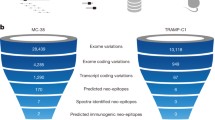
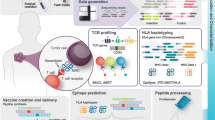
Abstract
Our immune system plays a key role in health and disease as it is capable of responding to foreign antigens as well as acquired antigens from cancer cells. Latter are caused by somatic mutations, the so-called neoepitopes, and might be recognized by T cells if they are presented by HLA molecules on the surface of cancer cells. Personalized mutanome vaccines are a class of customized immunotherapies, which is dependent on the detection of individual cancer-specific tumor mutations and neoepitope (i.e., prediction, followed by a rational vaccine design, before on-demand production. The development of next generation sequencing (NGS) technologies and bioinformatic tools allows a large-scale analysis of each parameter involved in this process. Here, we provide an overview of the bioinformatic aspects involved in the design of personalized, neoantigen-based vaccines, including the detection of mutations and the subsequent prediction of potential epitopes, as well as methods for associated biomarker research, such as high-throughput sequencing of T-cell receptors (TCRs), followed by data analysis and the bioinformatics quantification of immune cell infiltration in cancer samples.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1):57–70
Britten CM, Singh-Jasuja H, Flamion B et al (2013) The regulatory landscape for actively personalized cancer immunotherapies. Nat Biotechnol 31(10):880–882. https://doi.org/10.1038/nbt.2708
Castle JC, Kreiter S, Diekmann J et al (2012) Exploiting the mutanome for tumor vaccination. Cancer Res 72(5):1081–1091. https://doi.org/10.1158/0008-5472.CAN-11-3722
Kreiter S, Vormehr M, van de Roemer N et al (2015) Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520(7549):692–696. https://doi.org/10.1038/nature14426
Sahin U, Derhovanessian E, Miller M et al (2017) Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547(7662):222–226. https://doi.org/10.1038/nature23003
Sahin U, Türeci Ö (2018) Personalized vaccines for cancer immunotherapy. Science 359(6382):1355–1360. https://doi.org/10.1126/science.aar7112
Riaz N, Morris L, Havel JJ et al (2016) The role of neoantigens in response to immune checkpoint blockade. Int Immunol 28(8):411–419. https://doi.org/10.1093/intimm/dxw019
Park Y-J, Kuen D-S, Chung Y (2018) Future prospects of immune checkpoint blockade in cancer: from response prediction to overcoming resistance. Exp Mol Med 50(8):109. https://doi.org/10.1038/s12276-018-0130-1
Darvin P, Toor SM, Sasidharan Nair V et al (2018) Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 50(12):165. https://doi.org/10.1038/s12276-018-0191-1
McGranahan N, Furness AJS, Rosenthal R et al (2016) Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351(6280):1463–1469. https://doi.org/10.1126/science.aaf1490
Löwer M, Renard BY, de Graaf J et al (2012) Confidence-based somatic mutation evaluation and prioritization. PLoS Comput Biol 8(9):e1002714. https://doi.org/10.1371/journal.pcbi.1002714
Jurtz VI, Olsen LR (2019) Computational methods for identification of T cell neoepitopes in tumors. Methods Mol Biol 1878:157–172. https://doi.org/10.1007/978-1-4939-8868-6_9
Xu H, DiCarlo J, Satya RV et al (2014) Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics 15:244. https://doi.org/10.1186/1471-2164-15-244
Vormehr M, Schrörs B, Boegel S et al (2015) Mutanome engineered RNA immunotherapy: towards patient-centered tumor vaccination. J Immunol Res 2015:595363. https://doi.org/10.1155/2015/595363
Kim S, Scheffler K, Halpern AL et al (2018) Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 15(8):591–594. https://doi.org/10.1038/s41592-018-0051-x
Cibulskis K, Lawrence MS, Carter SL et al (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3):213–219. https://doi.org/10.1038/nbt.2514
Poplin R, Chang P-C, Alexander D et al (2018) A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol 36(10):983–987. https://doi.org/10.1038/nbt.4235
Kawaguchi S, Higasa K, Shimizu M et al (2017) HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum Mutat 38(7):788–797. https://doi.org/10.1002/humu.23230
Boegel S, Löwer M, Schäfer M et al (2012) HLA typing from RNA-Seq sequence reads. Genome Med 4(12):102. https://doi.org/10.1186/gm403
Jurtz V, Paul S, Andreatta M et al (2017) NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 199(9):3360–3368. https://doi.org/10.4049/jimmunol.1700893
Bjerregaard A-M, Nielsen M, Jurtz V et al (2017) An analysis of natural T cell responses to predicted tumor neoepitopes. Front Immunol 8:1566. https://doi.org/10.3389/fimmu.2017.01566
Ghorani E, Rosenthal R, McGranahan N et al (2018) Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann Oncol 29(1):271–279. https://doi.org/10.1093/annonc/mdx687
Duan F, Duitama J, Al Seesi S et al (2014) Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med 211(11):2231–2248. https://doi.org/10.1084/jem.20141308
Karosiene E, Rasmussen M, Blicher T et al (2013) NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 65(10):711–724. https://doi.org/10.1007/s00251-013-0720-y
Abelin JG, Keskin DB, Sarkizova S et al (2017) Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46(2):315–326. https://doi.org/10.1016/j.immuni.2017.02.007
