HCV through B-cell infection and deregulation has been proposed since the virus is lymphotropic, but this has never been proved[75]. The observation of HCV viral genome or proteins in only a subset of the neoplastic cells of HCV-associated NHLs, whereas frequently detection of viral genome and proteins in the stromal cells surrounding the neoplastic cells suggest that specific B-cell clones proliferate as a consequence of the chronic antigenic stimulation sustained by HCV. The immunoglobin variable region genes expressed by B-NHL cells from HCV-positive patients show somatic mutations suggestive of an antigen selection process and the amino acid sequences of B-cell receptors in HCV-associated lymphoproliferations has been reported to have a similarity with anti-HCV antibodies[75-78]. Moreover, the histologic presentation of many B-NHL cells from HCV positive patients are characteristic of germinal center (GC) and post-GC B-cells, suggesting the occurrence of lymphomagenesis at the time of B-cells proliferation in response to a viral antigen[76].
Neoplastic transformation may also be the result of direct anti-apoptotic pathways activated by HCV within B-cells. In fact, HCV sequences have been detected in lymph node biopsy specimens from patients with B-NHL[79] and the presence of HCV-associated proteins within lymphoma cells has also been demonstrated[80]. Moreover, studies in severe combined immunodeficiency mice have provided evidence of the persistence and low-rate multiplication of HCV infection in human mononuclear cells[81]. Finally, some HCV proteins have been shown to exert anti-apoptotic effects in infected cells in transgenic mice[82,83].
HCV related lymphoproliferation is thought to be due to binding of HCV to receptors on the surface of B-lymphocytes lowering their threshold for antigen response, inducing DNA mutations, increased frequency of abnormal chromosomal translocations associated with NHL[84,85]. The identification of the specific binding of HCV E2 protein to the ubiquitously abundant B-cell surface molecule CD81, has supported the hypothesis that a consistent polyclonal B-cell response to viral antigens favour the development of lymphoproliferative disorders[27,84]. Although HCV can infect lymphocytes, there is lack of evidence for its direct lymphomagenic role[75]. Different studies have showed a significant association between HCV infection and Bcl-2 rearrangement t(14;18 translocation)[85-89]. In these patients, clonal expansion of B-cells harboring translocation t(14;18) was demonstrated, with overexpression of the anti-apoptotic Bcl-2 protein, resulting in higher Bcl-2/bax ratio[87,90]. Fascinatingly, the observation that treatment with interferon-α of HCV positive patients with splenic marginal zone NHL, results in HCV clearance, reduced frequency of translocation t(14;18) and regression of NHL clearly implies the causal role of HCV infection in at least certain subsets of NHLs[29,31,90-92]. Different possible modes of NHL causation by viral infection are schematically presented in Figure 2.
Recently, Machida and colleagues have demonstrated that HCV infection resulted in a 5 to 10-fold increase in somatic gene mutation frequency in IgHC, BCL-6, p53, and β-catenin genes of in vitro HCV-infected B-cell lines and HCV-associated peripheral blood mononuclear cells, lymphomas, and HCC, and proposed that this mutator phenotype of HCV leads to selection and amplification of deleterious mutations in the protooncogenes or tumor-suppressor genes in tumors[93]. They also suggested two different mechanisms of mutation, based on the observation that nucleotide-substitution pattern of p53 and β-catenin is different from that of IgHC. Very interestingly this mutator function of HCV was found to be unique among oncogenic viruses, as similar amplification of protooncogene mutations in HCV-associated lymphomas and HCCs were not detectable in other types of tumors, lymphomas not related to HCV, HBV-associated HCCs, and HCCs of nonviral origin. Furthermore, the ability of HCV to induce high mutation frequency of cellular genes suggests that HCV may cause tumor formation by a hit-and-run mechanism.
Quite obviously, being of multi-factorial aetiology, there are a number of possible mechanisms through which HCV can induce lymphomagenesis, some elucidated, rest remain to be explored. Compared to HCV, HBV was discovered much earlier and its lymphotropic properties have been documented long back. Despite this fact, ironically, HBV related research has not been properly focused on its lymphomagenic properties. In contrast, HGV/GBV-C being relatively newly discovered, studies are in progress to describe different properties and mechanisms of pathogenesis. It is assumed that being a member of the Flaviviridae family, as HCV does, HGV/GBV-C might utilize similar mechanisms.
MicroRNAs (miRNA) are a group of lately discovered, highly conserved small noncoding RNAs arising from eukaryote genomes, that play an extremely important role in post-transcriptional regulation and are involved in a wide variety of biological pathways[94-96]. They control gene expression through base-pairing with the 3’-UTRs of target mRNAs, inducing mRNA degradation or suppressing translation, depending on the perfection of base-pairing[97-99]. One miRNA is capable of regulating the expression of multiple target genes; computational analyses have indicated that expression of more than 30% human genes is regulated by miRNAs[100]. MiRNAs show a highly tissue specific expression pattern, having important role in organ development, cellular differentiation, homeostasis, immune response, apoptosis and carcinogenesis also[95,101,102]. Very interestingly, apart from eukaryotes, a number of DNA viruses have also been shown to encode miRNAs, which probably help these viruses in modulating host gene expression favourable for its own replication[96]. Among the human DNA viruses, mainly Herpesviruses (EBV; human cytomegalovirus, hCMV; herpes simplex virus, HSV; Kaposi’s sarcoma-associated herpesvirus, KHSV, etc.), Polyomaviruses, and Adenoviruses (human adenovirus, hAV) have been reported to encode and express miRNAs[96]. Using computational approaches, recently, Jin and colleagues found that HBV (Hepadnavirus family, DNA virus) could putatively encode atleast one candidate pre-miRNA, which could target one of its own viral mRNA, but could not target any of the cellular transcripts[103]. However this data remains to be experimentally validated at the cellular level. In contrast, using standard sequencing or advanced sequencing techniques, no viral miRNAs have been identified HPV or in RNA viruses such as Lentiviruses (human T-cell leukemia virus I, HTLV-1) or Flaviviruses (HCV; dengue virus, DENV, etc.), which led to a general thought that RNA viruses might not encode or express similar regulatory small RNAs[95,104-106]. Interestingly, recently, Andrew Fire’s group studied infection of six different RNA viruses (including HCV, DENV, West Nile viruses, WNV, etc.) in 41 experimentally susceptible and resistant host systems, and reported identification of a class of RNA virus-derived small RNAs, termed as “vsRNAs”, 99.97% of them showing perfect homology only with the infecting virus genomes. The authors also found that the cellular short RNA apparatus was capable to employ these vsRNAs as vsRNA-primed Ago (Argonaute) complexes, but the degree to which these complexes executed silencing of functional viral RNAs and modulation of cellular miRNAs require further investigations[107].
