Abstract
The Up-and-Coming-Scientist section of the current issue of the Journal of Membrane Biology features the invited essay by Dr. Mercedes Alfonso-Prieto, Assistant Professor at the Forschungszentrum Jülich (FZJ), Germany, and the Heinrich-Heine University Düsseldorf, Vogt Institute for Brain Research. Dr. Alfonso-Prieto completed her doctoral degree in chemistry at the Barcelona Science Park, Spain, in 2009, pursued post-doctoral research in computational molecular sciences at Temple University, USA, and then, as a Marie Curie post-doctoral fellow at the University of Barcelona, worked on computations of enzyme reactions and modeling of photoswitchable ligands targeting neuronal receptors. In 2016, she joined the Institute for Advanced Science and the Institute for Computational Biomedicine at the FZJ, where she pursues research on modeling and simulation of chemical senses. The invited essay by Dr. Alfonso-Prieto discusses state-of-the-art modeling of molecular receptors involved in chemical sensing – the senses of taste and smell. These receptors, and computational methods to study them, are the focus of Dr. Alfonso-Prieto’s research. Recently, Dr. Alfonso-Prieto and colleagues have presented a new methodology to predict ligand binding poses for GPCRs, and extensive computations that deciphered the ligand selectivity determinants of bitter taste receptors. These developments inform our current understanding of how taste occurs at the molecular level.
Graphic Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
In contrast with vision or hearing, the senses of taste and smell (or olfaction) involve the detection of chemicals present in the environment and thus are considered as “chemical senses”. They inform about the aroma and flavor of food and beverages and act as warning system for toxic substances. Hence, these two chemical senses strongly affect human well-being, food acceptance and intake (Tepper et al. 2020; Boesveldt and Parma 2021), as well as drug compliance (Pawar and Kumar 2002; Menella et al. 2013), especially in children. In addition, taste and smell impact key brain processes (Sullivan et al. 2015; Boesveldt and de Graaf 2017; Sabiniewicz et al. 2021), such as memory, emotional responses or behavior.
Taste and olfaction impairments strongly affect the quality of life, social interactions and dietary habits (Mainland et al. 2020). Moreover, the loss of taste and smell is a common symptom of Parkinson’s and Alzheimer’s diseases (Robert et al. 2016; Tarakad and Jankovic 2017; Oppo et al. 2020) and has been recently shown to be one of the symptoms of COVID-19 infection (Parma et al. 2020; Gerkin et al. 2021; Pierron et al. 2020). Therefore, taste and olfaction are also clinically relevant.
The molecules responsible for taste and smell (tastants and odorants, respectively) are extremely chemically diverse (Malnic et al. 1999; Meyerhof et al. 2010). Consequently, the human genome contains a large number of membrane proteins dedicated to recognize them (Buck and Axel 1991; Adler et al. 2000; Chandrashekar et al. 2006). In particular, the two largest chemosensory families correspond to olfactory receptors (ORs) and taste 2 (or bitter taste) receptors (TAS2Rs). Although originally identified in nose and mouth, these receptors have been later shown to be expressed also in other parts of the body (Behrens and Meyerhof 2011; Massberg and Hatt 2018; Dalesio et al. 2018; Behrens and Meyerhof 2019). This extraoral and extranasal expression indicates that ORs and TAS2Rs can also play a role in other physiological and pathological processes, besides bitter taste and smell perception, and thus opens the way for new therapeutical interventions (Lee et al. 2019; Di Pizio et al. 2019).
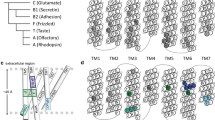
Both TAS2Rs and ORs are G-protein coupled receptors (GPCRs). The GPCR superfamily is the largest in the human genome, with approx. 800 genes, of which half correspond to chemosensory receptors (Venter et al., 2001; Alexander et al., 2019). Within the class A-D system (Fredriksson et al. 2003; Schioth and Fredriksson 2005; Lagerstrom and Schioth 2008), ORs are part of class A, whereas the classification of TAS2Rs is still under debate (as either class A, or class F or a new class T) (Nordstrom et al., 2011; de March et al. 2015; Di Pizio et al. 2016; Munk et al., 2016a). Regardless of their classification, TAS2Rs and ORs share the same topology, with a seven transmembrane (TM) helix bundle (see Fig. 1). Bitter tastants and odorants are recognized by their corresponding receptor by binding in a cavity located in the extracellular part of the TM bundle. Ligand binding triggers a conformational change of the receptor that promotes binding of the associated G-protein in the intracellular part. This, in turn, activates the G-protein, which acts as a transducer, initiating an intracellular signaling cascade that results in a cellular response (Alexander et al., 2019).
Computational model of TAS2R46 in complex with the bitter tastant strychnine, showing the two-site architecture explored in the simulations. The ligand pose in the vestibular site is colored in red, whereas the pose in the orthosteric site is in blue. Data were taken from reference (Sandal et al. 2015) (Color figure online)
ORs and TAS2Rs can identify a wide range of ligands and thus are considered promiscuous receptors. In humans, the approx. 400 ORs can recognize up to one trillion different odorants (Bushdid et al. 2014), whereas the 25 TAS2Rs detect around 1000 bitter tastants (Behrens and Meyerhof 2018; Dagan-Wiener et al. 2019). One receptor may be able to bind several molecules and the same ligand can be recognized by multiple receptors (Malnic et al., 1999; Krautwurst 2008; Meyerhof et al. 2010; Ji et al., 2014; Dunkel et al. 2014; Di Pizio and Niv 2015). This complex combinatorial code is still not fully understood, hindering identification of new ligands and receptor deorphanization.
The GPCR superfamily is highly pharmacologically relevant, with approx. 110 GPCRs being targeted by one third of all the FDA-approved drugs (Hauser et al. 2017, 2018). Similarly, ORs and TAS2Rs appear to be promising candidates for future drug design efforts (Lee et al. 2019; Di Pizio et al. 2019; Ahmad et al. 2020). Some examples include bronchodilators targeting TAS2Rs expressed in the airways (Nayak et al. 2019) or the sandalwood odorant used as hair loss therapy by stimulating OR2AT4 expressed in human hair follicles (Cheret 2018). Nonetheless, further therapeutical applications will require a more comprehensive characterization of the extranasal and extraoral roles of these receptors, as well as better understanding of the binding determinants of chemosensory receptors (Lee et al. 2019; Di Pizio et al. 2019; Ahmad et al. 2020).
