Abstract
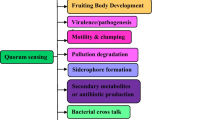
Quorum sensing (QS), a microbial cell-to-cell communication process, dynamically regulates a variety of metabolism and physiological activities. In this review, we provide an update on QS applications based on autoinducer molecules including acyl-homoserine lactones (AHLs), auto-inducing peptides (AIPs), autoinducer 2 (AI-2) and indole in population-level control of bacteria, and highlight the potential in developing novel clinical therapies. We summarize the development in the combination of various genetic circuits such as genetic oscillators, toggle switches and logic gates with AHL-based QS devices in Gram-negative bacteria. An overview is then offered to the state-of-the-art of much less researched applications of AIP-based QS devices with Gram-positive bacteria, followed by a review of the applications of AI-2 and indole based QS for interspecies communication among microbial communities. Building on these general-purpose QS applications, we highlight the disruptions and manipulations of QS devices as potential clinical therapies for diseases caused by biofilm formation, antibiotic resistance and the phage invasion. The last part of reviewed literature is dedicated to mathematical modelling for QS applications. Finally, the key challenges and future perspectives of QS applications in monoclonal synthetic biology and synthetic ecology are discussed.









Similar content being viewed by others
Abbreviations
- [A]:
-
Intracellular AHL concentration (mM)
- [C]:
-
Intracellular CI protein concentration (mM)
- [E]:
-
Intracellular CcdB protein concentration (mM)
- [L]:
-
Intracellular LacR concentration (mM)
- [LuxR]:
-
Intracellular LuxR concentration (mM)
- [R]:
-
Intracellular AHL/LuxR complex concentration (mM)
- N :
-
The cell density (CFU ml−1)
- N m :
-
The maximum cell density (CFU ml−1)
- F pfk :
-
The fractional Pfk-1 activity (U/mg)
- Kd:
-
The cumulative dissociation constant
- X :
-
Biomass concentration (g L−1)
- n1, n2 :
-
Transcription factor cooperativity/multimerization
- α C :
-
CI protein synthesis rate constant (μM min−1)
- αL1, αL2 :
-
LacR protein synthesis rate constants (μM min−1)
- β C :
-
CI repression coefficient (mM)
- β L :
-
LacR repression coefficient (mM)
- d :
-
Cell death rate (nM−1 h−1)
- dA, dE :
-
AHL and CcdB protein decay constant (min−1)
- d C :
-
CI protein decay constant (min−1)
- dL, dR :
-
LacR and LuxR–AHL complex decay constants (min−1)
- k :
-
Growth rate (h−1)
- k E :
-
CcdB protein production rate constant (h−1)
- v A :
-
AHL production rate constant (nM mL h−1)
- θ R :
-
LuxR/AHL activation coefficient (mM)
- ρ R :
-
LuxR/AHL dimerization constant (μM−3 min−1)
References
Homer CM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, Moresco JJ, Toffaletti D, Upadhya R, Caradonna I, Petnic S, Pessino V, Cuomo CA, Lodge JK, Perfect J, Yates JR 3rd, Nielsen K, Craik CS, Madhani HD (2016) Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe 19:849–864
Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S (2017) Communication between viruses guides lysis-lysogeny decisions. Nature 541:488–493
Melissa B, Miller BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55(1):165–199
Whiteley M, Diggle SP, Greenberg EP (2017) Progress in and promise of bacterial quorum sensing research. Nature 551:313–320
Chubukov V, Gerosa L, Kochanowski K, Sauer U (2014) Coordination of microbial metabolism. Nat Rev Microbiol 12:327–340
Holtz WJ, Keasling JD (2010) Engineering static and dynamic control of synthetic pathways. Cell 140:19–23
Venayak N, Anesiadis N, Cluett WR, Mahadevan R (2015) Engineering metabolism through dynamic control. Curr Opin Biotechnol 34:142–152
Gupta A, Reizman IM, Reisch CR, Prather KL (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35:273–279
Liu D, Evans T, Zhang F (2015) Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng 31:35–43
Soma Y, Hanai T (2015) Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 30:7–15
Doong SJ, Gupta A, Prather KLJ (2018) Layered dynamic regulation for improving metabolic pathway productivity in escherichia coli. Proc Natl Acad Sci USA 115(12):2964–2969
Liu X, Li XB, Jiang J, Liu ZN, Qiao B, Li FF, Cheng JS, Sun X, Yuan YJ, Qiao J (2018) Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng 47:243–253
Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403(6767):339–342
Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403:335–338
Win MN, Smolke CD (2007) A modular and extensible rna-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA 104:14283–14288