Whether virus encoded regulatory small RNAs are detectable or not in the infected cells, viral infection has been shown to modulate the host cellular miRNA expression, altering the cellular environment, leading to pathogenesis[96]. Fascinatingly, it is well established that certain viruses such as EBV, Vesicular Stomatitis virus (VSV) induces host miR-155 expression which is one of the typical multifunctional miRNAs involved in B-cell differentiation and proliferation, and has been shown to be overexpressed in Hodgkin and non-Hodgkin lymphomas, chronic lymphocytic leukaemia[95,108-114]. MiR-155 is involved in numerous physiological and pathological processes including innate and adaptive immunity, inflammation and tumorigenesis[115]. It has been shown to be important for immunoglobulin class switching and to prevent potentially oncogenic chromosomal aberrations through regulation of activation induced cytidine deaminase[116,117]. In addition to miR-155, EBV also induces miR-146a expression in B-cells[118]. MiR-146a can target TNF receptor associated factor 6 and Interleukin-1 receptor-associated kinase 1, of the Toll-like Receptor signaling pathway, suggesting a negative-feedback loop to limit innate immune responses[119,120]. Similarly, E6 protein of HPV downregulates the expression of miR-34a (a p53-regulated miRNA) and miR-218, that leads to increase in cell growth and tumorigenicity[121,122]. In contrast, viruses may also induce miRNAs that restrict viral replication. For example, HCV is known to upregulate IFN-β, which induces cellular miR-196, miR-296, miR-351, miR-431, and miR-448, etc., which in turn attenuate viral replication and viral accumulation[123].
Apart from modulating the cellular miRNA expression, viruses can even encode mimics of host miRNAs, the later been hypothesized to cause pathogenesis[95,96]. Support to this thought comes from the fact that the chicken oncogenic Marek’s disease virus type 1 (MDV-1), expresses a miR-155-mimic, while the non-oncogenic MDV-2 does not express miR-155. Viral mimic of cellular miR-155, known as miR-K12-11 has also been reported from KSHV[124]. More interestingly, viruses have been recognized to utilize certain cellular miRNA for tissue specific infection also. HCV has been demonstrated to exploit a liver tissue specific miR-122 to positively regulate RNA replication[125]. Mammalian miR-122 expression is generally confined to the liver, and it helps maintain liver tissue identity by regulating fatty acid and cholesterol biosynthesis, pathways[95,96]. The liver-specific expression of miR-122 and its positive effect on HCV replication has been associated with hepatic tissue tropism of HCV and it has been hypothesized that tissue specific expression of certain miRNAs subject viruses to selective pressures, and viruses get optimized or evolved for replication in certain tissues (target tissue), while certain other tissues with different miRNAs might pose significant hurdles for viral replication, rendering them non-target tissues for the given virus[95]. This hypothesis has been firmly supported by experimental findings that engineering cellular miRNA target sites in the viral genomes can alter tissue tropism[126]. Even though further investigations are needed to conclude, but this might be a consistent explanation for the compartmentalization of different variants/genotypes of the same virus in different tissues of the same subject, a phenomenon very frequently observed in a number of viral infection[127].
Although the miRNA expression patterns have been widely studied in different human cancers, but similar studies on NHL is relatively scarce, despite its immense importance. Recently some studies have focused on the role of miRNA in development of NHL[128-131]. Generally, up-regulation of miR-155 has been described in NHLs, as well as in several other solid and hematologic malignancies. Another miR-17-92 cluster has been found to be frequently amplified in malignant B-cell lymphomas, and is over-expressed in 65% of B-cell lymphoma patients[132]. MiR-143 and miR-145 expression is shown to be reduced in B-cell malignancies[133]. A recent study showed that enforced expression of the miR-17-92 cluster accelerated MYC induced lymphomagenesis[134]. Unfortunately however, there is hardly any published data on the miRNA expression patterns among NHL patients infected with HBV/HCV or HGV/GBV-C. A recent study clearly demonstrated the existence of HBV- and HCV specific differential cellular miRNAs expression profiles in HBV and HCV infected liver samples as well in Huh 7.5 cell culture models, revealing that entirely different pathways were modulated in HBV or HCV infected liver[135]. These results clearly depict the contribution of the hepatitis viruses (at the focus of this review mainly HBV and HCV) in cellular miRNA deregulation leading to liver pathogenesis. Similarly, it is plausible to assume that similar to hepatocytes, these hepatitis viruses may also alter the miRNA expression patterns in lymphocytes.