Another common feature between GPCRs and ORs/TAS2Rs is the presence of genetic variants across the human population. In the case of GPCR drug targets, these variants can affect the pharmacological response of the receptor (Hauser et al. 2018). For chemosensory receptors, they have been mostly studied in the context of bitter taste/smell sensitivity, but they can also affect health status and propensity to certain diseases (Shaw et al. 2011; Mainland et al. 2014; Logan 2014; Chakraborty et al. 2019; Risso et al. 2021). One of the most well characterized examples is TAS2R38. After the serendipitous discovery of differences in the individual sensitivity to phenylthiocarbamide (PTC, a TAS2R38 ligand) (Fox 1932; Blakeslee 1932), a large-scale study (Blakeslee and Fox 1932) showed two main categories in the population, non-tasters and super-tasters. Later, a genome-wide linkage analysis (Kim et al. 2003) confirmed that sequence variants in the TAS2R38 gene have a direct influence in the observed PTC taste sensitivity. In addition to affecting food preferences (Robino et al. 2014), TAS2R38 polymorphisms have been recently shown to influence respiratory innate immunity mechanisms and susceptibility to chronic rhinosinusitis ((Jeruzal-Swiatecka et al. 2020) and references within).
Computational Structural Methods
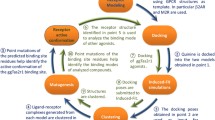
Understanding the molecular determinants of odorant or bitter tastant binding to their corresponding receptor requires structural information (Di Pizio and Niv 2014; de March et al. 2015; Fierro et al. 2017; Behrens et al. 2018a, b; Alfonso-Prieto et al. 2019a, b). X-ray or cryo-EM structures have been solved for only ~ 10% of human GPCRs (Munk et al. 2019; Bender et al. 2020; Kooistra et al. 2021) and, in particular, experimental structures of human ORs and TAS2Rs are still missing (https:// gpcrdb.org/structure/statistics; version 2021-01-27). Hence, computational methods have been used to fill the structural gap for chemosensory receptors, in particular homology modeling, molecular docking and molecular dynamics (MD) simulations (Di Pizio and Niv 2014; de March et al. 2015; Behrens et al. 2018a, b; Alfonso-Prieto et al. 2019a, b).
Building a high-resolution receptor homology model requires the identification of a template with sequence identity above 35% (Chothia and Lesk, 1986; Olivella et al. 2013; Piccoli et al. 2013). This is often not straightforward for the large and heterogeneous GPCR superfamily: only 10% of the GPCRs of unknown experimental structure have a closely related template with sequence identity above the 35% threshold (Zhang et al. 2006; Bender et al. 2020). In the case of ORs and TAS2Rs, their sequence identity with any of the available templates is unfortunately lower (< 20%) and thus the resulting homology models are low resolution (Fierro et al. 2017). Nonetheless, several computational approaches have been proposed to overcome this limitation, including homology modeling based on multiple templates or generation of an ensemble of models (Biarnés et al. 2010; de March et al. 2015; Di Pizio et al. 2017; Dagan-Wiener et al. 2019; Nowak et al. 2018; Bushdid et al. 2019; Spaggiari et al. 2020).
Molecular docking is then used in combination with these homology models to predict the binding mode of known ligands and, in some cases, identify new compounds. However, the accuracy of the docking results is limited by the low quality of the initial receptor models. In particular, the receptor-ligand interactions depend on the orientation of the amino acid side chains, which is uncertain in such low resolution models (Fierro et al. 2017). Hence, strategies aiming at enhancing the sampling of the conformational space of the receptor-ligand complex have been applied to overcome this limitation. These include flexible docking approaches and experimental data-driven model refinement (Di Pizio et al. 2017; Nowak et al. 2018; Bushdid et al. 2018; Bushdid et al. 2019; Di Pizio et al. 2020a, b), as well as molecular dynamics (MD) simulations (Gelis et al. 2012; Charlier et al. 2012; Charlier et al. 2013; Topin et al. 2014; Marchiori et al. 2013; Sandal et al. 2015; Li et al. 2016; Fierro et al. 2017; Ahmed et al. 2018; Fierro et al. 2019; Alfonso-Prieto et al. 2019a, 2019b; Haag et al., 2020; Schneider et al. 2020).
The thus-generated receptor-ligand complex models are validated by comparison with experimental data. Site-directed mutagenesis combined with functional experiments can be used to verify the predicted receptor binding residues (Munk et al. 2016b). In addition, the predicted ligand binding mode(s) can be compared against structure–activity relationship data (Vaas et al. 2018). Therefore, the interplay between computational and experimental data is crucial to obtain an accurate molecular picture of bitter taste and olfaction (Behrens et al. 2018a, b; Alfonso-Prieto et al. 2019a; Spaggiari et al. 2020).
The above described computational molecular modeling approaches have offered crucial insights into the ligand promiscuity of chemosensory receptors. Both TAS2Rs and ORs can recognize a broad range of ligands, yet they are selective. Based on mutagenesis data on TAS2Rs, it was proposed that such discriminating promiscuity could be achieved by a so-called “access control” that is able to dismiss the wrong compounds (Brockhoff et al. 2010). The molecular basis of such mechanism has been revealed using MD. Simulations showed that bitter tastants or odorants can explore not only one but two binding pockets in the corresponding chemosensory receptor (Sandal et al. 2015; Bushdid et al. 2019). This two-site architecture (Fig. 1) acts as a two-step verification system: the vestibular site (located close to the extracellular loops) filters out the receptor cognate ligands, which can then move downwards to the orthosteric site (situated inside the seven TM helix bundle), in order to trigger receptor activation. The interplay of the two binding sites is further validated by the observation that mutations of residues identified in either site affect the receptor response to its ligands. Moreover, the position of the orthosteric site in TAS2Rs and ORs coincides with that observed in experimental structures of other class A GPCRs in complex with their ligands (Venkatakrishnan et al. 2013; Latorraca et al. 2017). Similarly, the vestibular site overlaps with the extracellular allosteric site of other class A GPCRs (Thal et al. 2018; Latorraca et al. 2017). Further computational and experimental studies on other chemosensory receptors are needed to confirm whether this two-site architecture is conserved across the TAS2R and OR families.
Several FDA-approved drugs taste bitter (Dagan-Wiener et al. 2017; Di Pizio et al. 2019) and others affect olfactory perception (Lötsch et al. 2012). Moreover, TAS2Rs and ORs are also expressed in other parts of the body outside of the tongue and the nose, respectively, where they are involved in (yet not fully characterized) physiological and pathological processes (Lee et al. 2019; Di Pizio et al. 2019; Ahmad et al. 2020). Taken together, this suggests that bitter taste and odor molecules may be potential drug candidates (Di Pizio et al. 2019). A few computational molecular modeling studies have been carried out to explore the pharmacological potential of TAS2Rs and ORs (Tong et al. 2017; Nowak et al. 2018; Di Pizio et al. 2020a, b). Besides generating structural models of the receptor/ligand pairs already known (Levit et al. 2014; Tong et al. 2017; Nowak et al. 2018), computational approaches can also be used to design chemical modifications to improve ligand-receptor affinity and other drug-like properties (Di Pizio et al. 2020a, b). For instance, TAS2R14 has been recently studied as potential drug target against respiratory infections (Di Pizio et al. 2020a, b) due to its association with innate immune responses (Hariri et al. 2017) and its ability to bind clinical drugs that taste bitter, such as flufenamic acid (Levit et al. 2014; Behrens et al. 2018a, b). By integrating experimental mutagenesis data, homology modeling and molecular docking, an initial structural model of TAS2R14 in complex with flufenamic acid was generated (Levit et al. 2014; Nowak et al. 2018). Then, a combinatorial library of flufenamic acid derivatives was virtually screened against this model and the best candidate compounds were selected based on their docking score and visual inspection. These ligands, as well as additional analogs designed using medicinal chemistry concepts, were synthesized and subsequently tested with in vitro functional assays, resulting in the identification of new TAS2R14 agonists with nanomolar potency. Moreover, these experimental data were further used to refine the initial TAS2R14 model and obtain a better molecular description of the ligand binding modes (Di Pizio et al. 2020a, b). Altogether, this success story shows the potential of integrated experimental-computational approaches for ligand design for TAS2Rs and eventually ORs and opens the way to exploit the largely untapped pharmacological potential of these chemosensory receptors.
