Abstract
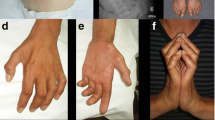
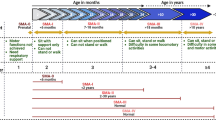
Distal spinal muscular atrophy type 1 (DSMA1) is caused by mutations in the immunoglobulin μ-binding protein 2 (IGHMBP2) gene. Patients with DSMA1 present between 6 weeks and 6 months of age with progressive muscle weakness and respiratory failure due to diaphragmatic palsy. Contrary to this “classic” infantile disease, we have previously described a DSMA1 patient with juvenile disease onset. In this paper, we present (1) a second juvenile case and (2) the first study of DSMA1 on protein level in patients with infantile (n = 3) as well as juvenile (n = 2) disease onset observing elevated residual steady-state IGHMBP2 protein levels in the patients with late onset DSMA1 as compared to those with classic DSMA1. Mutation screening in IGHMBP2 revealed two patients compound heterozygous for a novel missense mutation (c.1478C→T; p.T493I) and another previously described mutation. In lymphoblastoid cells of both patients, steady-state IGHMBP2 protein levels were reduced. In comparison to wild-type IGHMBP2, the p.T493I variant protein had an increased tendency to aggregate and spontaneously degrade in vitro. We verified a change in the physicochemical properties of the p.T493I variant which may explain the pathogenicity of this mutation. Our data further suggest that the age of onset of DSMA1 is variable, and we discuss the effect of residual IGHMBP2 protein levels on the clinical course and the severity of the disease.





Similar content being viewed by others
References
Grohmann K, Varon R, Stolz P, Schuelke M, Janetzki C, Bertini E, Bushby K, Muntoni F, Ouvrier R, Van ML, Goemans NM, Lochmuller H, Eichholz S, Adams C, Bosch F, Grattan-Smith P, Navarro C, Neitzel H, Polster T, Topaloglu H, Steglich C, Guenther UP, Zerres K, Rudnik-Schoneborn S, Hubner C (2003) Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1). Ann Neurol 54:719–724
Guenther UP, Varon R, Schlicke M, Dutrannoy V, Volk A, Hubner C, von Au K, Schuelke M (2007) Clinical and mutational profile in spinal muscular atrophy with respiratory distress (SMARD): defining novel phenotypes through hierarchical cluster analysis. Hum Mutat 28:808–815
Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, Bertini E, Leonhardt-Horti H, Muntoni F, Ouvrier R, Pfeufer A, Rossi R, Van ML, Wilmshurst JM, Wienker TF, Sendtner M, Rudnik-Schoneborn S, Zerres K, Hubner C (2001) Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet 29:75–77
Koonin EV (1992) A new group of putative RNA helicases. Trends Biochem Sci 17:495–497
Biswas EE, Nagele RG, Biswas S (2001) A novel human hexameric DNA helicase: expression, purification and characterization. Nucleic Acids Res 29:1733–1740
Molnar GM, Crozat A, Kraeft SK, Dou QP, Chen LB, Pardee AB (1997) Association of the mammalian helicase MAH with the pre-mRNA splicing complex. Proc Natl Acad Sci U S A 94:7831–7836
Cheng Z, Muhlrad D, Lim MK, Parker R, Song H (2007) Structural and functional insights into the human Upf1 helicase core. EMBO J 26:253–264
Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8:251–262
Fukita Y, Mizuta TR, Shirozu M, Ozawa K, Shimizu A, Honjo T (1993) The human S mu bp-2, a DNA-binding protein specific to the single-stranded guanine-rich sequence related to the immunoglobulin mu chain switch region. J Biol Chem 268:17463–17470
Grohmann K, Rossoll W, Kobsar I, Holtmann B, Jablonka S, Wessig C, Stoltenburg-Didinger G, Fischer U, Hubner C, Martini R, Sendtner M (2004) Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1). Hum Mol Genet 13:2031–2042
Shieh SY, Stellrecht CM, Tsai MJ (1995) Molecular characterization of the rat insulin enhancer-binding complex 3b2. Cloning of a binding factor with putative helicase motifs. J Biol Chem 270:21503–21508
Maddatu TP, Garvey SM, Schroeder DG, Hampton TG, Cox GA (2004) Transgenic rescue of neurogenic atrophy in the nmd mouse reveals a role for Ighmbp2 in dilated cardiomyopathy. Hum Mol Genet 13:1105–1115
Maystadt I, Zarhrate M, Landrieu P, Boespflug-Tanguy O, Sukno S, Collignon P, Melki J, Verellen-Dumoulin C, Munnich A, Viollet L (2004) Allelic heterogeneity of SMARD1 at the IGHMBP2 locus. Hum Mutat 23:525–526