NHL comprises diverse subtypes of lymphoproliferative malignancies with distinct epidemiologic, etiologic, morphologic, genetic/molecular, clinical, immunologic and histological features[136,137]. Compared to HL, different NHLs have a higher tendency for extranodal sites. Like most other cancers, prognosis of NHL largely depends on the accuracy of histological classification, stage at detection and response to treatment[138,139]. A number of classification systems have been proposed and updated in the past decades, systems such as Rappaport and Lukes Collins classification system (developed in 1966, modified by Lukes Collins in 1974), Kiel classification system (developed in 1974), Working Formulation (developed in 1982), and the Revised European-American Lymphoid neoplasms classification (REAL, published in 1994) are the important ones. Earlier, classification was solely based on morphological characteristics, but later immunological, cytogenetic and molecular features were incorporated, that facilitate precise classification[138]. Presently, the World Health Organization classification (based on the principles of the REAL system, published in 2001, updated in 2008) is widely accepted and practised[138,140,141]. However, for selecting a therapeutic strategy, Ann Arbor anatomic staging system is presently in practice[138,142,143].
Being highly heterogeneous, different NHL subtypes are associated with a variety of typical and atypical clinical manifestations. However, more than 7o% of the patients do not present classic symptoms, and are incidentally diagnosed while undergoing treatment for other nonspecific complaints. Usually NHL may present atypically with fever, fatigue, loss of appetite, drenching night sweats, weight loss, red patchy skin, etc. These general symptoms are termed as “B symptoms” and are generally associated with increased cancer “burden” due to delayed detection and poor prognosis. In contrast to Hodgkin’s disease, most patients with NHL present with advanced stage III or IV disease. The most typical clinical presentation of NHL is lymphadenopathy or extranodal mass in one or more lymph nodes. NHL involving abdominal lymphatic tissues often present with swelling of belly and lymphoma involving thymus is frequently presented with chest pain, or respiratory problems, etc. Sites often involved in NHL include skin thyroid, breast, gastrointestinal tract, brain, and ovaries or testes[138]. NHL may also involve unusual sites, such as epitrochlear or popliteal nodes or Waldeyer’s ring (nasopharynx).
Generally, NHLs are divided into two prognostic groups, the indolent lymphomas (mostly nodular or follicular in morphology) have a relatively good prognosis and the aggressive lymphomas have a shorter natural history. Early-stages (I and II) of indolent lymphomas are effectively treated with radiation therapy, but almost incurable in advanced stages. However, a considerable number of patients with aggressive lymphoma are curable with intensive combination chemotherapy.
Significant improvements in the field of NHL management made in the recent years have resulted in cure of 30% to 60% patients with aggressive NHL. Decision on treatment depends largely on lymphoma sub-type, stage, age and overall health condition of the patient, etc. often, in certain cases of indolent lymphomas, treatment delayed till the manifestation, technically known as "watchful waiting". Nevertheless, majority of the patients require treatment consisting of chemotherapy, radiation therapy, and bone marrow/stem cell transplantation, alone or in a combination.
Chemotherapy is the most imperative treatment and often includes anti-cancer drug combinations[138,144,145]. Methotrexate, Doxorubicin Hydrochloride, Chlorambucil, Nelarabine, Tositumomab, Bleomycin, Cyclophosphamide, Liposomal Cytarabine, Pralatrexate, Romidepsin, Rituximab, Vinblastine Sulfate, Vorinostat, etc., are some of the drugs used for chemotherapy. Conversely, CHOP (Cyclophosphamide, Hydroxydauno-rubicin/doxorubicin, Oncovin, Prednisone or prednisolone), rituximab with CHOP (R-CHOP), cyclophosphamide, oncovin, procarbazine and prednisone, Etoposide, doxorubicin, vincristine, prednisone, and cycolophosphamide, ifosfamide, carboplatin and etoposide are some of the recipe used for combination chemotherapy. Although chemotherapy alone can cure several high-grade lymphomas, but sometimes patients with recurring or drug irresponsive lymphomas are considered for higher dose of chemotherapy followed by autologous bone marrow transplant. The most widespread therapy for NHL is the R-CHOP combination therapy and consists of the CHOP chemotherapy along with Rituximab immunotherapy (a chimeric monoclonal antibody against CD20 protein, principally found on B cells surface), which distinctively targets certain lymphoma cells and selectively kills them.
Apart from chemo-immune therapy, radio-immuno conjugates (radio actives conjugated to monoclonal antibodies) are also used for selective destruction of cancerous cells. This therapy is among the best treatment for NHL, as NHL is highly radiosensitive. In this therapy, Iodine-131 (Tositumomab) or the Yttrium-90 (Ibritumomab tiuxetan) is conjugated to antibodies against CD20, increasing the specificity, while diminishing off-target damage[145]. In addition, conjugation of immunotherapeutic monoclonal antibodies with immunomodulators (such as α-Interferon) is also under clinical trials[145]. Other treatment approaches for NHL, drug regimen, efficacy and results of different clinical trials may be found elsewhere in more details[138,139,144,145]. In addition, novel therapeutic approaches, target pathways and potential small molecule inhibitors for treatment of NHL resistant to conventional treatment have also been reviewed recently[146].
The available literature clearly signifies that hepatitis viruses (HBV, HCV and HGV/GBV-C) have strong lymphotropic properties and most of the published data corroborate a causal association between these viruses and NHL. Additionally, prevailing data showing rapid increase in incidences and deaths due to NHL highlight the magnitude of disease burden in developed as well as less developed areas. Despite its significance, hepatitis virus associated NHL remains poorly understood. Although, mechanism of HCV related NHL has been studied in some details, but the other two viruses have remained poorly studied from the perspective of their involvement in NHL development. It is also evident that there is scarcity of data related to miRNA regulation patterns in HBV, HCV and HGV/GBV-C related NHL. The availability of miRNA regulation data might help reveal important facets of lymphomagenic mechanisms. Therefore research efforts focused on hepatitis virus induced NHL is essential to properly understand the virus induced lymphomagenic mechanisms, in order to develop effective intervention strategies and to reduce the disease burden.