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27(8):753–759
Danino T, Mondragon-Palomino O, Tsimring L, Hasty J (2010) A synchronized quorum of genetic clocks. Nature 463:326–330
Papenfort K, Bassler BL (2016) Quorum sensing signal–response systems in gram-negative bacteria. Nat Rev Microbiol 14:576–588
Diggle SP, Griffin AS, Campbell GS, West SA (2007) Cooperation and conflict in quorum-sensing bacterial populations. Nature 450:411–414
Modi SR, Collins JJ, Relman DA (2014) Antibiotics and the gut microbiota. J Clin Investig 124:4212–4218
Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575
Yan J, Bassler BL (2019) Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26:15–21
Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK (2012) Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat Commun 3:613–620
Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626
Kalia VC (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31:224–245
Ma S, Zhou Z (2017) Recent advances in the discovery of pqsd inhibitors as antimicrobial agents. ChemMedChem 12:420–425
Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB (2015) Manipulation of the quorum sensing signal ai-2 affects the antibiotic-treated gut microbiota. Cell Rep 10:1861–1871. https://doi.org/10.1016/j.celrep.2015.02.049
Abedon ST (2012) Spatial vulnerability: bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses 4:663–687
Semenova E, Severinov K (2016) Come together: Crispr-cas immunity senses the quorum. Mol Cell 64:1013–1015
Hawver LA, Jung SA, Ng WL (2016) Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40:738–752
Monnet V, Gardan R (2015) Quorum-sensing regulators in gram-positive bacteria: ‘Cherchez le peptide’. Mol Microbiol 97:181–184
Monnet V, Juillard V, Gardan R (2014) Peptide conversations in gram-positive bacteria. Crit Rev Microbiol 42:339–351
Lee JH, Wood TK, Lee J (2015) Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol 23:707–718
Hmelo LR (2017) Quorum sensing in marine microbial environments. Ann Rev Mar Sci 9:257–281
Zhou L, Zhang LH, Camara M, He YW (2017) The dsf family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol 25:293–303
Xu P (2017) Production of chemicals using dynamic control of metabolic fluxes. Curr Opin Biotechnol 53:12–19
Shong J, Diaz MRJ, Collins CH (2012) Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol 23:798–802
Song H, Ding MZ, Jia XQ, Ma Q, Yuan YJ (2014) Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev 43:6954–6981
Dolinsek J, Goldschmidt F, Johnson DR (2016) Synthetic microbial ecology and the dynamic interplay between microbial genotypes. FEMS Microbiol Rev 40:961–979
Asfahl KL, Schuster M, Gibbs K (2017) Social interactions in bacterial cell–cell signaling. FEMS Microbiol Rev 41:92–107
Choudhary S, Schmidt-Dannert C (2010) Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol 86:1267–1279
Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L (2017) Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740–755
Mukherjee S, Bassler BL (2019) Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17:371–382
Goryachev AB (2011) Understanding bacterial cell–cell communication with computational modeling. Chem Rev 111:238–250
Hense BA, Schuster M (2015) Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev 79:153–169
Pérez-Velázquez J, Gölgeli M, García-Contreras R (2016) Mathematical modelling of bacterial quorum sensing: a review. Bull Math Biol 78:1–55
Boyer M, Wisniewski-Dye F (2009) Cell–cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol 70:1–19
An JH, Goo E, Kim H, Seo YS, Hwang I (2014) Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci USA 111:14912–14917
Goo E, Majerczyk CD, An JH, Chandler JR, Seo YS, Ham H, Lim JY, Kim H, Lee B, Jang MS, Greenberg EP, Hwang I (2012) Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci USA 109:19775–19780
You L, Cox RS 3rd, Weiss R, Arnold FH (2004) Programmed population control by cell–cell communication and regulated killing. Nature 428:868–871
Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L (2008) A synthetic escherichia coli predator–prey ecosystem. Mol Syst Biol 4:187–194
Wu F, Lopatkin AJ, Needs DA, Lee CT, Mukherjee S, You L (2019) A unifying framework for interpreting and predicting mutualistic systems. Nat Commun 10:242–251
Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN (2016) Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536:81–85