Vang YS, Xie X (2017) HLA class I binding prediction via convolutional neural networks. Bioinformatics 33(17):2658–2665. https://doi.org/10.1093/bioinformatics/btx264
Liu Z, Cui Y, Xiong Z et al (2019) DeepSeqPan, a novel deep convolutional neural network model for pan-specific class I HLA-peptide binding affinity prediction. Sci Rep 9(1):794. https://doi.org/10.1038/s41598-018-37214-1
Woodsworth DJ, Castellarin M, Holt RA (2013) Sequence analysis of T-cell repertoires in health and disease. Genome Med 5(10):98. https://doi.org/10.1186/gm502
Wieland A, Kamphorst AO, Adsay NV et al (2018) T cell receptor sequencing of activated CD8 T cells in the blood identifies tumor-infiltrating clones that expand after PD-1 therapy and radiation in a melanoma patient. Cancer Immunol Immunother 67(11):1767–1776. https://doi.org/10.1007/s00262-018-2228-7
Lin K-R, Pang D-M, Jin Y-B et al (2018) Circulating CD8+ T-cell repertoires reveal the biological characteristics of tumors and clinical responses to chemotherapy in breast cancer patients. Cancer Immunol Immunother 67(11):1743–1752. https://doi.org/10.1007/s00262-018-2213-1
Jin Y-B, Luo W, Zhang G-Y et al (2018) TCR repertoire profiling of tumors, adjacent normal tissues, and peripheral blood predicts survival in nasopharyngeal carcinoma. Cancer Immunol Immunother 67(11):1719–1730. https://doi.org/10.1007/s00262-018-2237-6
Rosati E, Dowds CM, Liaskou E et al (2017) Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 17(1):61. https://doi.org/10.1186/s12896-017-0379-9
Klausen MS, Anderson MV, Jespersen MC et al (2015) LYRA, a webserver for lymphocyte receptor structural modeling. Nucleic Acids Res 43(W1):W349–W355. https://doi.org/10.1093/nar/gkv535
Jurtz VI, Jessen LE, Bentzen AK et al (2018) NetTCR: sequence-based prediction of TCR binding to peptide-MHC complexes using convolutional neural networks. Preprint available on bioRxiv. https://doi.org/10.1101/433706
Han A, Glanville J, Hansmann L et al (2014) Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol 32(7):684–692. https://doi.org/10.1038/nbt.2938
Friedman AA, Letai A, Fisher DE et al (2015) Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 15(12):747–756. https://doi.org/10.1038/nrc4015
Petitprez F, Sun C-M, Lacroix L et al (2018) Quantitative analyses of the tumor microenvironment composition and orientation in the era of precision medicine. Front Oncol 8:390. https://doi.org/10.3389/fonc.2018.00390
Sturm G, Finotello F, Petitprez F et al (2019) Comprehensive evaluation of computational cell-type quantification methods for immuno-oncology. Bioinformatics 35(14):436–445. https://doi.org/10.1093/bioinformatics/btz363
Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18(1):220. https://doi.org/10.1186/s13059-017-1349-1
Becht E, Giraldo NA, Lacroix L et al (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 17(1):218. https://doi.org/10.1186/s13059-016-1070-5
Finotello F, Trajanoski Z (2018) Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother 67(7):1031–1040. https://doi.org/10.1007/s00262-018-2150-z
Newman AM, Liu CL, Green MR et al (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5):453–457. https://doi.org/10.1038/nmeth.3337
Finotello F, Mayer C, Plattner C et al (2019) Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 11(1):34. https://doi.org/10.1186/s13073-019-0638-6
Li B, Severson E, Pignon J-C et al (2016) Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol 17(1):174. https://doi.org/10.1186/s13059-016-1028-7
Forbes SA, Beare D, Gunasekaran P et al (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43(Database issue):D805–D811. https://doi.org/10.1093/nar/gku1075
Wala JA, Bandopadhayay P, Greenwald NF et al (2018) SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res 28(4):581–591. https://doi.org/10.1101/gr.221028.117
Rausch T, Zichner T, Schlattl A et al (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. https://doi.org/10.1093/bioinformatics/bts378
Shen S, Park JW, Huang J et al (2012) MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40(8):e61. https://doi.org/10.1093/nar/gkr1291
Rogers MF, Thomas J, Reddy AS et al (2012) SpliceGrapher: detecting patterns of alternative splicing from RNA-Seq data in the context of gene models and EST data. Genome Biol 13(1):R4. https://doi.org/10.1186/gb-2012-13-1-r4
Bjerregaard A-M, Nielsen M, Hadrup SR et al (2017) MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother 66(9):1123–1130. https://doi.org/10.1007/s00262-017-2001-3
Kim S, Kim HS, Kim E et al (2018) Neopepsee: accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann Oncol 29(4):1030–1036. https://doi.org/10.1093/annonc/mdy022
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Holtsträter, C., Schrörs, B., Bukur, T., Löwer, M. (2020). Bioinformatics for Cancer Immunotherapy. In: Boegel, S. (eds) Bioinformatics for Cancer Immunotherapy. Methods in Molecular Biology, vol 2120. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0327-7_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0327-7_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0326-0
Online ISBN: 978-1-0716-0327-7
eBook Packages: Springer Protocols