Peer reviewer: Preeti Bharaj, PhD, 10344 Falcon Parc Blvd, Orlando, FL. PhD, Diabetes and Obesity, Sanford-Burnham MRI, 6400 Sanger Road, Orlando, FL 32827, United States
S- Editor Wang JL L- Editor A E- Editor Zheng XM
| 1. | Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1217] [Cited by in F6Publishing: 1269] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 2. | Dean M. Cancer as a complex developmental disorder--nineteenth Cornelius P. Rhoads Memorial Award Lecture. Cancer Res. 1998;58:5633-5636. [PubMed] [Cited in This Article: ] |
| 3. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (4)] |
| 4. | Emerging Links between Chronic Disease and Environmental Exposure. Non-Hodgkin’s Lymphoma. Physicians for Social Responsibility. Available from: www.psr.org/assets/pdfs/emerging-links-non-hodgkins.pdf. [Cited in This Article: ] |
| 5. | Robbins SL, Cotran RS, Kumar V. Pathologic basis of disease. 2nd ed. Philadelphia: WB Saunders 1995; . [Cited in This Article: ] |
| 6. | Arber DA. Molecular diagnostic approach to non-Hodgkin's lymphoma. J Mol Diagn. 2000;2:178-190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Matasar MJ, Zelenetz AD. Overview of lymphoma diagnosis and management. Radiol Clin North Am. 2008;46:175-98, vii. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 8. | Boffetta P. Epidemiology of adult non-Hodgkin lymphoma. Ann Oncol. 2011;22 Suppl 4:iv27–iv31 [doi: 10.1093/annonc/mdr167]. [Cited in This Article: ] |
| 9. | American Cancer Society. Global Cancer Facts & Figures. 2nd ed. Atlanta: American Cancer Society 2011; . [Cited in This Article: ] |
| 10. | Marcucci F, Mele A. Hepatitis viruses and non-Hodgkin lymphoma: epidemiology, mechanisms of tumorigenesis, and therapeutic opportunities. Blood. 2011;117:1792-1798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 188] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 11. | Ye BH. BCL-6 in the pathogenesis of non-Hodgkin's lymphoma. Cancer Invest. 2000;18:356-365. [PubMed] [Cited in This Article: ] |
| 12. | Lan Q, Wang SS, Menashe I, Armstrong B, Zhang Y, Hartge P, Purdue MP, Holford TR, Morton LM, Kricker A. Genetic variation in Th1/Th2 pathway genes and risk of non-Hodgkin lymphoma: a pooled analysis of three population-based case-control studies. Br J Haematol. 2011;153:341-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 13. | Kinlen L. Immunosuppressive therapy and acquired immunological disorders. Cancer Res. 1992;52:5474s-5476s. [PubMed] [Cited in This Article: ] |
| 14. | Tirelli U, Spina M, Gaidano G, Vaccher E, Franceschi S, Carbone A. Epidemiological, biological and clinical features of HIV-related lymphomas in the era of highly active antiretroviral therapy. AIDS. 2000;14:1675-1688. [PubMed] [Cited in This Article: ] |
| 15. | Fisher SG, Fisher RI. The epidemiology of non-Hodgkin's lymphoma. Oncogene. 2004;23:6524-6534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 277] [Cited by in F6Publishing: 270] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 16. | Dich J, Zahm SH, Hanberg A, Adami HO. Pesticides and cancer. Cancer Causes Control. 1997;8:420-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 17. | Dich J; IARC. Polychlorinated dibenzo-para-dioxins and polychlorinated dibenzofurans. Lyon: International Agency for Research on Cancer 1997; . [Cited in This Article: ] |
| 18. | Zahm SH, Blair A. Pesticides and non-Hodgkin's lymphoma. Cancer Res. 1992;52:5485s-5488s. [PubMed] [Cited in This Article: ] |
| 19. | Pearce N, Bethwaite P. Increasing incidence of non-Hodgkin's lymphoma: occupational and environmental factors. Cancer Res. 1992;52:5496s-5500s. [PubMed] [Cited in This Article: ] |
| 20. | Alexander DD, Mink PJ, Adami HO, Chang ET, Cole P, Mandel JS, Trichopoulos D. The non-Hodgkin lymphomas: a review of the epidemiologic literature. Int J Cancer. 2007;120 Suppl 12:1-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 255] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 21. | zur Hausen H. Viruses in human cancers. Science. 1991;254:1167-1173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 513] [Cited by in F6Publishing: 486] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 22. | Hjalgrim H, Engels EA. Infectious aetiology of Hodgkin and non-Hodgkin lymphomas: a review of the epidemiological evidence. J Intern Med. 2008;264:537-548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 23. | Engels EA. Infectious agents as causes of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:401-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 24. | McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782:127-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 225] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 25. | Michalak TI. Occult persistence and lymphotropism of hepadnaviral infection: insights from the woodchuck viral hepatitis model. Immunol Rev. 2000;174:98-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 68] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Pham TN, Michalak TI. Occult persistence and lymphotropism of hepatitis C virus infection. World J Gastroenterol. 2008;14:2789-2793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 28] [Cited by in F6Publishing: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Zignego AL, Giannini C, Monti M, Gragnani L. Hepatitis C virus lymphotropism: lessons from a decade of studies. Dig Liver Dis. 2007;39 Suppl 1:S38-S45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141-1157. [PubMed] [Cited in This Article: ] |
| 29. | Hermine O, Lefrère F, Bronowicki JP, Mariette X, Jondeau K, Eclache-Saudreau V, Delmas B, Valensi F, Cacoub P, Brechot C. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347:89-94. [PubMed] [Cited in This Article: ] |