Viollet L, Zarhrate M, Maystadt I, Estournet-Mathiaut B, Barois A, Desguerre I, Mayer M, Chabrol B, LeHeup B, Cusin V, Billette D V, Bonneau D, Saugier-Veber P, Touzery-De VA, Delaubier A, Kaplan J, Jeanpierre M, Feingold J, Munnich A (2004) Refined genetic mapping of autosomal recessive chronic distal spinal muscular atrophy to chromosome 11q13.3 and evidence of linkage disequilibrium in European families. Eur J Hum Genet 12:483–488
Guenther UP, Schuelke M, Bertini E, D'Amico A, Goemans N, Grohmann K, Hubner C, Varon R (2004) Genomic rearrangements at the IGHMBP2 gene locus in two patients with SMARD1. Hum Genet 115:319–326
Neitzel H (1986) A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet 73:320–326
den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11
Desai-Mehta A, Cerosaletti KM, Concannon P (2001) Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol 21:2184–2191
Cox GA, Mahaffey CL, Frankel WN (1998) Identification of the mouse neuromuscular degeneration gene and mapping of a second site suppressor allele. Neuron 21:1327–1337
Sambrook J, Russel DW (2001) In vitro mutagenesis using double-stranded DNA templates: selection of mutants with DpnI. Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Schwannhäusser B, Gossen M, Dittmar G, Selbach M (2008) Global analysis of cellular protein translation by pulsed SILAC. Proteomics (in press)
Pitt M, Houlden H, Jacobs J, Mok Q, Harding B, Reilly M, Surtees R (2003) Severe infantile neuropathy with diaphragmatic weakness and its relationship to SMARD1. Brain 126:2682–2692
Collins JS, Schwartz CE (2002) Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet 71:1251–1252
DePristo MA, Weinreich DM, Hartl DL (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6:678–687
Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899
Obminska-Mrukowicz B, Szczypka M, Gaweda B (2006) Modulatory effects of chitosan adipate on the T and B lymphocyte subsets in mice. J Vet Sci 7:157–160
Hur DY, Lee MH, Kim JW, Kim JH, Shin YK, Rho JK, Kwack KB, Lee WJ, Han BG (2005) CD19 signalling improves the Epstein–Barr virus-induced immortalization of human B cell. Cell Prolif 38:35–45
Helmken C, Hofmann Y, Schoenen F, Oprea G, Raschke H, Rudnik-Schoneborn S, Zerres K, Wirth B (2003) Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet 114:11–21
Acknowledgments
We thank the patients and their families for participation in this study. G. Koch, Children’s Hospital Hagen, and A. Hahn, Department of Neuropediatrics, Kiel University, contributed to the medical management of one patient. We thank Véronique Dutrannoy for providing HPRT primers and help in the RT-PCR TaqMan assay and Dr. Dietrich Carlhoff from GE Healthcare for his continuous help and counseling regarding the ÄKTA FPLC system. The work was supported by grants of the Deutsche Forschungsgemeinschaft (HU 408/3-2 and HU 408/3-3), the Charité (Rahel-Hirsch-Fellowship to K.v.A.), and by a generous donation of the parents’ self help group “Helft dem muskelkranken Kind, Hamburg e.V.”. The Protein Sample Production Facility at the Max Delbrück Center is funded by the Helmholtz Association of German Research Centers.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the image is a link to a high resolution version.
ESM Fig. 1
Pedigrees of the two families with segregation of the various mutations. All mutations are numbered according to the IGHMBP2 protein sequence NP_002171 and the nomenclature suggested by den Dunnen et al. [17] (GIF 71 KB).
ESM Fig. 2
Multiple alignment of IGHMBP2 orthologs shows the high evolutionary conservation of the amino acid p.T493. The p.T493I mutation is located within the helicase domain, but outside of the canonical helicase motifs. Homologous sequences were retrieved from Genbank (http://www.ncbi.nlm.nih.gov/) and the Ensembl database (http://www.ensembl.org) with the following accession numbers: NP_012908.1; NP_565299; P40694; ENSPTRP00000006865; ENSMMUP00000013244; ENSBTAT0000002993; ENSCAFP00000015816; ENSGALP00000006890; ENSCINP00000010526; ENSDARP00000060290; ENSXETP00000043180 (GIF 141 KB).
Rights and permissions
About this article
Cite this article
Guenther, UP., Handoko, L., Varon, R. et al. Clinical variability in distal spinal muscular atrophy type 1 (DSMA1): determination of steady-state IGHMBP2 protein levels in five patients with infantile and juvenile disease. J Mol Med 87, 31–41 (2009). https://doi.org/10.1007/s00109-008-0402-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-008-0402-7