Mays ZJ, Nair NU (2018) Synthetic biology in probiotic lactic acid bacteria: at the frontier of living therapeutics. Curr Opin Biotechnol 53:224–231
Scott SR, Din MO, Bittihn P, Xiong L, Tsimring LS, Hasty J (2017) A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol 2:17083–17091
Woods ML, Leon M, Perezcarrasco R, Barnes CP (2016) A statistical approach reveals designs for the most robust stochastic gene oscillators. ACS Synth Biol 5:459–470
Jörg DJ, Morelli LG, Jülicher F (2018) Chemical event chain model of coupled genetic oscillators. Phys Rev E 97(3–1):1–11 (032409)
Potvin-Trottier L, Lord ND, Vinnicombe G, Paulsson J (2016) Synchronous long-term oscillations in a synthetic gene circuit. Nature 538:514–517
Tu BP, Mcknight SL (2006) Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol 7:696–701
Mondragon-Palomino O, Danino T, Selimkhanov J, Tsimring L, Hasty J (2011) Entrainment of a population of synthetic genetic oscillators. Science 333:1315–1319
Purcell O, Savery NJ, Grierson CS, Bernardo MD (2010) A comparative analysis of synthetic genetic oscillators. J R Soc Interface 7(52):1503–1524
Barkai N, Leibler S (2000) Circadian clocks limited by noise. Nature 403(6767):267–268
Hasty J, Dolnik M, Rottschäfer V, Collins JJ (2002) Synthetic gene network for entraining and amplifying cellular oscillations. Phys Rev Lett 88(14):1–4 (148101)
Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J (2008) A fast, robust and tunable synthetic gene oscillator. Nature 456:516–519
Kellogg R, Tay S (2015) Noise facilitates transcriptional control under dynamic inputs. Cell 160:381–392
Tokuda IT, Okamoto A, Matsumura R, Takumi T, Akashi M (2017) Potential contribution of tandem circadian enhancers to nonlinear oscillations in clock gene expression. Mol Biol Cell 28(17):2333–2342
Wintermute EH, Silver PA (2010) Dynamics in the mixed microbial concourse. Genes Dev 24:2603–2614
Xavier JB (2011) Social interaction in synthetic and natural microbial communities. Mol Syst Biol 7:483–493
Chuang JS (2012) Engineering multicellular traits in synthetic microbial populations. Curr Opin Chem Biol 16:370–378
Scott SR, Hasty J (2016) Quorum sensing communication modules for microbial consortia. ACS Synth Biol 5:969–977
Mcmillen D, Kopell N, Hasty J, Collins JJ (2002) Synchronizing genetic relaxation oscillators by intercell signaling. Proc Natl Acad Sci USA 99:679–684
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Slager J, Kjos M, Attaiech L, Veening JW (2014) Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157:395–406
Baumgart L, Mather W, Hasty J (2017) Synchronized DNA cycling across a bacterial population. Nat Genet 49(8):1282–1285
Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J (2012) A sensing array of radically coupled genetic/’biopixels/’. Nature 481:39–44
Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J (2014) Rapid and tunable post-translational coupling of genetic circuits. Nature 508:387–391
Levchenko I, Seidel M, Sauer RT, Baker TA (2000) A specificity-enhancing factor for the clpxp degradation machine. Science 289:2354–2356
Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR (2015) Emergent genetic oscillations in a synthetic microbial consortium. Science 349(6251):986–989
Feist AM, Palsson BO (2010) The biomass objective function. Curr Opin Microbiol 13:344–349
Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T (2014) Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng 23:175–184
Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ (2004) Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA 101:8414–8419
Anesiadis N, Cluett WR, Mahadevan R (2008) Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng 10:255–266
Anesiadis N, Kobayashi H, Cluett WR, Mahadevan R (2013) Analysis and design of a genetic circuit for dynamic metabolic engineering. ACS Synth Biol 2:442–452
Honjo H, Iwasaki K, Soma Y, Tsuruno K, Hamada H, Hanai T (2019) Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab Eng 55:268–275
Wang EX, Liu Y, Ma Q, Dong XT, Ding MZ, Yuan YJ (2019) Synthetic cell–cell communication in a three-species consortium for one-step vitamin c fermentation. Biotechnol Lett 41:951–961
Tsoi R, Dai Z, You L (2019) Emerging strategies for engineering microbial communities. Biotechnol Adv 37(6):1–9 (107372)
Nandagopal N, Elowitz MB (2011) Synthetic biology: integrated gene circuits. Science 333:1244–1248
Benenson Y (2012) Biomolecular computing systems: principles, progress and potential. Nat Rev Genet 13(7):455–468
Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA (2012) Genetic programs constructed from layered logic gates in single cells. Nature 491:249–253
Baig H, Madsen J (2017) Simulation approach for timing analysis of genetic logic circuits. ACS Synth Biol 6(7):1169–1179
Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA (2016) Genetic circuit design automation. Science 352(aac7341):1–11
Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420:224–230
Li Z, Rosenbaum MA, Venkataraman A, Tam TK, Katz E, Angenent LT (2011) Bacteria-based and logic gate: a decision-making and self-powered biosensor. Chem Commun 47:3060–3062
Arugula MA, Shroff N, Katz E, He Z (2012) Molecular and logic gate based on bacterial anaerobic respiration. Chem Commun 48:10174–10176
Tamsir A, Tabor JJ, Voigt CA (2010) Robust multicellular computing using genetically encoded nor gates and chemical /`wires/’. Nature 469:212–215
Yokobayashi Y, Weiss R, Arnold F (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci USA 99:16587–16591
Miyamoto T, Razavi S, Derose R, Inoue T (2013) Synthesizing biomolecule-based boolean logic gates. ACS Synth Biol 2:72–82
Shong J, Collins CH (2014) Quorum sensing-modulated and-gate promoters control gene expression in response to a combination of endogenous and exogenous signals. ACS Synth Biol 3:238–246
Hu Y, Yang Y, Katz E, Song H (2015) Programming the quorum sensing-based and gate in shewanella oneidensis for logic gated-microbial fuel cells. Chem Commun 51:4184–4187
He X, Chen Y, Liang Q, Qi Q (2017) An autoinduced and-gate controlling metabolic pathway dynamically in response to microbial communities and cell physiological state. ACS Synth Biol 6:463–470
Takayanagi Y, Tanaka K, Takahashi H (1994) Structure of the 5′ upstream region and the regulation of the rpos gene of Escherichia coli. Mol Genet Genomics 243:525–531
Keller L, Surette MG (2006) Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol 4:249–258
Papadimitriou K, Alegria A, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zuniga M, Tsakalidou E, Kok J (2016) Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80:837–890
Cook LC, Federle MJ (2014) Peptide pheromone signaling in streptococcus and enterococcus. FEMS Microbiol Rev 38:473–492
Marchand N, Collins CH (2016) Synthetic quorum sensing and cell–cell communication in gram-positive bacillus megaterium. ACS Synth Biol 5:597–606
Cooksley CM, Davis IJ, Winzer K, Chan WC, Peck MW, Minton PN (2010) Regulation of neurotoxin production and sporulation by a putative agrbd signaling system in proteolytic clostridium botulinum. Appl Environ Microbiol 76:4448–4460
Steiner E, Scott J, Minton NP, Winzer K (2012) An agr quorum sensing system that regulates granulose formation and sporulation in clostridium acetobutylicum. Appl Environ Microbiol 78:1113–1122
Vivant AL, Garmyn D, Gal L, Piveteau P (2014) The agr communication system provides a benefit to the populations of listeria monocytogenes in soil. Front Cell Infect Microbiol 4(160):1–7
Ma M, Li J, Mcclane BA (2015) Structure-function analysis of peptide signaling in the clostridium perfringens agr-like quorum sensing system. J Bacteriol 197:1807–1818
Yang T, Talgan Y, Paharik AE, Horswill AR, Blackwell HE (2016) Structure-function analyses of a staphylococcus epidermidis autoinducing peptide reveals motifs critical for agrc-type receptor modulation. ACS Chem Biol 11:1982–1991
Yu Q, Lepp D, Mehdizadeh GI, Wu T, Zhou H, Yin X, Yu H, Prescott JF, Nie SP, Xie MY (2017) The agr-like quorum sensing system is required for necrotic enteritis pathogenesis in poultry caused by clostridium perfringens. Infect Immun 85:e00975-16
Fontaine L, Boutry C, Guédon E, Guillot A, Ibrahim M, Grossiord B, Hols P (2007) Quorum-sensing regulation of the production of blp bacteriocins in Streptococcus thermophilus. J Bacteriol 189:7195–7205
Håvarstein LS (2010) Increasing competence in the genus streptococcus. Mol Microbiol 78:541–544
Mirouze N, Bergé MA, Soulet AL, Mortierbarrière I, Quentin Y, Fichant G, Granadel C, Noirotgros MF, Noirot P, Polard P (2013) Direct involvement of dpra, the transformation-dedicated reca loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci USA 110:352–361
Reck M, Tomasch J, Wagner-Dobler I (2015) The alternative sigma factor sigx controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in streptococcus mutans. PLoS Genet 11:e1005353
Moreno-GãmS Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening JW (2017) Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8:854–865
Gallego del Sol F, Marina A (2013) Structural basis of rap phosphatase inhibition by phr peptides. PLoS Biol 11:e1001511
Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, Lemy C, Aumont-Nicaise M, Deutscher J, Gohar M, Nessler S (2011) A cell–cell communication system regulates protease production during sporulation in bacteria of the bacillus cereus group. Mol Microbiol 82:619–633
Zouhir S, Perchat S, Nicaise M, Perez J, Guimaraes B, Lereclus D, Nessler S (2013) Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor nprr. Nucleic Acids Res 41:7920–7933
Grenha R, Slamti L, Nicaise M, Refes Y, Lereclus D, Nessler S (2013) Structural basis for the activation mechanism of the plcr virulence regulator by the quorum-sensing signal peptide papr. Proc Natl Acad Sci USA 110:1047–1052
Slamti L, Perchat S, Huillet E, Lereclus D (2014) Quorum sensing in bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins 6:2239–2255
Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu WS, Dunny GM (2017) Mechanisms of peptide sex pheromone regulation of conjugation in enterococcus faecalis. Microbiologyopen 6(e492):1–13
Mashburn-Warren L, Morrison DA, Federle MJ (2010) A novel double-tryptophan peptide pheromone controls competence in streptococcus spp. via an rgg regulator. Mol Microbiol 78:589–606
Fleuchot B, Guillot A, Mézange C, Besset C, Chambellon E, Monnet V, Gardan R (2013) Rgg-associated shp signaling peptides mediate cross-talk in streptococci. PLoS One 8:e66042
Parashar V, Aggarwal C, Federle MJ, Neiditch MB (2015) Rgg protein structure-function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci USA 112:5177–5182
Haustenne L, Bastin G, Hols P, Fontaine L (2015) Modeling of the comrs signaling pathway reveals the limiting factors controlling competence in Streptococcus thermophilus. Front Microbiol 6(1413):1–20
Talagas A, Fontaine L, Ledesma-García L, Mignolet J, Li de la Sierra-Gallay I, Lazar N, Aumont-Nicaise M, Federle MJ, Prehna G, Hols P, Nessler S (2016) Structural insights into streptococcal competence regulation by the cell-to-cell communication system comrs. PLoS Pathog 12:e1005980
Underhill SAM, Shields RC, Kaspar JR, Haider M, Burne RA, Hagen SJ (2018) Intracellular signaling through the comrs system in streptococcus mutans genetic competence. mSphere 3(5):e00444-18
Garcíacontreras R, Nuñezlópez L, Jassochávez R, Kwan BW, Belmont JA, Rangelvega A, Maeda T, Wood TK (2014) Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J 9(1):115–125
Bassler BL, Wright M, Showalter RE, Silverman MR (1993) Intercellular signalling in vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 9:773–786
Chen X (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549
Sedlmayer F, Hell D, Muller M, Auslander D, Fussenegger M (2018) Designer cells programming quorum-sensing interference with microbes. Nat Commun 9(1822):1–13
Lee JH, Lee J (2010) Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444
Wang D, Ding X, Rather PN (2001) Indole can act as an extracellular signal in Escherichia coli. J Bacteriol 183:4210–4216
Bassler BL, Losick R (2006) Bacterially speaking. Cell 125:237–246
De Keersmaecker SC, Sonck K, Vanderleyden J (2006) Let luxs speak up in ai-2 signaling. Trends Microbiol 14:114–119
Pereira CS, Thompson JA, Xavier KB (2013) Ai-2-mediated signalling in bacteria. FEMS Microbiol Rev 37:156–181
Xavier KB, Bassler BL (2005) Regulation of uptake and processing of the quorum-sensing autoinducer ai-2 in Escherichia coli. J Bacteriol 187:238–248
Xavier KB, Bassler BL (2005) Interference with ai-2-mediated bacterial cell–cell communication. Nature 437:750–753
Armbruster CE, Hong W, Pang B, Weimer KED, Juneau RA, Turner J, Swords WE (2010) Indirect pathogenicity of haemophilus influenzae and moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. Mbio 1:119–121
Newton WA, Snell EE (1965) Formation and interrelationships of tryptophanase and tryptophan synthetases in Escherichia coli. J Bacteriol 89:355–364
Lee J, Jayaraman A, Wood TK (2007) Indole is an inter-species biofilm signal mediated by sdia. BMC Microbiol 7:1–15
Chant EL, Summers DK (2007) Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol 63:35–43
Lee HH, Molla MN, Cantor CR, Collins JJ (2010) Bacterial charity work leads to population-wide resistance. Nature 467:82–85
Jintae L, Can A, Cirillo SLG, Cirillo JD, Wood TK (2010) Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol 2:75–90
Vega NM, Allison KR, Khalil AS, Collins JJ (2011) Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431
Kim YG, Lee JH, Cho MH, Lee J (2011) Indole and 3-indolylacetonitrile inhibit spore maturation in paenibacillus alvei. BMC Microbiol 11:119
Catalin C, Field CM, Silvia PF, Keyser UF, Summers DK (2012) Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta Biomembr 1818:1590–1594
Yee DC, Maynard JA, Wood TK (1998) Rhizoremediation of trichloroethylene by a recombinant, root-colonizing pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl Environ Microbiol 64:112–118
Han TH, Cho MH, Lee J (2014) Indole oxidation enhances electricity production in an E. coli-catalyzed microbial fuel cell. Biotechnol Bioprocess E 19:126–131
Lee JH, Kim YG, Kim CJ, Lee JC, Cho MH, Lee J (2012) Indole-3-acetaldehyde from Rhodococcus sp. Bfi 332 inhibits Escherichia coli o157:h7 biofilm formation. Appl Microbiol Biotechnol 96:1071–1078
Lee JH, Cho HS, Kim Y, Kim JA, Banskota S, Cho MH, Lee J (2013) Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl Microbiol Biotechnol 97:4543–4552
Lee JH, Kim YG, Baek KH, Cho MH, Lee J (2015) The multifaceted roles of the interspecies signalling molecule indole in Agrobacterium tumefaciens. Environ Microbiol 17:1234–1244
Chu W, Zere TR, Weber MM, Wood TK, Whiteley M, Hidalgo-Romano B Jr, Valenzuela E, Mclean RJ (2012) Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl Environ Microbiol 78:411–419
Lee JH, Kim YG, Kim M, Kim E, Choi H, Kim Y, Lee J (2017) Indole-associated predator–prey interactions between the nematode Caenorhabditis elegans and bacteria. Environ Microbiol 19:1776–1790
Tomberlin JK, Crippen TL, Wu G, Griffin AS, Wood TK, Kilner RM (2017) Indole: an evolutionarily conserved influencer of behavior across kingdoms. BioEssays 39(1600203):1–12
Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ (1987) Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464
Van AH, Van DP, Coenye T (2014) Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333
Lebeaux D, Ghigo JM, Beloin C (2014) Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543
Parsek MR, Greenberg EP (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33
Deng YY, Wu JE, Tao F, Zhang LH (2011) Listening to a new language: Dsf-based quorum sensing in gram-negative bacteria. Chem Rev 111:160–173
Lebeer S, Claes IJ, Verhoeven TL, Shen C, Lambrichts I, Ceuppens JL, Vanderleyden J, De Keersmaecker SC (2008) Impact of luxs and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus gg. Appl Environ Microbiol 74:4711–4718
Sun Z, He X, Brancaccio VF, Yuan J, Riedel CU (2014) Bifidobacteria exhibit luxs-dependent autoinducer 2 activity and biofilm formation. PLoS One 9:e88260
Li H, Li X, Wang Z, Fu Y, Ai Q, Dong Y, Yu J (2015) Autoinducer-2 regulates pseudomonas aeruginosa pao1 biofilm formation and virulence production in a dose-dependent manner. BMC Microbiol 15:1–8
Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K (2015) Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379
Xue T, Ni J, Shang F, Chen X, Zhang M (2015) Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in staphylococcus epidermidis rp62a. Microbes Infect 17:345–352
Laganenka L, Colin R, Sourjik V (2016) Corrigendum: chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun 7:13979
Papenfort K, Silpe JE, Schramma KR, Cong JP, Seyedsayamdost MR, Bassler BL (2017) A vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat Chem Biol 13:551–557
Liu L, Wu R, Zhang J, Shang N, Li P (2017) D-ribose interferes with quorum sensing to inhibit biofilm formation of Lactobacillus paraplantarum l-zs9. Front Microbiol 8:1860
Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM (2008) Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68:75–86
Dean SN, Chung MC, van Hoek ML (2015) Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol 81:7057–7066
An SQ, Tang JL (2018) Diffusible signal factor signaling regulates multiple functions in the opportunistic pathogen Stenotrophomonas maltophilia. BMC Res Notes 11:569–575
Krzyzek P, Gosciniak G (2018) A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of Helicobacter pylori. Turk J Gastroenterol 29:7–13
Deng YY, Lim A, Lee J, Chen SH, An SW, Dong YH, Zhang LH (2014) Diffusible signal factor (dsf) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol 14:51–59
Barel V, Chalupowicz L, Barash I, Sharabani G, Reuven M, Dror O, Burdman S, Manulis-Sasson S (2015) Virulence and in planta movement of Xanthomonas hortorum pv. Pelargonii are affected by the diffusible signal factor (dsf)-dependent quorum sensing system. Mol Plant Pathol 16:710–723
Dow JM (2017) Diffusible signal factor-dependent quorum sensing in pathogenic bacteria and its exploitation for disease control. J Appl Microbiol 122:2–11
Allen RC, Popat R, Diggle SP, Brown SP (2014) Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol 12:300–308
Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH (2001) Quenching quorum-sensing-dependent bacterial infection by an n-acyl homoserine lactonase. Nature 411:813–817
Shen G, Rajan R, Zhu J, Bell CE, Pei D (2006) Design and synthesis of substrate and intermediate analogue inhibitors of s-ribosylhomocysteinase. J Med Chem 49:3003–3011
Zhang M, Jiao X, Hu Y, Sun L (2009) Attenuation of edwardsiella tarda virulence by small peptides that interfere with luxs/autoinducer type 2 quorum sensing. Appl Environ Microbiol 75:3882–3890
Ni N, Li M, Wang J, Wang B (2009) Inhibitors and antagonists of bacterial quorum sensing. Med Res Rev 29:65–124
Brackman G, Defoirdt T, Miyamoto C, Bossier P, Calenbergh SV, Nelis H, Coenye T (2008) Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in vibriospp. By decreasing the DNA-binding activity of the quorum sensing response regulator luxr. BMC Microbiol 8:149
Brackman G, Cos P, Maes L, Nelis HJ, Coenye T (2011) Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother 55:2655–2661
Christensen QH, Grove TL, Booker SJ, Greenberg EP (2013) A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci USA 110:13815–13820
O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL (2013) A quorum-sensing inhibitor blocks pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA 110:17981–17986
Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A (2014) Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog 10:e1004321
Ouyang J, Sun F, Feng W, Sun Y, Qiu X, Xiong L, Liu Y, Chen Y (2016) Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol 120:966–974
Zerfaß C, Chen J, Soyer OS (2018) Engineering microbial communities using thermodynamic principles and electrical interfaces. Curr Opin Biotechnol 50:121–127
McCardell RD, Huang S, Green LN, Murray RM (2017) Control of bacterial population density with population feedback and molecular sequestration. https://doi.org/10.1101/225045
Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SS, Poh CL, Chang MW (2011) Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol 7(521):1–11
Gupta S, Bram EE, Weiss R (2013) Genetically programmable pathogen sense and destroy. ACS Synth Biol 2:715–723
Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW (2014) Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol 3:228–237
Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW (2017) Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun 8:15028
Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J (2018) Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156
Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L (2017) Salt-responsive gut commensal modulates th17 axis and disease. Nature 551:585–589
Crotty MP, Jackson PJ (2017) Terminal room disinfection: how much betr can it get? Lancet 389:765–766
Routy B, Le CE, Derosa L, Cpm D, Alou MT, DaillãRe R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP (2018) Gut microbiome influences efficacy of pd-1-based immunotherapy against epithelial tumors. Science 359:91–97
Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, Kumar MV, Gewirtz AT (2018) Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring il-22-mediated colonic health. Cell Host Microbe 23:41–53
Sun Z, Grimm V, Riedel CU (2015) Ai-2 to the rescue against antibiotic-induced intestinal dysbiosis? Trends Microbiol 23:327–328
Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G (2012) Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336:1325–1329
Dandekar AA, Greenberg EP (2012) Bacterial quorum sensing and metabolic incentives to cooperate. Science 338:264–266
Bongaerts GP, Severijnen RS (2016) A reassessment of the propatria study and its implications for probiotic therapy. Nat Biotechnol 34:55–63
Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, Destefano J, Meier MF, Muegge BD (2014) Persistent gut microbiota immaturity in malnourished bangladeshi children. Nature 510:417–421
Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL Jr, Petri WA, Haque R, Ahmed T, Gordon JI (2014) Members of the human gut microbiota involved in recovery from vibrio cholerae infection. Nature 515:423–426
Daniel R, Rubens JR, Sarpeshkar R, Lu TK (2013) Synthetic analog computation in living cells. Nature 497:619–623
Bivar Xavier K (2018) Bacterial interspecies quorum sensing in the mammalian gut microbiota. C R Biol 341:297–299
Gilmore MS, Lebreton F, Van SW (2013) Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16
Borrero J, Chen Y, Dunny GM, Kaznessis YN (2015) Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth Biol 4:299–306
Arias CA, Murray BE (2012) The rise of the enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278
Coyte KZ, Rakoff-Nahoum S (2019) Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol 29:538–544
Li Q, Ren Y, Fu X (2019) Inter-kingdom signaling between gut microbiota and their host. Cell Mol Life Sci 76:2383–2389
Mirzaei MK, Maurice CF (2017) Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15:397–408
Bondydenomy J, Pawluk A, Maxwell KL, Davidson AR (2013) Bacteriophage genes that inactivate the crispr/cas bacterial immune system. Nature 493:429–432
Patterson AG, Yevstigneyeva MS, Fineran PC (2017) Regulation of crispr-cas adaptive immune systems. Curr Opin Microbiol 37:1–7
Patterson A, Jackson S, Taylor C, Evans G, Salmond GC, Przybilski R, Staals RJ, Fineran P (2016) Quorum sensing controls adaptive immunity through the regulation of multiple crispr-cas systems. Mol Cell 64:1102–1108
Brophy JA, Voigt CA (2014) Principles of genetic circuit design. Nat Methods 11:508–520
Buescher JM (2012) Global network reorganization during dynamic adaptations of bacillus subtilis metabolism. Science 335:1099–1103
Woolston BM, Edgar S, Stephanopoulos G (2013) Metabolic engineering: past and future. Annu Rev Chem Biomol Eng 4:259–288
Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nat Biotechnol 28:245–248
Meadows AL, Karnik R, Lam H, Forestell S, Snedecor B (2010) Application of dynamic flux balance analysis to an industrial escherichia coli fermentation. Metab Eng 12:150–160
Hill A (1913) The combinations of haemoglobin with oxygen and with carbon monoxide. I. Biochem J 7:577–586
Schmidt SK, Simkins S, Alexander M (1985) Models for the kinetics of biodegradation of organic compounds not supporting growth. Appl Environ Microbiol 50:323–331
English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS (2006) Ever-fluctuating single enzyme molecules: Michaelis-menten equation revisited. Nat Chem Biol 2:87–94
Basu Subhayu, Gerchman Yoram, Collins Cynthia H, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nature 434:1130–1134
Song H, Stephen P, Meagan G, You L (2009) Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol 5:929–935
Kong W, Meldgin DR, Collins JJ, Lu T (2018) Designing microbial consortia with defined social interactions. Nat Chem Biol 14:821–829
Mads K, Elston TC, Blake WJ, Collins JJ (2005) Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6:451–464
Tian T, Burrage K (2006) Stochastic models for regulatory networks of the genetic toggle switch. Proc Natl Acad Sci USA 103:8372–8377
Prindle A, Liu J, Asally M, Ly S, Garciaojalvo J, Süel GM (2015) Ion channels enable electrical communication within bacterial communities. Nature 527:59–63
Popkin G (2017) Bacteria use brainlike bursts of electricity to communicate. Quanta Magazine, New York
Liu J, Zhou J, Wang L, Ma Z, Zhao G, Ge Z, Zhu H, Qiao J (2017) Improving nitrogen source utilization from defatted soybean meal for nisin production by enhancing proteolytic function of Lactococcus lactis f44. Sci Rep 7:6189
Wu H, Song S, Tian K, Zhou D, Wang B, Liu J, Zhu H, Qiao J (2018) A novel small rna s042 increases acid tolerance in Lactococcus lactis f44. Biochem Biophys Res Commun 500:544–549
Song AA, In LLA, Lim SHE, Rahim RA (2017) A review on Lactococcus lactis: from food to factory. Microb Cell Fact 16:55
Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Mao T, Donia MS, Nayfach S, Pollard KS (2017) Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 168:517–526
Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S (2017) The evolution of the host microbiome as an ecosystem on a leash. Nature 548:43–51
Fernandez L, Rodriguez A, Garcia P (2018) Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J 12:1171–1179
Acknowledgements
This study was supported by the National Key Research and Development Project of China (2017YFD0201400), the National Natural Science Foundation of China (31570089, 31170076), and the Funds for Creative Research Groups of China (21621004), Dr. Jianjun Qiao was supported by The New Century Outstanding Talent Support Program, Education Ministry of China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, S., Liu, J., Liu, C. et al. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343 (2020). https://doi.org/10.1007/s00018-019-03326-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03326-8