| 30. | Vallisa D, Bernuzzi P, Arcaini L, Sacchi S, Callea V, Marasca R, Lazzaro A, Trabacchi E, Anselmi E, Arcari AL. Role of anti-hepatitis C virus (HCV) treatment in HCV-related, low-grade, B-cell, non-Hodgkin's lymphoma: a multicenter Italian experience. J Clin Oncol. 2005;23:468-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 192] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 31. | Zuckerman E, Zuckerman T, Sahar D, Streichman S, Attias D, Sabo E, Yeshurun D, Rowe JM. The effect of antiviral therapy on t(14; 18) translocation and immunoglobulin gene rearrangement in patients with chronic hepatitis C virus infection. Blood. 2001;97:1555-1559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 94] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Dal Maso L, Franceschi S. Hepatitis C virus and risk of lymphoma and other lymphoid neoplasms: a meta-analysis of epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2006;15:2078-2085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 208] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 33. | Hausfater P, Cacoub P, Sterkers Y, Thibault V, Amoura Z, Nguyen L, Ghillani P, Leblond V, Piette JC. Hepatitis C virus infection and lymphoproliferative diseases: prospective study on 1,576 patients in France. Am J Hematol. 2001;67:168-171. [PubMed] [Cited in This Article: ] |
| 34. | Collier JD, Zanke B, Moore M, Kessler G, Krajden M, Shepherd F, Heathcote J. No association between hepatitis C and B-cell lymphoma. Hepatology. 1999;29:1259-1261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 88] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 35. | Negri E, Little D, Boiocchi M, La Vecchia C, Franceschi S. B-cell non-Hodgkin's lymphoma and hepatitis C virus infection: a systematic review. Int J Cancer. 2004;111:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 36. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [PubMed] [Cited in This Article: ] |
| 37. | Shi L, Zhang SL, Li K, Hong Y, Wang Q, Li Y, Guo J, Fan WH, Zhang L, Cheng J. NS5ATP9, a gene up-regulated by HCV NS5A protein. Cancer Lett. 2008;259:192-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | Schmitz U, Tan SL. NS5A--from obscurity to new target for HCV therapy. Recent Pat Antiinfect Drug Discov. 2008;3:77-92. [PubMed] [Cited in This Article: ] |
| 39. | Gisbert JP, García-Buey L, Pajares JM, Moreno-Otero R. Prevalence of hepatitis C virus infection in B-cell non-Hodgkin's lymphoma: systematic review and meta-analysis. Gastroenterology. 2003;125:1723-1732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 203] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 40. | Marcucci F, Mele A, Spada E, Candido A, Bianco E, Pulsoni A, Chionne P, Madonna E, Cotichini R, Barbui A. High prevalence of hepatitis B virus infection in B-cell non-Hodgkin's lymphoma. Haematologica. 2006;91:554-557. [PubMed] [Cited in This Article: ] |
| 41. | Galun E, Ilan Y, Livni N, Ketzinel M, Nahor O, Pizov G, Nagler A, Eid A, Rivkind A, Laster M. Hepatitis B virus infection associated with hematopoietic tumors. Am J Pathol. 1994;145:1001-1007. [PubMed] [Cited in This Article: ] |
| 42. | Park SC, Jeong SH, Kim J, Han CJ, Kim YC, Choi KS, Cho JH, Lee M, Jung HH, Ki SS. High prevalence of hepatitis B virus infection in patients with B-cell non-Hodgkin's lymphoma in Korea. J Med Virol. 2008;80:960-966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 43. | Kim JH, Bang YJ, Park BJ, Yoo T, Kim CW, Kim TY, Heo DS, Lee HS, Kim NK. Hepatitis B virus infection and B-cell non-Hodgkin's lymphoma in a hepatitis B endemic area: a case-control study. Jpn J Cancer Res. 2002;93:471-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 44. | Chen MH, Hsiao LT, Chiou TJ, Liu JH, Gau JP, Teng HW, Wang WS, Chao TC, Yen CC, Chen PM. High prevalence of occult hepatitis B virus infection in patients with B cell non-Hodgkin's lymphoma. Ann Hematol. 2008;87:475-480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Wang F, Xu RH, Han B, Shi YX, Luo HY, Jiang WQ, Lin TY, Huang HQ, Xia ZJ, Guan ZZ. High incidence of hepatitis B virus infection in B-cell subtype non-Hodgkin lymphoma compared with other cancers. Cancer. 2007;109:1360-1364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 46. | Lim ST, Fei G, Quek R, Lim LC, Lee LH, Yap SP, Loong S, Tao M. The relationship of hepatitis B virus infection and non-Hodgkin's lymphoma and its impact on clinical characteristics and prognosis. Eur J Haematol. 2007;79:132-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Kuniyoshi M, Nakamuta M, Sakai H, Enjoji M, Kinukawa N, Kotoh K, Fukutomi M, Yokota M, Nishi H, Iwamoto H. Prevalence of hepatitis B or C virus infections in patients with non-Hodgkin's lymphoma. J Gastroenterol Hepatol. 2001;16:215-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 48. | Anderson LA, Pfeiffer R, Warren JL, Landgren O, Gadalla S, Berndt SI, Ricker W, Parsons R, Wheeler W, Engels EA. Hematopoietic malignancies associated with viral and alcoholic hepatitis. Cancer Epidemiol Biomarkers Prev. 2008;17:3069-3075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 49. | Ulcickas Yood M, Quesenberry CP, Guo D, Caldwell C, Wells K, Shan J, Sanders L, Skovron ML, Iloeje U, Manos MM. Incidence of non-Hodgkin's lymphoma among individuals with chronic hepatitis B virus infection. Hepatology. 2007;46:107-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 50. | Nath A, Agarwal R, Malhotra P, Varma S. Prevalence of hepatitis B virus infection in non-Hodgkin lymphoma: a systematic review and meta-analysis. Intern Med J. 2010;40:633-641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 51. | Bréchot C, Thiers V, Kremsdorf D, Nalpas B, Pol S, Paterlini-Bréchot P. Persistent hepatitis B virus infection in subjects without hepatitis B surface antigen: clinically significant or purely "occult". Hepatology. 2001;34:194-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 414] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 52. | Rehermann B, Ferrari C, Pasquinelli C, Chisari FV. The hepatitis B virus persists for decades after patients' recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med. 1996;2:1104-1108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 624] [Cited by in F6Publishing: 588] [Article Influence: 21.0] [Reference Citation Analysis (1)] |
| 53. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [PubMed] [Cited in This Article: ] |
| 54. | Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 183] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 55. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 56. | Cheung RC, Keeffe EB, Greenberg HB. Hepatitis G virus: is it a hepatitis virus. West J Med. 1997;167:23-33. [PubMed] [Cited in This Article: ] |