In addition, computational molecular modeling has been useful to understand the effect of genetic variants of TASR2s and ORs on ligand sensitivity (Biarnés et al. 2010; Marchiori et al. 2013; Geithe et al. 2017; March et al., 2015; Cierco-Jiménez et al. 2021). In the aforementioned case of TAS2R38, an ensemble of receptor homology models was generated, followed by molecular docking of PTC and further refinement with multiscale MD simulations (Biarnés et al. 2010; Marchiori et al. 2013). The resulting TAS2R38/PTC complex models allowed the identification of the residues putatively involved in binding, which were subsequently validated using mutagenesis and functional assays. Moreover, such models showed that amino acid 296, which varies between super-taster and non-taster variants, is not involved in ligand binding. Instead, residue 296 (located in TM7 at position 7.52 in the Ballesteros and Weinstein (1995) numbering scheme) faces F255 in the adjacent TM6, a helix which moves significantly during GPCR activation (Tehan et al. 2014; Venkatakrishnan et al. 2016; Weis and Kobilka 2018; Filipek 2019). Therefore, the interaction between these residues 296 and 255 (or lack thereof) can have an effect on receptor activation (Biarnés et al. 2010). The TAS2R38 variant present in the super-taster haplotype contains Val at position 296 and thus a hydrophobic interaction V296-F255 can be formed. This correlates with the in vitro activity measurements and the higher PTC sensitivity phenotype. On the contrary, the TAS2R38 variant present in the non-taster haplotypes contains Ile at position 296, a bulkier amino acid that will disrupt such interaction. This is in agreement with the lack of or reduced activity of the Ile296-containing TAS2R38 mutants and the lower PTC sensitivity phenotype. This hypothesis was further confirmed by creating a double swap-mutant F255V/V296F, which shows normal activity, consistent with the recovery of the proposed interaction (Biarnés et al. 2010). Interestingly, a subsequent bioinformatics analysis showed that, besides Val296 in TAS2R38, a hydrophobic residue at the equivalent position 7.52 is conserved for most TAS2Rs, as well as class A GPCRs. Additional comparisons against active-inactive pairs of experimental structures for class A GPCRs, together with mutagenesis data, further suggest that this position might be a not-yet-characterized activation microswitch for class A GPCRs (Fierro et al. 2017).
Data Science Approaches
Given the wide chemical space covered by odorants and bitter tastants, as well as the large number of ORs and TAS2Rs, it is not surprising that several online resources have been developed to compile the vast amount of data associated to these chemosensory receptors. Moreover, the combinatorial nature of bitter taste and olfaction is perfectly suited for the application of data-driven approaches, in particular machine learning (Lötsch et al., 2019).
For TAS2Rs, BitterDB contains information about approx. 1000 molecules that have been reported as bitter in humans, as well as other species (Wiener et al. 2012; Dagan-Wiener et al. 2019). Moreover, the database lists, if available, which TAS2Rs bind these bitter compounds, along with mutations that can affect receptor response and frequently present genetic variants (Wiener et al. 2012; Dagan-Wiener et al. 2019). Furthermore, already precomputed homology models for TAS2Rs are provided, facilitating future structure-based computational studies (Dagan-Wiener et al. 2019; Di Pizio et al. 2020a, b). Ligand-based studies have already been carried out. Chemoinformatics analysis have been applied to the list of bitter compounds in BitterDB to investigate the promiscuity, toxicity or drug-like properties of bitter compounds (Di Pizio and Niv 2015; Nissim et al. 2017; Di Pizio et al. 2020a, b). In addition, machine learning algorithms have been trained to predict bitterness (Dagan-Wiener et al. 2017; Zheng et al. 2018; Banerjee and Preissner 2018; Margulis et al., 2020).
For ORs, the amount of the data is significantly larger (especially when considering not only the ~ 400 human ORs, but also ORs from mouse and other species) and is distributed among several databases (Marenco et al. 2016; Di Pizio et al. 2020a, b). For instance, ORDB (Crastro et al. 2002), OlfactionDB (Modena et al. 2011), OdorDB (Marenco et al. 2013) and the Leibniz-LSB@TUM Odorant Database (Dunkel et al. 2014; Kreissl et al. 2019) contain information about odorant molecules and/or their cognate olfactory receptors. Complementarily, HORDE (Olender et al. 2013) is dedicated to olfactory receptor SNPs and haplotypes and their frequency in the population, whereas hORMdb (Cierco-Jiménez et al. 2021) additionally maps the sequence variants onto known topological positions of class A GPCRs to predict the functional impact of such mutations. As in the case of bitter tastants, machine learning approaches have also been developed for odorants (Lötsch et al. 2019). These algorithms aim at predicting either new ligand-receptor pairs (Liu et al. 2011; Audouze et al. 2014; Bushdid et al. 2018; Caballero-Vidal et al. 2020; Cong et al. 2020) or smells (Keller et al. 2017; Poivet et al. 2018; Nozaki and Nakamoto 2018; Chacko et al. 2020; Sharma et al. 2021), based on chemical features of the odorants.
Future Directions
The interplay of experimental and computational approaches has enabled a deeper molecular characterization of bitter taste and olfaction. However, the lack of experimental structures of bitter taste and olfactory receptors is still a hurdle that limits the accuracy of the computational structural models that can be generated. Moreover, a more extensive characterization of the physiological and pathological roles of extraoral TAS2Rs and extranasal ORs is required to exploit their potential as novel drug targets. In addition to computational molecular modeling and data science approaches, systems biology is expected to contribute to further understand the connection between ligand-receptor recognition and the subcellular response of the corresponding type II taste cell or olfactory sensory neuron.
References
Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS (2000) A novel family of mammalian taste receptors. Cell 100:693–702. https://doi.org/10.1016/s0092-8674(00)80705-9
Ahmad R, Dalziel JE (2020) G protein-coupled receptors in taste physiology and pharmacology. Front Pharmacol. https://doi.org/10.3389/fphar.2020.587664
Ahmed L, Zhang Y, Block E, Buehl M, Corr MJ, Cormanich RA, Gundala S, Matsunami H, O’Hagan D, Ozbil M, Pan Y, Sekharan S, Ten N, Wang M, Yang M, Zhang Q, Zhang R, Batista VS, Zhuang H (2018) Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc Natl Acad Sci USA 115(17):E3950–E3958. https://doi.org/10.1073/pnas.1713026115
Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Sharman JL, Southan C, Davies JA, Collaborators CGTP (2019) The concise guide to pharmacology 2019/20: G protein-coupled receptors. Br J Pharmacol 176(Suppl 1):S21–S141. https://doi.org/10.1111/bph.14748
Alfonso-Prieto M, Navarini L, Carloni P (2019a) Understanding ligand binding to G-protein coupled receptors using multiscale simulations. Front Mol Biosci 6:29. https://doi.org/10.3389/fmolb.2019.00029
Alfonso-Prieto M, Giorgetti A, Carloni P (2019b) Multiscale simulations on human Frizzled and Taste2 GPCRs. Curr Opin Struct Biol 55:8–16. https://doi.org/10.1016/j.sbi.2019.02.009
Audouze K, Tromelin A, Le Bon AM, Belloir C, Petersen RK, Kristiansen K, Brunak S, Taboureau O (2014) Identification of odorant-receptor interactions by global mapping of the human odorome. PLoS ONE 9(4):e93037. https://doi.org/10.1371/journal.pone.0093037
Ballesteros JA, Weinstein H (1995) [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences, Academic Press 25:366–428. https://doi.org/10.1016/S1043-9471(05)80049-7
Banerjee P, Preissner R (2018) BitterSweetForest: A random forest based binary classifier to predict bitterness and sweetness of chemical compounds. Front Chem 6:93. https://doi.org/10.3389/fchem.2018.00093
Behrens M, Meyerhof W (2011) Gustatory and extragustatory functions of mammalian taste receptors. Physiol Behav 105(1):4–13. https://doi.org/10.1016/j.physbeh.2011.02.010
Behrens M, Meyerhof W (2018) Vertebrate bitter taste receptors: keys for survival in changing environments. J Agric Food Chem 66(10):2204–2213. https://doi.org/10.1021/acs.jafc.6b04835
Behrens M, Meyerhof W (2019) A role for taste receptors in (neuro)endocrinology? J Neuroendocrinol 31:e12691. https://doi.org/10.1111/jne.12691
Behrens M, Briand L, de March CA, Matsunami H, Yamashita A, Meyerhof W, Weyand S (2018a) Structure-function relationships of olfactory and taste receptors. Chem Senses 43(2):81–87. https://doi.org/10.1093/chemse/bjx083
Behrens M, Gu M, Fan S, Huang C, Meyerhof W (2018b) Bitter substances from plants used in traditional Chinese medicine exert biased activation of human bitter taste receptors. Chem Biol Drug Des 91(2):422–433. https://doi.org/10.1111/cbdd.13089
Bender BJ, Marlow B, Meiler J (2020) Improving homology modeling from low-sequence identity templates in Rosetta: a case study in GPCRs. PLoS Comput Biol 16(10):e1007597. https://doi.org/10.1371/journal.pcbi.1007597
Biarnés X, Marchiori A, Giorgetti A, Lanzara C, Gasparini P, Carloni P, Born S, Brockhoff A, Behrens M, Meyerhof W (2010) Insights into the binding of phenyltiocarbamide (PTC) agonist to its target human TAS2R38 bitter receptor. PLoS ONE 5(8):e12394. https://doi.org/10.1371/journal.pone.0012394
Blakeslee AF (1932) Genetics of sensory thresholds: taste for phenylthiocarbamide. Proc Natl Acad Sci USA 18(1):120–130. https://doi.org/10.1073/pnas.18.1.120
Blakeslee AF, Fox AL (1932) Our different taste worlds: PTC as a demonstration of genetic differences in taste. J Hered 23(3):97–107. https://doi.org/10.1093/oxfordjournals.jhered.a103585
Boesveldt S, de Graaf K (2017) The differential role of smell and taste for eating behavior. Perception 46(3–4):307–319. https://doi.org/10.1177/0301006616685576
Boesveldt S, Parma V (2021) The importance of the olfactory system in human well-being, through nutrition and social behavior. Cell Tissue Res 383(1):559–567. https://doi.org/10.1007/s00441-020-03367-7
Brockhoff A, Behrens M, Niv MY, Meyerhof W (2010) Structural requirements of bitter taste receptor activation. Proc Natl Acad Sci USA 107(24):11110–11115. https://doi.org/10.1073/pnas.0913862107
Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65(1):175–187. https://doi.org/10.1016/0092-8674(91)90418-x
Bushdid C, Magnasco MO, Vosshall LB, Keller A (2014) Humans can discriminate more than 1 trillion olfactory stimuli. Science 343(6177):1370–1372. https://doi.org/10.1126/science.1249168
Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J (2018) Agonists of G-protein-coupled odorant receptors are predicted from chemical features. J Phys Chem Lett 9(9):2235–2240. https://doi.org/10.1021/acs.jpclett.8b00633
Bushdid C, de March CA, Topin J, Do M, Matsunami H, Golebiowski J (2019) Mammalian class I odorant receptors exhibit a conserved vestibular-binding pocket. Cell Mol Life Sci 76(5):995–1004. https://doi.org/10.1007/s00018-018-2996-4
Caballero-Vidal G, Bouysset C, Grunig H, Fiorucci S, Montagné N, Golebiowski J, Jacquin-Joly E (2020) Machine learning decodes chemical features to identify novel agonists of a moth odorant receptor. Sci Rep 10(1):1655. https://doi.org/10.1038/s41598-020-58564-9
Chacko R, Jain D, Patwardhan M, Puri A, Karande S, Rai B (2020) Data based predictive models for odor perception. Sci Rep 10(1):17136. https://doi.org/10.1038/s41598-020-73978-1
Chakraborty S, Dakle P, Sinha A, Vishweswaraiah S, Nagori A, Salimath S, Prakash YS, Lodha R, Kabra SK, Ghosh B, Faruq M, Mahesh PA, Agrawal A (2019) Genetic variations in olfactory receptor gene OR2AG2 in a large multigenerational family with asthma. Sci Rep 9(1):19029. https://doi.org/10.1038/s41598-019-54718-6
Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS (2006) The receptors and cells for mammalian taste. Nature 444(7117):288–294. https://doi.org/10.1038/nature05401
Charlier L, Topin J, Ronin C, Kim SK, Goddard WA, Efremov R, Golebiowski J (2012) How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell Mol Life Sci 69(24):4205–13. https://doi.org/10.1007/s00018-012-1116-0
Charlier L, Topin J, de March CA, Lai PC, Crasto CJ, Golebiowski J (2013) Molecular modelling of odorant/olfactory receptor complexes. Methods Mol Biol 1003:53–65. https://doi.org/10.1007/978-1-62703-377-0_4
Cheret J, Bertolini M, Ponce L, Lehmann J, Tsai T, Alam M, Hatt H (2018) Paus R (2018) Olfactory receptor OR2AT4 regulates human hair growth. Nat Commun 9(1):3624. https://doi.org/10.1038/s41467-018-05973-0
Chothia C, Lesk AM (1986) The relation between the divergence of sequence and structure in proteins. EMBO J 5:823–826. https://doi.org/10.1002/j.1460-2075.1986.tb04288.x
Cierco-Jiménez R, Casajuana-Martin N, García-Recio A, Alcántara L, Pardo L, Campillo M, Gonzalez A (2021) The mutational landscape of human olfactory G protein-coupled receptors. BMC Biol 19(1):21. https://doi.org/10.1186/s12915-021-00962-0
Cong X, Ren W, Pacalon J, Claire A, Xu L, Matsunami H, Yu Y and Golebiowski J (2020) Functions of olfactory receptors are decoded from their sequence. bioRxiv 2020.01.06.895540. Doi:https://doi.org/10.1101/2020.01.06.895540
Crasto C, Marenco L, Miller P, Shepherd G (2002) Olfactory Receptor Database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res 30(1):354–360. https://doi.org/10.1093/nar/30.1.354
Dagan-Wiener A, Nissim I, Ben Abu N, Borgonovo G, Bassoli A, Niv MY (2017) Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci Rep 7(1):12074. https://doi.org/10.1038/s41598-017-12359-7
Dagan-Wiener A, Di Pizio A, Nissim I, Bahia MS, Dubovski N, Margulis E, Niv MY (2019) BitterDB: taste ligands and receptors database in 2019. Nucleic Acids Res 47(D1):D1179–D1185. https://doi.org/10.1093/nar/gky974
Dalesio NM, Barreto Ortiz SF, Pluznick JL, Berkowitz DE (2018) Olfactory, taste, and photo sensory receptors in non-sensory organs: it just makes sense. Front Physiol 9:1673. https://doi.org/10.3389/fphys.2018.01673
de March CA, Kim SK, Antonczak S, Goddard WA 3rd, Golebiowski J (2015) G protein-coupled odorant receptors: From sequence to structure. Protein Sci 24(9):1543–1548. https://doi.org/10.1002/pro.2717
Di Pizio A, Niv MY (2014) Computational studies of smell and taste receptors. Isr J Chem 54(8–9):1205–1218. https://doi.org/10.1002/ijch.201400027
Di Pizio A, Niv MY (2015) Promiscuity and selectivity of bitter molecules and their receptors. Bioorg Med Chem 23(14):4082–4091. https://doi.org/10.1016/j.bmc.2015.04.025
Di Pizio A, Levit A, Slutzki M, Behrens M, Karaman R, Niv MY (2016) Comparing Class A GPCRs to bitter taste receptors: Structural motifs, ligand interactions and agonist-to-antagonist ratios. Methods Cell Biol 132:401–427. https://doi.org/10.1016/bs.mcb.2015.10.005
Di Pizio A, Kruetzfeldt LM, Cheled-Shoval S, Meyerhof W, Behrens M, Niv MY (2017) Ligand binding modes from low resolution GPCR models and mutagenesis: chicken bitter taste receptor as a test-case. Sci Rep 7(1):8223. https://doi.org/10.1038/s41598-017-08344-9
Di Pizio A, Behrens M, Krautwurst D (2019) Beyond the flavour: the potential druggability of chemosensory G protein-coupled receptors. Int J Mol Sci 20(6):1402. https://doi.org/10.3390/ijms20061402
Di Pizio A, Behr J, Krautwurst D (2020) 3.39 - Toward the digitalization of olfaction. In: Fritsch B (ed) The senses: a comprehensive reference, 2nd edn. Elsevier, Amsterdam, pp 758–768
Di Pizio A, Waterloo LAW, Brox R, Löber S, Weikert D, Behrens M, Gmeiner P, Niv MY (2020a) Rational design of agonists for bitter taste receptor TAS2R14: from modeling to bench and back. Cell Mol Life Sci 77(3):531–542. https://doi.org/10.1007/s00018-019-03194-2
Dunkel A, Steinhaus M, Kotthoff M, Nowak B, Krautwurst D, Schieberle P, Hofmann T (2014) Nature’s chemical signatures in human olfaction: a foodborne perspective for future biotechnology. Angew Chem Int Ed Engl 53(28):7124–7143. https://doi.org/10.1002/anie.201309508
Fierro F, Suku E, Alfonso-Prieto M, Giorgetti A, Cichon S, Carloni P (2017) Agonist binding to chemosensory receptors: a systematic bioinformatics analysis. Front Mol Biosci 4:63. https://doi.org/10.3389/fmolb.2017.00063
Fierro F, Giorgetti A, Carloni P, Meyerhof W, Alfonso-Prieto M (2019) Dual binding mode of “bitter sugars” to their human bitter taste receptor target. Sci Rep 9(1):8437. https://doi.org/10.1038/s41598-019-44805-z
Filipek S (2019) Molecular switches in GPCRs. Curr Opin Struct Biol 55:114–120. https://doi.org/10.1016/j.sbi.2019.03.017
Fox AL (1932) The relationship between chemical constitution and taste. Proc Natl Acad Sci USA 18(1):115–120. https://doi.org/10.1073/pnas.18.1.115
Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272. https://doi.org/10.1124/mol.63.6.1256
Geithe C, Protze J, Kreuchwig F, Krause G, Krautwurst D (2017) Structural determinants of a conserved enantiomer-selective carvone binding pocket in the human odorant receptor OR1A1. Cell Mol Life Sci 74(22):4209–4229. https://doi.org/10.1007/s00018-017-2576-z
Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K (2012) Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew Chem Int Ed Engl 51(5):1274–1278. https://doi.org/10.1002/anie.201103980
Gerkin RC, Ohla K, Veldhuizen MG, Joseph PV, Kelly CE, Bakke AJ, Steele KE, Farruggia MC, Pellegrino R, Pepino MY, Bouysset C, Soler GM, Pereda-Loth V, Dibattista M, Cooper KW, Croijmans I, Di Pizio A, Ozdener MH, Fjaeldstad AW, Lin C, Sandell MA, Singh PB, Brindha VE, Olsson SB, Saraiva LR, Ahuja G, Alwashahi MK, Bhutani S, D’Errico A, Fornazieri MA, Golebiowski J, Dar Hwang L, Öztürk L, Roura E, Spinelli S, Whitcroft KL, Faraji F, Fischmeister FPS, Heinbockel T, Hsieh JW, Huart C, Konstantinidis I, Menini A, Morini G, Olofsson JK, Philpott CM, Pierron D, Shields VDC, Voznessenskaya VV, Albayay J, Altundag A, Bensafi M, Bock MA, Calcinoni O, Fredborg W, Laudamiel C, Lim J, Lundström JN, Macchi A, Meyer P, Moein ST, Santamaría E, Sengupta D, Rohlfs Dominguez P, Yanik H, Hummel T, Hayes JE, Reed DR, Niv MY, Munger SD, Parma V, GCCR Group Author (2021) Recent smell loss is the best predictor of COVID-19 among individuals with recent respiratory symptoms. Chem Senses. https://doi.org/10.1093/chemse/bjaa081
Haag F, Ahmed L, Reiss K, Block E, Batista VS, Krautwurst D (2020) Copper-mediated thiol potentiation and mutagenesis-guided modeling suggest a highly conserved copper-binding motif in human OR2M3. Cell Mol Life Sci 77(11):2157–2179. https://doi.org/10.1007/s00018-019-03279-y
Hariri BM, McMahon DB, Chen B, Freund JR, Mansfield CJ, Doghramji LJ, Adappa ND, Palmer JN, Kennedy DW, Reed DR, Jiang P, Lee RJ (2017) Flavones modulate respiratory epithelial innate immunity: Anti-inflammatory effects and activation of the T2R14 receptor. J Biol Chem 292(20):8484–8497. https://doi.org/10.1074/jbc.M116.771949
Hauser AS, Attwood MM, Rask-Andersen M, Gloriam SHB, DE, (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Disc 16:829–842. https://doi.org/10.1038/nrd.2017.178
Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE, Babu MM (2018) Pharmacogenomics of GPCR drug targets. Cell 172:41–54. https://doi.org/10.1016/j.cell.2017.11.033
Jeruzal-Swiatecka J, Fendler W, Pietruszewska W (2020) Clinical role of extraoral bitter taste receptors. Int J Mol Sci 21(14):5156. https://doi.org/10.3390/ijms21145156
Ji M, Su X, Su X, Chen Y, Huang W, Zhang J, Gao Z, Li C, Lu X (2014) Identification of novel compounds for human bitter taste receptors. Chem Biol Drug Des 84(1):63–74. https://doi.org/10.1111/cbdd.12293
Keller A, Gerkin RC, Guan Y, Dhurandhar A, Turu G, Szalai B, Mainland JD, Ihara Y, Yu CW, Wolfinger R, Vens C, Schietgat L, De Grave K, Norel R, DREAM Olfaction Prediction Consortium, Stolovitzky G, Cecchi GA, Vosshall LB, Meyer P (2017) Predicting human olfactory perception from chemical features of odor molecules. Science 355(6327):820–826. https://doi.org/10.1126/science.aal2014
Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D (2003) Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299(5610):1221–1225. https://doi.org/10.1126/science.1080190
Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, Keserű GM, Gloriam DE (2021) GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res 49(D1):D335–D343. https://doi.org/10.1093/nar/gkaa1080
Krautwurst D (2008) Human olfactory receptor families and their odorants. Chem Biodivers 5(6):842–852. https://doi.org/10.1002/cbdv.200890099
Kreissl J, Mall V, Steinhaus P, Steinhaus M (2019) Leibniz-LSB@TUM Odorant Database, version 1.0. Leibniz-Institute for Food Systems Biology at the Technical University of Munich, Freising, Germany. https://www.leibniz-lsb.de/en/databases/leibniz-lsbtum-odorant-database. Accessed 11 March 2021
Lagerstrom MC, Schioth HB (2008) Structural diversity of G protein- coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357. https://doi.org/10.1038/nrd2518
Latorraca NR, Venkatakrishnan AJ, Dror RO (2017) GPCR dynamics: structures in motion. Chem Rev 117(1):139–155. https://doi.org/10.1021/acs.chemrev.6b00177
Lee S-J, Depoortere I, Hatt H (2019) Therapeutic potential of ectopic olfactory and taste receptors. Nat Rev Drug Discov 18(2):116–138. https://doi.org/10.1038/s41573-018-0002-3
Levit A, Nowak S, Peters M, Wiener A, Meyerhof W, Behrens M, Niv MY (2014) The bitter pill: clinical drugs that activate the human bitter taste receptor TAS2R14. FASEB J 28(3):1181–1197. https://doi.org/10.1096/fj.13-242594
Li S, Ahmed L, Zhang R, Pan Y, Matsunami H, Burger JL, Block E, Batista VS, Zhuang H (2016) Smelling sulfur: copper and silver regulate the response of human odorant receptor OR2T11 to low-molecular-weight thiols. J Am Chem Soc 138(40):13281–13288. https://doi.org/10.1021/jacs.6b06983
Liu X, Su X, Wang F, Huang Z, Wang Q, Li Z, Zhang R, Wu L, Pan Y, Chen Y, Zhuang H, Chen G, Shi T, Zhang J (2011) ODORactor: a web server for deciphering olfactory coding. Bioinformatics 27(16):2302–2303. https://doi.org/10.1093/bioinformatics/btr385
Logan DW (2014) Do you smell what I smell? Genetic variation in olfactory perception. Biochem Soc Trans 42(4):861–865. https://doi.org/10.1042/bst20140052
Lötsch J, Geisslinger G, Hummel T (2012) Sniffing out pharmacology: interactions of drugs with human olfaction. Trends Pharmacol Sci 33(4):193–199. https://doi.org/10.1016/j.tips.2012.01.004
Lötsch J, Kringel D, Hummel T (2019) Machine learning in human olfactory research. Chem Senses 44(1):11–22. https://doi.org/10.1093/chemse/bjy067
Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, Zhan S, Lee SS, Lin A, Matsunami H (2014) The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci 17(1):114–120. https://doi.org/10.1038/nn.3598
Mainland JD, Barlow LA, Munger SD, Millar SE, Vergara MN, Jiang P, Schwob JE, Goldstein BJ, Boye SE, Martens JR, Leopold DA, Bartoshuk LM, Doty RL, Hummel T, Pinto JM, Trimmer C, Kelly C, Pribitkin EA, Reed DR (2020) Identifying treatments for taste and smell disorders: gaps and opportunities. Chem Senses 45(7):493–502. https://doi.org/10.1093/chemse/bjaa038
Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96(5):713–723. https://doi.org/10.1016/s0092-8674(00)80581-4
Marchiori A, Capece L, Giorgetti A, Gasparini P, Behrens M, Carloni P, Meyerhof W (2013) Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: experimentally-validated detailed structural prediction of agonist binding. PLoS ONE. https://doi.org/10.1371/journal.pone.0064675
Marenco LN, Bahl G, Hyland L, Shi J, Wang R, Lai PC, Miller PL, Shepherd GM, Crasto CJ (2013) Databases in SenseLab for the genomics, proteomics, and function of olfactory receptors. Methods Mol Biol 1003:3–22. https://doi.org/10.1007/978-1-62703-377-0_1
Marenco L, Wang R, McDougal R, Olender T, Twik M, Bruford E, Liu X, Zhang J, Lancet D, Shepherd G, Crasto C (2016) ORDB, HORDE ODORactor and other on-line knowledge resources of olfactory receptor-odorant interactions. Database (Oxford). https://doi.org/10.1093/database/baw132
Margulis E, Dagan-Wiener A, Ives RS, Jaffari S, Siems K, Niv MY (2020) Intense bitterness of molecules: Machine learning for expediting drug discovery. Comput Struct Biotechnol J 19:568–576. https://doi.org/10.1016/j.csbj.2020.12.030
Maßberg D, Hatt H (2018) Human olfactory receptors: novel cellular functions outside of the nose. Physiol Rev 98(3):1739–1763. https://doi.org/10.1152/physrev.00013.2017
Mennella JA, Spector AC, Reed DR, Coldwell SE (2013) The bad taste of medicines: overview of basic research on bitter taste. Clin Ther 35(8):1225–1246. https://doi.org/10.1016/j.clinthera.2013.06.007
Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M (2010) The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses 35(2):157–170. https://doi.org/10.1093/chemse/bjp092
Modena D, Trentini M, Corsini M, Bombaci A, Giorgetti A (2011) OlfactionDB: a database of olfactory receptors and their ligands. Adv Lif Sci 1(1):1–5. https://doi.org/10.5923/j.als.20110101.01
Munk C, Isberg V, Mordalski S, Harpsøe K, Rataj K, Hauser AS, Kolb P, Bojarski AJ, Vriend G, Gloriam DE (2016) GPCRdb: the G protein-coupled receptor database – an introduction. Br J Pharmacol 173:2195–2207. https://doi.org/10.1111/bph.13509
Munk C, Harpsøe K, Hauser AS, Isberg V, Gloriam DE (2016) Integrating structural and mutagenesis data to elucidate GPCR ligand binding. Curr Opin Pharmacol 30:51–58. https://doi.org/10.1016/j.coph.2016.07.003
Munk C, Mutt E, Isberg V, Nikolajsen LF, Bibbe JM, Flock T, Hanson MA, Stevens RC, Deupi X, Gloriam DE (2019) An online resource for GPCR structure determination and analysis. Nat Methods 16(2):151–162. https://doi.org/10.1038/s41592-018-0302-x
Nayak AP, Shah SD, Michael JV, Deshpande DA (2019) Bitter taste receptors for asthma therapeutics. Front Physiol 10:884. https://doi.org/10.3389/fphys.2019.00884
Nissim I, Dagan-Wiener A, Niv MY (2017) The taste of toxicity: A quantitative analysis of bitter and toxic molecules. IUBMB Life 69(12):938–946. https://doi.org/10.1002/iub.1694
Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB (2011) Independent HHsearch, Needleman-Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol 28(9):2471–2480. https://doi.org/10.1093/molbev/msr061
Nowak S, Di Pizio A, Levit A, Niv MY, Meyerhof W, Behrens M (2018) Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim Biophys Acta Gen Subj. https://doi.org/10.1016/j.bbagen.2018.07.009
Nozaki Y, Nakamoto T (2018) Predictive modeling for odor character of a chemical using machine learning combined with natural language processing. PLoS ONE 13(6):e0198475. https://doi.org/10.1371/journal.pone.0198475
Olender T, Nativ N, Lancet D (2013) HORDE: comprehensive resource for olfactory receptor genomics. Methods Mol Biol 1003:23–38. https://doi.org/10.1007/978-1-62703-377-0_2
Olivella M, Gonzalez A, Pardo L, Deupi X (2013) Relation between sequence and structure in membrane proteins. Bioinformatics 29(13):1589–1592. https://doi.org/10.1093/bioinformatics/btt249
Oppo V, Melis M, Melis M, Tomassini Barbarossa I, Cossu G (2020) “Smelling and Tasting” Parkinson’s Disease: Using Senses to Improve the Knowledge of the Disease. Front Aging Neurosci 12:43. https://doi.org/10.3389/fnagi.2020.00043
Parma V, Ohla K, Veldhuizen MG, Niv MY, Kelly CE, Bakke AJ, Cooper KW, Bouysset C, Pirastu N, Dibattista M, Kaur R, Liuzza MT, Pepino MY, Schöpf V, Pereda-Loth V, Olsson SB, Gerkin RC, Rohlfs Domínguez P, Albayay J, Farruggia MC, Bhutani S, Fjaeldstad AW, Kumar R, Menini A, Bensafi M, Sandell M, Konstantinidis I, Di Pizio A, Genovese F, Öztürk L, Thomas-Danguin T, Frasnelli J, Boesveldt S, Saatci Ö, Saraiva LR, Lin C, Golebiowski J, Hwang LD, Ozdener MH, Guàrdia MD, Laudamiel C, Ritchie M, Havlícek J, Pierron D, Roura E, Navarro M, Nolden AA, Lim J, Whitcroft KL, Colquitt LR, Ferdenzi C, Brindha EV, Altundag A, Macchi A, Nunez-Parra A, Patel ZM, Fiorucci S, Philpott CM, Smith BC, Lundström JN, Mucignat C, Parker JK, van den Brink M, Schmuker M, Fischmeister FPS, Heinbockel T, Shields VDC, Faraji F, Santamaría E, Fredborg WEA, Morini G, Olofsson JK, Jalessi M, Karni N, D’Errico A, Alizadeh R, Pellegrino R, Meyer P, Huart C, Chen B, Soler GM, Alwashahi MK, Welge-Lüssen A, Freiherr J, de Groot JHB, Klein H, Okamoto M, Singh PB, Hsieh JW, GCCR Group Author, Reed DR, Hummel T, Munger SD, Hayes JE (2020) More than smell - COVID-19 is associated with Severe impairment of smell, taste, and chemesthesis. Chem Senses 45(7):609–622. https://doi.org/10.1093/chemse/bjaa041
Pawar S, Kumar A (2002) Issues in the formulation of drugs for oral use in children: role of excipients. Paediatr Drugs 4(6):371–379. https://doi.org/10.2165/00128072-200204060-00004
Piccoli S, Suku E, Garonzi M, Giorgetti A (2013) Genome-wide membrane protein dtructure prediction. Curr Genomics 14(5):324–9. https://doi.org/10.2174/13892029113149990009
Pierron D, Pereda-Loth V, Mantel M, Moranges M, Bignon E, Alva O, Kabous J, Heiske M, Pacalon J, David R, Dinnella C, Spinelli S, Monteleone E, Farruggia MC, Cooper KW, Sell EA, Thomas-Danguin T, Bakke AJ, Parma V, Hayes JE, Letellier T, Ferdenzi C, Golebiowski J, Bensafi M (2020) Smell and taste changes are early indicators of the COVID-19 pandemic and political decision effectiveness. Nat Commun 11(1):5152. https://doi.org/10.1038/s41467-020-18963-y
Poivet E, Tahirova N, Peterlin Z, Xu L, Zou DJ, Acree T, Firestein S (2018) Functional odor classification through a medicinal chemistry approach. Sci Adv. https://doi.org/10.1126/sciadv.aao6086
Risso D, Drayna D, Tofanelli S, Morini G (2021) Open questions in sweet, umami, and bitter taste genetics. Curr Op Physiol 20:174–179. https://doi.org/10.1016/j.cophys.2020.12.007
Roberts RO, Christianson TJ, Kremers WK, Mielke MM, Machulda MM, Vassilaki M, Alhurani RE, Geda YE, Knopman DS, Petersen RC (2016) Association between olfactory dysfunction and amnestic mild cognitive impairment and Alzheimer disease dementia. JAMA Neurol 73(1):93–101. https://doi.org/10.1001/jamaneurol.2015.2952
Robino A, Mezzavilla M, Pirastu N, Dognini M, Tepper BJ, Gasparini P (2014) A population-based approach to study the impact of PROP perception on food liking in populations along the Silk Road. PLoS ONE 9(3):e91716. https://doi.org/10.1371/journal.pone.0091716
Sabiniewicz A, Heyne F, Hummel T (2021) Odors modify emotional responses. Flavour Fragr J 36(2):256–263. https://doi.org/10.1002/ffj.3640
Sandal M, Behrens M, Brockhoff A, Musiani F, Giorgetti A, Carloni P, Meyerhof W (2015) Evidence for a transient additional ligand binding site in the TAS2R46 bitter taste receptor. J Chem Theory Comput 11(9):4439–4449. https://doi.org/10.1021/acs.jctc.5b00472
Schiöth HB, Fredriksson R (2005) The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen Comp Endocrinol 142(1–2):94–101. https://doi.org/10.1016/j.ygcen.2004.12.018
Schneider J, Korshunova K, Si Chaib Z, Giorgetti A, Alfonso-Prieto M, Carloni P (2020) Ligand pose predictions for human G protein-coupled receptors: insights from the Amber-based hybrid Molecular Mechanics/Coarse-Grained approach. J Chem Inf Model 60(10):5103–5116. https://doi.org/10.1021/acs.jcim.0c00661
Sharma A, Kumar R, Ranjta S, Varadwaj PK (2021) SMILES to smell: decoding the structure-odor relationship of chemical compounds using the deep neural network approach. J Chem Inf Model 61(2):676–688. https://doi.org/10.1021/acs.jcim.0c01288
Shaw CA, Li Y, Wiszniewska J, Chasse S, Zaidi SN, Jin W, Dawson B, Wilhelmsen K, Lupski JR, Belmont JW, Doody RS, Szigeti K (2011) Olfactory copy number association with age at onset of Alzheimer disease. Neurology 76(15):1302–1309. https://doi.org/10.1212/WNL.0b013e3182166df5
Spaggiari G, Di Pizio A, Cozzini P (2020) Sweet, umami and bitter taste receptors: State of the art of in silico molecular modeling approaches. Trends Food Sci Technol 96:21–29. https://doi.org/10.1016/j.tifs.2019.12.002
Sullivan RM, Wilson DA, Ravel N, Mouly AM (2015) Olfactory memory networks: from emotional learning to social behaviors. Front Behav Neurosci 9:36. https://doi.org/10.3389/fnbeh.2015.00036
Tarakad A, Jankovic J (2017) Anosmia and Ageusia in Parkinson’s Disease. Int Rev Neurobiol 133:541–556. https://doi.org/10.1016/bs.irn.2017.05.028
Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS (2014) Unifying family A GPCR theories of activation. Pharmacol Ther 143(1):51–60. https://doi.org/10.1016/j.pharmthera.2014.02.004
Tepper BJ, Barbarossa IT (2020) Taste, Nutrition, and Health. Nutrients 12(1):155. https://doi.org/10.3390/nu12010155
Thal DM, Glukhova A, Sexton PM, Christopoulos A (2018) Structural insights into G-protein-coupled receptor allostery. Nature 559(7712):45–53. https://doi.org/10.1038/s41586-018-0259-z
Tong T, Ryu SE, Min Y, de March CA, Bushdid C, Golebiowski J, Moon C, Park T (2017) Olfactory receptor 10J5 responding to α-cedrene regulates hepatic steatosis via the cAMP-PKA pathway. Sci Rep 7(1):9471. https://doi.org/10.1038/s41598-017-10379-x
Topin J, de March CA, Charlier L, Ronin C, Antonczak S, Golebiowski J (2014) Discrimination between olfactory receptor agonists and non-agonists. Chemistry 20(33):10227–10230. https://doi.org/10.1002/chem.201402486
Vass M, Kooistra AJ, Yang D, Stevens RC, Wang MW, de Graaf C (2018) Chemical diversity in the G protein-coupled receptor superfamily. Trends Pharmacol Sci 39(5):494–512. https://doi.org/10.1016/j.tips.2018.02.004
Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM (2013) Molecular signatures of G-protein-coupled receptors. Nature 494(7436):185–194. https://doi.org/10.1038/nature11896
Venkatakrishnan AJ, Deupi X, Lebon G, Heydenreich FM, Flock T, Miljus T, Balaji S, Bouvier M, Veprintsev DB, Tate CG, Schertler GF, Babu MM (2016) Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature 536(7617):484–487. https://doi.org/10.1038/nature19107
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD (2001) The sequence of the human genome. Science 291(5507):1304–1351. https://doi.org/10.1126/science.1058040
Weis WI, Kobilka BK (2018) The molecular basis of G protein-coupled receptor activation. Annu Rev Biochem 87:897–919. https://doi.org/10.1146/annurev-biochem-060614-033910
Wiener A, Shudler M, Levit A, Niv MY (2012) BitterDB: a database of bitter compounds. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr755
Zhang Y, Devries ME, Skolnick J (2006) Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.0020013
Zheng S, Jiang M, Zhao C, Zhu R, Hu Z, Xu Y, Lin F (2018) e-Bitter: Bitterant prediction by the consensus voting from the machine-learning methods. Front Chem 6:82. https://doi.org/10.3389/fchem.2018.00082
Acknowledgements
I would like to thank Paolo Carloni and Alejandro Giorgetti for introducing me to the field of chemical senses. I am also grateful to Luciano Navarini, Wolfgang Meyerhof, Marc Spehr, Fabrizio Fierro, Giuliano Santarpia and Nicolas P. Müller for useful discussions along the years. This work was supported by the Ernesto Illy Foundation (Trieste, Italy).
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Ernesto Illy Foundation (Trieste, Italy).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alfonso-Prieto, M. Bitter Taste and Olfactory Receptors: Beyond Chemical Sensing in the Tongue and the Nose. J Membrane Biol 254, 343–352 (2021). https://doi.org/10.1007/s00232-021-00182-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-021-00182-1