| 57. | Radkowski M, Kubicka J, Kisiel E, Cianciara J, Nowicki M, Rakela J, Laskus T. Detection of active hepatitis C virus and hepatitis G virus/GB virus C replication in bone marrow in human subjects. Blood. 2000;95:3986-3989. [PubMed] [Cited in This Article: ] |
| 58. | Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71:7804-7806. [PubMed] [Cited in This Article: ] |
| 59. | Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol. 1998;79:705-714. [PubMed] [Cited in This Article: ] |
| 60. | Radkowski M, Wang LF, Cianciara J, Rakela J, Laskus T. Analysis of hepatitis G virus/GB virus C quasispecies and replication sites in human subjects. Biochem Biophys Res Commun. 1999;258:296-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 61. | Tucker TJ, Smuts HE, Eedes C, Knobel GD, Eickhaus P, Robson SC, Kirsch RE. Evidence that the GBV-C/hepatitis G virus is primarily a lymphotropic virus. J Med Virol. 2000;61:52-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 62. | Madejón A, Fogeda M, Bartolomé J, Pardo M, González C, Cotonat T, Carreńo V. GB virus C RNA in serum, liver, and peripheral blood mononuclear cells from patients with chronic hepatitis B, C, and D. Gastroenterology. 1997;113:573-578. [PubMed] [Cited in This Article: ] |
| 63. | Saito S, Tanaka K, Kondo M, Morita K, Kitamura T, Kiba T, Numata K, Sekihara H. Plus- and minus-stranded hepatitis G virus RNA in liver tissue and in peripheral blood mononuclear cells. Biochem Biophys Res Commun. 1997;237:288-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Ellenrieder V, Weidenbach H, Frickhofen N, Michel D, Prümmer O, Klatt S, Bernas O, Mertens T, Adler G, Beckh K. HCV and HGV in B-cell non-Hodgkin's lymphoma. J Hepatol. 1998;28:34-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 65. | De Renzo A, Persico E, de Marino F, di Giacomo Russo G, Notaro R, di Grazia C, Picardi M, Santoro L, Torella R, Rotoli B. High prevalence of hepatitis G virus infection in Hodgkin's disease and B-cell lymphoproliferative disorders: absence of correlation with hepatitis C virus infection. Haematologica. 2002;87:714-78; discussion 718. [PubMed] [Cited in This Article: ] |
| 66. | Krajden M, Yu A, Braybrook H, Lai AS, Mak A, Chow R, Cook D, Tellier R, Petric M, Gascoyne RD. GBV-C/hepatitis G virus infection and non-Hodgkin lymphoma: a case control study. Int J Cancer. 2010;126:2885-2892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 67. | Giannoulis E, Economopoulos T, Mandraveli K, Giannoulis K, Nikolaides C, Zervou E, Papageorgiou E, Zoulas D, Tourkantonis A, Giannopoulos G. The prevalence of hepatitis C and hepatitis G virus infection in patients with B cell non-Hodgkin lymphomas in Greece: a Hellenic Cooperative Oncology Group Study. Acta Haematol. 2004;112:189-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 68. | Wiwanitkit V. Individuals with HGV-RNA are at high risk of B cell non-Hodgkin's lymphoma development. Asian Pac J Cancer Prev. 2005;6:215-216. [PubMed] [Cited in This Article: ] |
| 69. | Wiwanitkit V. Hepatitis Virus Infection and Hodgkin's Lymphoma: A Review of the Literature. Hepatitis Mon. 2007;7:229-231. [Cited in This Article: ] |
| 70. | Kaya H, Polat MF, Erdem F, Gündogdu M. Prevalence of hepatitis C virus and hepatitis G virus in patients with non-Hodgkin's lymphoma. Clin Lab Haematol. 2002;24:107-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 71. | Rothman KJ, Greenland S. Causation and causal inference in epidemiology. Am J Public Health. 2005;95 Suppl 1:S144-S150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 695] [Cited by in F6Publishing: 620] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 72. | Zignego AL, Giannini C, Ferri C. Hepatitis C virus-related lymphoproliferative disorders: an overview. World J Gastroenterol. 2007;13:2467-2478. [PubMed] [Cited in This Article: ] |
| 73. | Stapleton JT, Chaloner K. GB virus C infection and non-Hodgkin lymphoma: important to know but the jury is out. Int J Cancer. 2010;126:2759-2761. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 74. | Pozzato G, Mazzaro C, Crovatto M, Modolo ML, Ceselli S, Mazzi G, Sulfaro S, Franzin F, Tulissi P, Moretti M. Low-grade malignant lymphoma, hepatitis C virus infection, and mixed cryoglobulinemia. Blood. 1994;84:3047-3053. [PubMed] [Cited in This Article: ] |
| 75. | Gasparotto D, De Re V, Boiocchi M. Hepatitis C virus, B-cell proliferation and lymphomas. Leuk Lymphoma. 2002;43:747-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 76. | De Re V, De Vita S, Marzotto A, Gloghini A, Pivetta B, Gasparotto D, Cannizzaro R, Carbone A, Boiocchi M. Pre-malignant and malignant lymphoproliferations in an HCV-infected type II mixed cryoglobulinemic patient are sequential phases of an antigen-driven pathological process. Int J Cancer. 2000;87:211-216. [PubMed] [Cited in This Article: ] |
| 77. | De Re V, De Vita S, Marzotto A, Rupolo M, Gloghini A, Pivetta B, Gasparotto D, Carbone A, Boiocchi M. Sequence analysis of the immunoglobulin antigen receptor of hepatitis C virus-associated non-Hodgkin lymphomas suggests that the malignant cells are derived from the rheumatoid factor-producing cells that occur mainly in type II cryoglobulinemia. Blood. 2000;96:3578-3584. [PubMed] [Cited in This Article: ] |
| 78. | Ivanovski M, Silvestri F, Pozzato G, Anand S, Mazzaro C, Burrone OR, Efremov DG. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood. 1998;91:2433-2442. [PubMed] [Cited in This Article: ] |
| 79. | Luppi M, Grazia Ferrari M, Bonaccorsi G, Longo G, Narni F, Barozzi P, Marasca R, Mussini C, Torelli G. Hepatitis C virus infection in subsets of neoplastic lymphoproliferations not associated with cryoglobulinemia. Leukemia. 1996;10:351-355. [PubMed] [Cited in This Article: ] |
| 80. | Mele A, Pulsoni A, Bianco E, Musto P, Szklo A, Sanpaolo MG, Iannitto E, De Renzo A, Martino B, Liso V. Hepatitis C virus and B-cell non-Hodgkin lymphomas: an Italian multicenter case-control study. Blood. 2003;102:996-999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 225] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 81. | Bronowicki JP, Loriot MA, Thiers V, Grignon Y, Zignego AL, Bréchot C. Hepatitis C virus persistence in human hematopoietic cells injected into SCID mice. Hepatology. 1998;28:211-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 85] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 82. | Majumder M, Ghosh AK, Steele R, Zhou XY, Phillips NJ, Ray R, Ray RB. Hepatitis C virus NS5A protein impairs TNF-mediated hepatic apoptosis, but not by an anti-FAS antibody, in transgenic mice. Virology. 2002;294:94-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Machida K, Tsukiyama-Kohara K, Seike E, Toné S, Shibasaki F, Shimizu M, Takahashi H, Hayashi Y, Funata N, Taya C. Inhibition of cytochrome c release in Fas-mediated signaling pathway in transgenic mice induced to express hepatitis C viral proteins. J Biol Chem. 2001;276:12140-12146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 86] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 84. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1572] [Cited by in F6Publishing: 1521] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 85. | Zignego AL, Giannelli F, Marrocchi ME, Mazzocca A, Ferri C, Giannini C, Monti M, Caini P, Villa GL, Laffi G. T(14; 18) translocation in chronic hepatitis C virus infection. Hepatology. 2000;31:474-479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 127] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 86. | Zignego AL, Giannelli F, Marrocchi ME, Giannini C, Gentilini P, Innocenti F, Ferri C. Frequency of bcl-2 rearrangement in patients with mixed cryoglobulinemia and HCV-positive liver diseases. Clin Exp Rheumatol. 1997;15:711-712. [PubMed] [Cited in This Article: ] |
| 87. | Zignego AL, Ferri C, Giannelli F, Giannini C, Caini P, Monti M, Marrocchi ME, Di Pietro E, La Villa G, Laffi G. Prevalence of bcl-2 rearrangement in patients with hepatitis C virus-related mixed cryoglobulinemia with or without B-cell lymphomas. Ann Intern Med. 2002;137:571-580. [PubMed] [Cited in This Article: ] |
| 88. | Kitay-Cohen Y, Amiel A, Hilzenrat N, Buskila D, Ashur Y, Fejgin M, Gaber E, Safadi R, Tur-Kaspa R, Lishner M. Bcl-2 rearrangement in patients with chronic hepatitis C associated with essential mixed cryoglobulinemia type II. Blood. 2000;96:2910-2912. [PubMed] [Cited in This Article: ] |
| 89. | Sasso EH, Martinez M, Yarfitz SL, Ghillani P, Musset L, Piette JC, Cacoub P. Frequent joining of Bcl-2 to a JH6 gene in hepatitis C virus-associated t(14; 18). J Immunol. 2004;173:3549-3556. [PubMed] [Cited in This Article: ] |
| 90. | Zuckerman E, Zuckerman T, Sahar D, Streichman S, Attias D, Sabo E, Yeshurun D, Rowe J. bcl-2 and immunoglobulin gene rearrangement in patients with hepatitis C virus infection. Br J Haematol. 2001;112:364-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 79] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 91. | Giannelli F, Moscarella S, Giannini C, Caini P, Monti M, Gragnani L, Romanelli RG, Solazzo V, Laffi G, La Villa G. Effect of antiviral treatment in patients with chronic HCV infection and t(14; 18) translocation. Blood. 2003;102:1196-1201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 92. | Mazzaro C, Franzin F, Tulissi P, Pussini E, Crovatto M, Carniello GS, Efremov DG, Burrone O, Santini G, Pozzato G. Regression of monoclonal B-cell expansion in patients affected by mixed cryoglobulinemia responsive to alpha-interferon therapy. Cancer. 1996;77:2604-2613. [PubMed] [Cited in This Article: ] |
| 93. | Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, Lai MM. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci USA. 2004;101:4262-4267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 272] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 94. | Ross JS, Carlson JA, Brock G. miRNA: the new gene silencer. Am J Clin Pathol. 2007;128:830-836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 95. | Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 317] [Cited by in F6Publishing: 342] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 96. | Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 494] [Cited by in F6Publishing: 525] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 97. | Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 98. | Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839-845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 824] [Cited by in F6Publishing: 870] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 99. | O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1160] [Cited by in F6Publishing: 1213] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 100. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8847] [Cited by in F6Publishing: 9072] [Article Influence: 477.5] [Reference Citation Analysis (0)] |
| 101. | Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343-351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 404] [Cited by in F6Publishing: 419] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 102. | Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396-400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in F6Publishing: 1040] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 103. | Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 104. | Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006;80:10890-10893. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 105. | Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218-12226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 106. | Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 900] [Cited by in F6Publishing: 854] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 107. | Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 207] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 108. | Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295-5306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 109. | Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226-6233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 110. | Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 512] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 111. | Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627-3632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1026] [Cited by in F6Publishing: 1022] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 112. | Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 407] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 113. | Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024-7029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 848] [Cited by in F6Publishing: 830] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 114. | Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944-4951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 376] [Cited by in F6Publishing: 401] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 115. | Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497-505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 530] [Cited by in F6Publishing: 570] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 116. | Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1145] [Cited by in F6Publishing: 1164] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 117. | Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621-629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 118. | Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946-1958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 241] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 119. | Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 804] [Cited by in F6Publishing: 823] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 120. | Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436-1442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 121. | Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575-2582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 270] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 122. | Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637-647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 123. | Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 686] [Cited by in F6Publishing: 727] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 124. | Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836-12845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 351] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 125. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1993] [Cited by in F6Publishing: 1929] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 126. | Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151-1164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 306] [Cited by in F6Publishing: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 127. | Datta S, Panigrahi R, Biswas A, Chandra PK, Banerjee A, Mahapatra PK, Panda CK, Chakrabarti S, Bhattacharya SK, Biswas K. Genetic characterization of hepatitis B virus in peripheral blood leukocytes: evidence for selection and compartmentalization of viral variants with the immune escape G145R mutation. J Virol. 2009;83:9983-9992. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 128. | Baltic V, Baltic M, Svircev Z, Jerant-Patic V. microRNA expression in non-Hodgkin’s lymphomas. Arch Oncol. 2008;16:59-68 [doi: 10.2298/AOO0804059B]. [Cited in This Article: ] |
| 129. | Fanini F, Vannini I, Fabbri M. MicroRNAs: tiny players with a big role in the pathogenesis of leukemias and lymphomas. Hematology Reviews. 2009;1:e8 [doi: 10.4081/hr.2009.e8]. [Cited in This Article: ] |
| 130. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5705] [Cited by in F6Publishing: 5849] [Article Influence: 324.9] [Reference Citation Analysis (0)] |
| 131. | Sandhu SK, Croce CM, Garzon R. Micro-RNA Expression and Function in Lymphomas. Adv Hematol. 2011;2011:347137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 132. | Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087-3095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 561] [Cited by in F6Publishing: 555] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 133. | Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 227] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 134. | He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2729] [Cited by in F6Publishing: 2776] [Article Influence: 146.1] [Reference Citation Analysis (0)] |
| 135. | Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 297] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 136. | Armitage JO, Berg AR, Purtilo DT. Adult non-Hodgkin’s lymphoma. Hematology: Clinical and Laboratory Practice. Louis: Mosby-Year Book Inc 1993; 875-893. [Cited in This Article: ] |
| 137. | Jaffe ES. Histopathology of the non-Hodgkin’s lymphomas and Hodgkin’s disease. The lymphomas. Philadelphia: WB Saunders Co 1998; 77-106. [Cited in This Article: ] |
| 138. | Skarin AT, Dorfman DM. Non-Hodgkin's lymphomas: current classification and management. CA Cancer J Clin. 1997;47:351-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 139. | Mounter PJ, Lennard AL. Management of non-Hodgkin's lymphomas. Postgrad Med J. 1999;75:2-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 140. | Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84:1361-1392. [PubMed] [Cited in This Article: ] |
| 141. | Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD. The World Health Organization classification of neoplastic diseases of the haematopoietic and lymphoid tissues: Report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November 1997. Histopathology. 2000;36:69-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 291] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 142. | Lister TA, Crowther D, Sutcliffe SB, Glatstein E, Canellos GP, Young RC, Rosenberg SA, Coltman CA, Tubiana M. Report of a committee convened to discuss the evaluation and staging of patients with Hodgkin's disease: Cotswolds meeting. J Clin Oncol. 1989;7:1630-1636. [PubMed] [Cited in This Article: ] |
| 143. | Armitage JO. Staging non-Hodgkin lymphoma. CA Cancer J Clin. 2005;55:368-376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 171] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 144. | Cerny T, Gillessen S. Advances in the treatment of non-Hodgkin's lymphoma. Ann Oncol. 2002;13 Suppl 4:211-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 145. | Vose JM, Chiu BC, Cheson BD, Dancey J, Wright J. Update on epidemiology and therapeutics for non-Hodgkin's lymphoma. Hematology Am Soc Hematol Educ Program. 2002;241-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 146. | Mahadevan D, Fisher RI. Novel therapeutics for aggressive non-Hodgkin's lymphoma. J Clin Oncol. 2011;29:1876-1884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |