Abstract
Vaccines are one of the most powerful tools for preventing infectious diseases. To effectively fight pathogens, vaccines should induce potent and long-lasting immune responses that are specific to the pathogens. However, not all vaccines can induce effective immune responses, and the responses vary greatly among individuals and populations. Although several factors, such as age, host genetics, nutritional status, and region, affect the effectiveness of vaccines, increasing data have suggested that the gut microbiota is critically associated with vaccine-induced immune responses. In this review, I discuss how gut microbiota affects vaccine effectiveness based on the clinical and preclinical data, and summarize possible underlying mechanisms related to the adjuvant effects of microbiota. A better understanding of the link between vaccine-induced immune responses and the gut microbiota using high-throughput technology and sophisticated system vaccinology approaches could provide crucial insights for designing effective personalized preventive and therapeutic vaccination strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vaccines are regarded as the most effective life-saving medical invention; and vaccines have been estimated to prevent 2–3 million deaths per year globally (Plotkin, 2014). Several pathogens, such as smallpox and poliovirus, have been eradicated or nearly eradicated after the introduction of vaccination (Rodrigues & Plotkin, 2020). However, vaccines against several pathogens, such as the hepatitis C virus, malaria, and human immunodeficiency virus, are not clinically available yet (Li et al., 2015), and the emergence of new infectious viruses requires continued development of vaccines. Furthermore, there is a growing need to develop therapeutic cancer vaccines (Guo et al., 2013). Therefore, the importance of vaccine development, which can induce effective and sustained immune responses, cannot be overemphasized. Vaccine-induced adaptive immune responses can be classifies into B cell/antibody-mediated and T cell-mediated immune responses (Clem, 2011). Antibodies, circulating in the blood or secreted into the mucosa, bind to pathogens or toxins and neutralize them. Thus, neutralizing antibody titers over certain thresholds are considered reliable predictors of the vaccine’s protective efficacy (Clem, 2011). T cells provide cytokines and co-stimulatory signals that promote the memory B cell generation and the production of high-affinity antibodies (Swain et al., 2012). Cytotoxic T cells secrete various cytotoxic molecules that directly kill virus-infected cells (Zhang & Bevan, 2011). Therefore, to develop effective protective vaccines, robust and long-lasting adaptive immune responses should be elicited via vaccination.
Interestingly, the immune responses induced by vaccines vary greatly among individuals and populations in different regions. For example, People who received the inactivated influenza vaccine showed ~ 100-fold variability in antibody levels, and those who received the pneumococcal and Hemophilus influenza type b (Hib) vaccines showed ~ 40-fold variability in antibody levels (Pulendran, 2009; Zimmermann & Curtis, 2019). Furthermore, cellular responses vary greatly among individuals; cytokine responses induced by Bacille Calmette-Guerin (BCG) vaccination show ~ 100-fold variability (Querec et al., 2009). Moreover, infants and the elderly show reduced vaccine-induced immune responses compared to healthy adults (Ciabattini et al., 2018; PrabhuDas et al., 2011). These data indicate that host factors affect the quality, quantity, and durability of vaccine-induced immune responses.
The human body is inhabited by numerous microorganisms including bacteria, viruses, and fungi, collectively termed microbiota. The human gastrointestinal tract contains an estimated 1014 microorganisms (Gill et al., 2006). The human microbiota is highly variable among individuals, and the number of microorganisms and composition of microbiota are also continuously changing within an individual, depending on the age, diet, and health status of the individual (DeJong et al., 2020; Ursell et al., 2012). Advances in immunology in recent decades have revealed that microbiota play a critical role in the regulation of the host immune system, and likewise, that the immune system can influence the composition and distribution of microorganisms (Zheng et al., 2020). Growing evidence has shown a link between human microbiota and diseases, including inflammatory bowel disease, arthritis, obesity, and autism (Cho & Blaser, 2012; Durack & Lynch, 2019). Similarly, several results from clinical and animal models demonstrated that the gut microbiota of the host affects vaccine effectiveness by modulating immune responses. In this review, I summarize the recent findings that provide evidence of the influence of microbiota on vaccine effectiveness and the possibility of microbiota acting as a vaccine adjuvant.
Dynamics of Gut Microbiota
Gut Microbiome Composition in Humans
Although the gut microbiota is composed of several microorganisms, such as bacteria, archaea, fungi, and viruses, bacteria are predominant. Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, Fusobacteria, and Verrucomicrobia are the main gut microbial phyla, and among them, Firmicutes and Bacteroidetes account for approximately 90% of the human gut microbiota (Rinninella et al., 2019). Representative genera of the Firmicutes phylum are Lactobacillus, Bacillus, Clostridium, Enterococcus, and Ruminicoccus (Rinninella et al., 2019). The dominant genera of the Bacteroidetes phylum are Bacterioides and Prevotella and the phylum Actinobacteria is mainly represented by the Bifidobacterium (Rinninella et al., 2019). The human microbiota is dynamic and influenced by various factors, including the type of delivery during birth, age, antibiotic treatment, and disease status.
Changes in Gut Microbiome According to Age and Health Status
At birth, humans acquire microbiota via contact with the surrounding environments. Thus, the type of delivery during birth can affect the composition of the microbiota. Interestingly, infants born vaginally possess increased abundance of Bacteroides, Bifidobacteria, and Lactobacillus and show more microbial diversity than infants born via cesarean delivery (C-section) (Coelho et al., 2021; Rutayisire et al., 2016). In contrast, infants born via C-section possess a less diversified microbiome composed primarily of Staphylococcus, Streptococcus, and Clostridium (Coelho et al., 2021). Furthermore, methods of milk feeding affect the composition of the microbiota. Breast-fed infants have a more diverse microbiota and a higher abundance of Bifidobacterium and lower abundance of E. coli in the gut microbiota than formula-fed infants (Roger et al., 2010). After the weaning period, the intake of solid food also changes the gut microbiota composition, and a shift from Bifidobacterium to Bacteroidetes and Firmicute species occurs (Fallani et al., 2011). Compared to infants and children, the microbiota diversity increases with aging (Lloyd-Price et al., 2017) and microbiomes in the adult are comparatively more stable and greatly influenced by the environment than by genetics (Rothschild et al., 2018).
Many clinical studies have compared the microbial composition between young and elderly people (Badal et al., 2020). One notable difference between the microbiota in healthy adults and the elderly is a reduction in microbial diversity (Lynch & Pedersen, 2016). This reduced diversity in the elderly is due to the increase in the content of specific groups. In general, the elderly have higher abundance of Enterobacteriaceae and lower abundance of Bifidobacterium and Lactobacillus compared to the young (Salazar et al., 2017). Furthermore, decreased microbial diversity has been associated with various diseases, such as inflammatory bowel disease, diabetes, rheumatoid arthritis, and allergy (Ding et al., 2019; Qiu et al., 2022). However, it is still unclear whether these changes in microbiota composition are the cause or consequence of aging or diseases. Furthermore, a significant diversity in microbiota exists even in individuals without diseases and is altered by diet, antibiotic treatment, infection, smoking, alcohol drinking, and exercise (Conlon & Bird, 2014; Lin et al., 2020; Lloyd-Price et al., 2016).
Possible Mechanisms by Which Microbiota Affects Vaccine Effectiveness
The effectiveness of vaccinations is determined by the mutual interplays of vaccine, gut microbiota, and the host’s immune system. Although the mechanisms by which microbiota affect the effectiveness of vaccines have not been elucidated, it has been suggested that microbiota could be a source of natural adjuvants that modulate the degree of innate and adaptive immune responses (Ciabattini et al., 2019). Furthermore, the type of vaccine (live-attenuated, inactivated, protein, viral vector, or mRNA), route of immunization, dosage, vaccination schedule, and vaccination strategy (homologous immunization or heterologous prime-boost) are critically important factors for determining the vaccine response (Zimmermann & Curtis, 2019).
The estimated mechanisms by which the intestinal microbiome affects the efficacy of vaccines are described as follows (Fig. 1).
Anticipated mechanisms of microbiota that could regulate vaccine effectiveness. A Pathogen-associated molecular patterns (PAMPs), such as LPS or peptidoglycan produced by microbiota, could be recognized by Pattern Recognition Receptors (PRR), such as TLRs and NOD2, expressed by immune cells. Signaling cascades activate transcription factors and induce gene expression, which could facilitate adjuvant effects to vaccines. B Microbiota-derived metabolites, such as SCFAs, could promote the differentiation of Th1 or Th17 cells and increase antibody production. C Epitopes from microbiota could cross-react with epitopes derived from target vaccine antigens
Act as an Adjuvant Through Sensing by Pattern Recognition Receptor
Unlike live-type vaccines, highly purified or inactivated vaccines elicit poor immune responses since they lack the signals to trigger sufficient innate immune responses that bring to robust and strong adaptive immune responses (Coffman et al., 2010). Adjuvants increase the magnitude and duration of vaccine-induced immune responses (Pulendran et al., 2021). Generally, they stimulate innate immune responses by providing pathogen-associated molecular pattern (PAMP)/damage-associated molecular pattern signaling and promoting antigen uptake (Pulendran et al., 2021). PAMP-type adjuvants include killed bacteria; microbial components such as lipopolysaccharide (LPS) (a TLR 4/2 ligand), flagellin (a TLR5 ligand), and DNA containing CpG motif (a TLR 9 ligand); analogs such as monophosphoryl lipid A (MPL); and synthetic TLR ligands. Since the microbiota enduringly releases immunostimulatory molecules, they can act as natural adjuvants.
Several studies using mouse models have revealed the pathways involved in the adjuvant activity of the microbiota. TLR5-deficient mice showed reduced antibody titers to a trivalent inactivated influenza vaccine, which is due to the failure of host microbiota sensing (Oh et al., 2014). Oral reconstitution with a flagellated strain of E. coli was shown to restore the impaired antibody response in germ-free or antibiotic-treated mice. (Oh et al., 2014). Microbiota sensing through TLR5 affected the antibody responses to inactivated polio vaccine. However, antibody responses induced by adjuvanted influenza or live attenuated yellow fever vaccines were not significantly reduced in TLR5-deficient mice (Oh et al., 2014).
Besides TLR5, interaction with the microbiota by nucleotide-binding oligomerization domain-containing (Nod) 2 also mediates the adjuvant activity. Compared to WT mice; Nod2 − / − , and receptor-interacting serine/threonine kinase (Ripk)-2–deficient mice (Ripk2 is an adaptor downstream of Nod2); and antibiotic-treated, germ-free mice showed significantly decreased IgG antibody responses to human serum albumin (HSA) when they were intranasally immunized with HSA and cholera toxin (Kim et al., 2016a). Reconstitution of germ-free mice with muramyl dipeptide (MDP), which is recognized by NOD2, or bacteria that activate the Nod2 pathway sufficiently restored antibody responses (Kim et al., 2016a). LPS, which is recognized by TLR2/4, is also a potent immunostimulatory molecule produced by the microbiota. Different microorganisms have different types of LPS, and each type exhibits different immunomodulatory functions (Vatanen et al., 2016). Currently, whether microbiota-derived LPS can affect vaccine-induced immune responses is unclear but is seems possible because immunization antibody with synthetic TLR4 agonist (MPL) induce more persistent germinal center reaction and antibody response (Kasturi et al., 2011).
The ability or degree of microbiota to provide vaccine adjuvant activity may be dependent on whether their components are restricted to the gut or released into the periphery. For example, when the gut is inflamed or under antibiotic treatment, an overgrowth of LPS-producing Enterobacteriaceae may increase LPS levels in the gut and periphery (Zeng et al., 2017). Additionally, gut integrity may affect the immune-modulating functions of the microbiota. If the gut epithelial barrier is damaged due to malnutrition, inflammation, or antibiotic treatment, it increases the permeability of the gut and leads to the influx of antigens or leakage of immunomodulatory molecules of the microbiota (Mu et al., 2017).
Microbiota-derived Metabolites
Gut microbiota produce several metabolites that have the potential to regulate immune responses. Short-chain fatty acids (SCFAs), such as acetate, butyrate, and propionate are generated by colonic bacteria through fermentation of fiber (Parada Venegas et al., 2019). SCFAs is one of the best-known metabolites that has immunomodulatory functions in various immune cells (Corrêa-Oliveira et al., 2016). SCFAs increased the building block for antibody generation by increasing acetyl-CoA, oxidative phosphorylation, glycolysis, and fatty acid synthesis (Kim et al., 2016b). Simultaneously, SCFAs regulated the gene expression profiles of molecules for plasma cell differentiation and antibody class switching. It was reported that defective pathogen specific antibody responses and increased susceptibility to Citrobacter rodentium infection were observed in mice with low SCFA production due to reduced dietary fiber or microbial deficiency (Kim et al., 2016b). However, another study reported that SCFAs reduced antibody responses to intragastrically administrated ovalbumin and autoantibody responses (Sanchez et al., 2020). Therefore, further investigations are needed to delineate the role of SCFAs in vaccine-induced antibody responses.
SCFAs also promote the differentiation of Th1 and Th17 cells in the intestine, spleen, and lymph nodes (Park et al., 2015). Furthermore, SCFAs increase the production of IL-22 by ILC3 and CD4+ T cells (Yang et al., 2020; Zhao et al., 2022), and IFN-γ production and cytotoxicity of CD8+ T cells (Luu et al., 2021). Thus, SCFAs may affect vaccine effectiveness by influencing the activity or differentiation of T-cells.
Besides SCFAs, the immunoregulatory functions of other metabolites or essential nutrients produced by microbiota, such as bile acids and metabolites of tryptophan, vitamins have been identified. Antibiotic treatment significantly reduced secondary bile acid and tryptophan metabolism, which was associated with an elevated inflammatory signature in subjects with impaired neutralizing antibody and IgA responses to the influenza vaccine (Hagan et al., 2019). However, studies on the effects of microbiota-derived metabolite on vaccine effectiveness are at an early stage. Thus, more research is required to clarify the association between metabolites and vaccine-induced immune response.
Cross Reaction of Microbiota-derived Antigens
The microbiota modulates immune responses by cross-reacting with self-antigens, tumor antigens, and pathogen-specific antigens. Roseburia intestinalis has proteins that cross-react with the self-antigen β2-glycoprotein I, and B. breve expresses peptides that cross-react with an artificially introduced tumor antigen in B16 melanoma cells (Zitvogel & Kroemer, 2021). In addition to self- or tumor antigens, pathogen-derived antigens can cross-react with microbiota-derived antigens.
Memory phenotype CD4+ T cells, specific to viruses, were abundant in individuals who had never been infected with those viruses (Su et al., 2013). One explanation for this observation is the cross-reactivity of the TCR with environmental antigens, and Su et al. (2013) found that microbial peptides were cross-recognized by HIV-1 and influenza-reactive T cells. Another study revealed that healthy individuals have abundant circulating and tissue-resident CD4+T cells that reacted with the intestinal microbiota, and the function of these CD4+T cells was changed during inflammation (Hegazy et al., 2017). Additionally, extensive bioinformatics analysis predicted that considerable amounts of the TCR epitope of the microbiota are shared with pathogenic bacteria (Bremel & Homan, 2015).
In addition, a recent study showed that antibodies specific to the S2 domain of SARS-CoV-2 could cross-react with commensal gut bacteria (Jia et al., 2022). Thus, these cross-reactive T or B cells may modulate immune responses to pathogens and vaccines. Another human study found that pre-existing cross-reactive CD4+ T cells are linked to lower influenza virus shedding and less severe illness (Wilkinson et al., 2012). Although this study did not identify the source of the cross-reactive T cells, it is possible that these cells were microbiota-reactive T cells because a previous study reported the sharing of their epitopes with influenza epitopes (Carrasco Pro et al., 2018). Despite of above findings, it is still unclear whether the epitope encoded by the microbiota enhances or dampens the vaccine-induced immune response; thus, further investigation is needed to identify the underlying mechanism.
Correlative Evidence of Microbiome and Vaccine Efficacy from Clinical Studies
Oral Vaccines
People in high-income countries are more responsive to oral vaccines (such as oral rotavirus and polio vaccines) than those in low-middle-income countries (Lewnard et al., 2020). Vaccine effectiveness is especially important in low- and middle-income countries because of the high susceptibility of individuals to infections and poor access to health care. Thus, the effect of microbiota on vaccine effectiveness could be of greater importance in low- and middle-income countries than in high-income countries. Several clinical studies in infants have found a correlation between gut microbiota and vaccine effectiveness.
Harris et al. (2017) analyzed the fecal microbiota of infants in Ghana who received an oral rotavirus vaccine (ORV). Of note, ORV responding infants (serum rotavirus-specific IgA level > 20 IU/ml) have a microbiota composition similar to that of Dutch infants, who generally show a good response to rotavirus vaccination. In particular, Streptococcus bovis was positively correlated, whereas Bacteroides and Prevotella species were negatively correlated with an enhanced rotavirus immune response. Additionally, the enterobacteria-Bacteroidetes ratio was significantly increased in ORV-responding infants in both Ghana and Dutch compared with that in non-responding infants in Ghana. Another infant study in Pakistan revealed that the immune response to ORV was positively associated with the prevalence of Clostridium cluster XI and Proteobacteria in fecal samples (Harris et al., 2018a). An increased ratio of Enterobacteriaceae to Bacteroides species was observed in the ORV responders in both studies. Another infant study in Zimbabwe showed that IgA titers positively correlated with the abundance of Bacteroides thetaiotaomicron (Robertson et al., 2021). Conversely, two other infant studies performed in India and Nicaragua found no notable correlation between fecal microbiota and response to ORV (Fix et al., 2020; Parker et al., 2018).
Regarding oral polio vaccine (OPV), an infant study in China reported a positive correlation of relative abundance of fecal Bifidobacterium with poliovirus-specific IgA levels (Zhao et al., 2020). However, another infant study performed in India showed no significant association between response to OPV and fecal microbiota (Praharaj et al., 2019). These discordant correlations between microbiota and vaccine efficacy in clinical studies might be due to the differences in microbiome composition in participants from each study because microbiome composition varies greatly depending on various genetic and environmental factors. Another possible explanation is the differences in vaccine antigens. As it has been reported that microbiota share cross-reactive epitopes with vaccine antigens, each strain of microbiota may have different cross-reactive epitope pools and might affect vaccine efficacy.
Non-oral and COVID-19 Vaccines
Some clinical studies have revealed an association between microbiota and responsiveness to non-orally administered vaccines. A prospective infant study in Bangladesh showed that abundance of fecal Bifidobacterium in infants is positively correlated with CD4+ T-cell and antibody responses to BCG, Tetanus toxoid, and hepatitis B virus vaccines (Huda et al., 2019). Therefore, an increased intestinal Bifidobacterium may increase the immunogenicity of vaccines, even if they are administered non-orally.
In addition, a few adult studies have shown an association between gut microbiota and vaccine effectiveness. Cholera vaccine responders with long-term IgG and IgA memory B cells to oral cholera vaccine showed a higher abundance of Clostridiales and lower abundance of Enterobacterales than non-responders at the time of vaccination (Chac et al., 2021). Interestingly, increased IL-1β and decreased IL-6 levels were observed in the feces of cholera vaccine responders. Thus, it is possible that Clostridiales either induced memory B cells that could cross-react with cholera antigen epitopes, or Clostridiales or their metabolites induced activation of the IL-1β cytokine pathway, which is important for Ag priming. Also, recently, the association between the microbiota and COVID-19 vaccine efficacy have been reported. Ng et al. (2022) found that gut microbiota composition is associated with immunogenicity as well as adverse effects of inactivated or mRNA type of COVID-19 vaccines. Subjects with high levels of neutralizing antibodies against the CoronaVac vaccine had high levels of Bifidobacterium adolescentis in the gut, and the gut microbiome was enriched in carbohydrate metabolism pathways (Ng et al., 2022). BNT162b2-induced neutralizing antibody levels were positively correlated with bacteria that have fimbriae and flagella, including Roseburia faecis (Ng et al., 2022). Notably, Prevotella copri and two Megamonas species were increased in subjects with fewer side effects. Thus, this study suggests that specific gut microbiota strains are associated with enhanced immune responses or fewer adverse effects of vaccines.
Effects of Antibiotics on Vaccine Effectiveness
To reveal the association between microbiota and vaccine-induced immune responses, several interventional studies using antibiotics were performed. A study of infants, enrolled in the ‘Melbourne Infant Study: BCG for Allergy and Infection Reduction,’ showed that treatment of antibiotics before vaccination did not significantly affect antibody levels and seroconversion to Diphtheria-Tetanus-Pertussis, Hepatitis B (HepB), Hib, poliovirus and Pneumococcal conjugate vaccine (PCV)13 vaccines (n = 29) (Zimmermann et al., 2020). Another double-blind, randomized infant study in India revealed that azithromycin treatment did not increase the immunogenicity of OPV (Grassly et al., 2016). Although azithromycin reduced the fecal biomarker of environmental enteropathy and frequency of pathogenic bacteria, which are associated with reduced immunogenicity of oral vaccine, it failed to enhance vaccine immunogenicity.
In contrast, a recent study where children aged 6–24 months (n = 560) were observed from 2006 to 2016 and analyzed retrospectively (Chapman et al., 2022), showed that children administered antibiotics at 9–12 months of age showed lower antibody levels to vaccines. Pre-booster antibody levels to diphtheria, tetanus, and acellular pertussis (DTaP); Hib; inactivated poliovirus (IPV); and PCV antigens decreased by 5.8%, 6.8%, 11.3%, and 10.4%, respectively (all P < 0.05), for each antibiotic course the child underwent, whereas antibody levels decreased by 18.1%, 21.3%, 18.9%, and 12.2%, respectively after boosting (Chapman et al., 2022).
Furthermore, some adult studies have reported the effects of antibiotics on vaccine immunogenicity. Adult participants who received a broad-spectrum antibiotic cocktail 36 h before ORV vaccination showed no significant changes in anti-rotavirus IgA levels, whereas some in the vancomycin-treated group exhibited a > twofold increase in the IgA levels 7 days after vaccination (Harris et al., 2018b).
Additionally, a study based on the system vaccinology approach assessed the effects of broad-spectrum antibiotics on influenza vaccine-induced immune response (Hagan et al., 2019). Healthy young adults who received antibiotics showed a temporary decrease in gut bacterial load and long-term decrease in bacteria diversity; however, significant changes in neutralizing antibodies were not detected. To identify whether pre-existing immunity affected the results, subjects with low baseline antibody levels against the influenza virus were re-analyzed. Notably, antibiotic treatment dramatically reduced H1N1-specific neutralizing IgG and IgA levels.
Recently, a prospective cohort study revealed that antibiotics treatment within six months before the COVID-19 vaccine (BNT162b2) immunization was associated with a lower rate of neutralizing antibodies seroconversion after priming. However, recent antibiotic use did not impair BNT162b immunogenicity after two doses (Cheung et al., 2022).
As such, the effects of antibiotic treatments on vaccine efficacy are controversial. This discrepancy may be due to the type of antibiotics, the immunogenicity of the vaccine, and the host’s health status. Depending on the immunogenicity of the vaccine, the effects of microbiota or antibiotic treatment on vaccine efficacy may be different. Furthermore, because not all bacterial strains have a response to antibiotics, the type of antibiotics and duration of antibiotic treatments would affect vaccine efficacy.
Effect of Probiotics on Vaccine Effectiveness
Clinical Data on the Effects of Probiotics on Vaccine Efficacy
Probiotics are live, non-pathogen microorganisms that provide health benefits to their hosts. Bifidobacteria (BIF), a probiotic, modulated lipid metabolism and alleviated allergic symptoms (Eslami et al., 2020). BIF also has beneficial effects on vaccine-induced immune responses. Probiotic feeding in the first 6 months of life increased IgG antibody responses to the HepB vaccine (Soh et al., 2010). Similarly, adults who take Bifidobacterium animalis ssp. lactis (BB-12®) and Lactobacillus paracasei (L. paracasei) ssp. for 6 weeks had increased antibody levels specific to the influenza vaccine after vaccination (Rizzardini et al., 2012). Furthermore, an infant study conducted in France showed that Bifidobacterium formula increases IgA response to IPV (Mullié et al., 2004). Although intramuscular injection of IPV elicited poor mucosal IgA response, anti-poliovirus IgA titers were increased in the Bifidobacterium formula group, and antibody titers correlated with Bifidobacterium longum/Bifidobacterium infantis and Bifidobacterium breve levels.
In addition to BIF, Lactobacillus and Lactococcus strains increased vaccine-induced immune responses.
Oral injection of Lactobacillus plantarum GUANKE significantly increased the COVID-19 vaccine-specific neutralizing antibodies in both serum and bronchoalveolar lavage in mice (Xu et al., 2021). Dietary supplements of Lactobacillus acidophilus W37 (LaW37) with long-chain inulin (lcITF) also enhanced vaccine efficacy against Salmonella typhimurium strains by twofold (Lépine et al., 2019).
Furthermore, a randomized, double-blind, placebo-controlled study revealed that Lactobacillus rhamnosus GG (L.GG) increased hemagglutinin inhibition titers of inactivated influenza vaccine (Davidson et al., 2011). Oral administration of Lacticaseibacillus casei (L. casei) strain with an ORV increased the ORV-specific IgM-secreting cells and IgA antibody levels (Isolauri et al., 1995). Also, oral administration of Lactococcus lactis for 7 days, together with oral attenuated typhoid vaccine, increased IgA responses. Neutrophils in those who received Lactococcus lactis along with their immunization expressed higher levels of complement receptor 3 than those in individuals from the placebo group (Fang et al., 2000). Oral administration of Lactobacillus paracasei (NCC 2461) for 4 months before influenza and pneumococcus vaccination increased NK cell activity and decreased influenza infections in subjects older than 70 years (Bunout et al., 2004). Besides, a recent study showed that individuals receiving Loigolactobacillus coryniformis have higher IgG levels specific to COVID-19 vaccines 81 days after the first vaccination and showed reduced vaccine-induced side effects (Rodriguez-Blanque et al., 2022).
However, not all studies have shown that probiotics improve the effectiveness of vaccine. For example, significant increases in antibody levels to the DPT-Hib (diphtheria and tetanus toxoids and whole-cell pertussis vaccine adsorbed with Hib conjugate vaccine) or pneumococcal vaccine were not detected in children who consumed milk fermented by Streptococcus thermophilus, L. casei, and Lactobacillus acidophilus (Pérez et al., 2010). In the elderly, daily intake of L. casei shirota did not significantly improve the protective efficacy of the influenza vaccine (Van Puyenbroeck et al., 2012). A recent study also examined the effect of L.GG uptake on influenza vaccine efficacy in children with type 1 diabetes (T1D) and found that there was no significant increase of humoral response in the L.GG uptake group (Bianchini et al., 2020). A randomized, double-blind, placebo-controlled study comprising 1104 healthy participants (aged 18–60 years) showed that the intake of L. casei 431 did not enhance the effectiveness of influenza vaccine, although the L. casei administered group showed a reduced period of upper respiratory symptoms (Jespersen et al., 2015). Such discrepancies between the studies may arise as a result of multiple factors, including the type of vaccine, type of probiotics, and the composition the host microbiota.
Alteration of the Diversity and Composition of Gut Microbiota
Given that probiotic treatments can induce alterations in both the diversity and composition of the gut microbiota, it is reasonable to speculate that probiotics could modify vaccine-induced responses by promoting changes in the composition of gut microbial communities. In this regard, the antimicrobial agents or metabolic compounds produced by probiotic strains could contribute to suppressing the growth of certain microorganisms or compete for the receptor and binding sites of particular microbes (Collado et al., 2007; Spinler et al., 2008). For example, L. reuteri has been established to produce the antimicrobial compound reuterin from glycerol, which has been shown to inhibit the growth of enteric bacteria (Spinler et al., 2008). Furthermore, 12 commercial, probiotic strains have been demonstrated to inhibit and disrupt the adhesion of Bacteroides, Clostridium, Staphylococcus, and Enterobacter to human mucus in vitro, with different strains of bacteria being inhibited by each of the different probiotic strains (Collado et al., 2007). Consequently, consideration should be given to the appropriate application of probiotics, depending on the purpose and target microorganisms.
Potential Mechanisms of Probiotics in Modulating Immune Responses
As previously mentioned, studies that seek to identify the potential mechanisms underlying the effects of probiotics in modulating vaccine-induced immune responses are currently ongoing. However, the findings of several studies conducted to date have offered certain clues in this regard. For example, orally administrated probiotics have been reported to promote the activation of the NF-κB and IRF signaling pathways in response to the stimulation of TLRs on both immune and epithelial cells (Peroni & Morelli, 2021). Intestinal dendritic cells in the lamina propria have been found to recognize microbes and thereby promote further signaling cascades. In addition, it has been reported that L. rhamnosus JB-1 can modulate the function of DCs via DC-SIGN and TLR2 (Konieczna et al., 2015). Interestingly, it has been established that probiotics can modulate the expression of TLRs. For example, Lactobacillus species have been observed to promote the down-regulation of TLR4 and up-regulation of TLR2 in C. albicans-infected macrophages (Matsubara et al., 2017). Moreover, Lactobacillus salivarius has been found to inhibit S. aureus-induced inflammatory responses, which is assumed to be mediated via TLR/PI3K/AKT/NF-κB signaling (Jia et al., 2020). In addition, probiotic secreted metabolites, including SCFA, have been established to have immunostimulatory effects, and as mentioned in the previous section, SCFAs can contribute to modulating the differentiation of T cells and antibody-producing cells, and as a key source of energy for the gut microbiota, can also influence the intestinal barriers by regulating gut gene expression (Kim et al., 2014).
Future Perspective
Over the past few years, interest in microbiota has increased exponentially. Microbiota undeniably plays critical roles in developing and fine-tuning innate and adaptive immune responses, and inversely, the immune system shapes the microbiota. Although increasing evidence supports that microbiota affects vaccine-induced immune responses and effectiveness, a detailed underlying mechanism has not been established yet. The results of current human studies show huge discrepancies due to various factors, including vaccine type, immunizing antigen, age, genetics of host, nutritional factors, and geographic region of study, which influence the microbiota composition, function, and status of immune cells. Furthermore, new types of vaccine are rapidly being developed. In response to the COVID-19 pandemic, mRNA- and viral vector-based vaccines were introduced, for the first time, into the clinic; the mRNA vaccines could elicit potent B and T cell immune responses compared to traditional subunit or inactivated vaccines (Chaudhary et al., 2021; Khoshnood et al., 2022). Owing to these characteristics, diverse research on mRNA therapeutic vaccines for cancer and autoimmune disease is actively being undertaken. Various types of the directly injectable mRNA vaccines, including those targeting dendritic cells, have been tested in clinical trials (Hogan & Pardi, 2022). Additionally, new vaccination strategies combining different types of vaccines and novel adjuvants targeting non-TLR, metabolic, and cell death pathways are actively being developed and tested.
Therefore, a precise and detailed analysis on the vaccine-induced immune responses and identifying the underlying mechanism of action for the new typed of vaccines, as well as adjuvants are required.
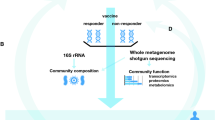
To overcome this, a multilateral study design on an increased sample size using multi-omics vaccinology techniques should be conducted (Pulendran, 2019; Wimmers & Pulendran, 2020). Analysis of gene expression, protein or lipid synthesis, and metabolite changes using multiple system vaccinology tools enables us to understand integrated networks and dynamic alteration of vaccine-induced immune responses (Fig. 2).
Future approaches to reveal the reciprocal interaction between microbiota and vaccine-induced immune responses. To decipher sophisticated cross-talk between microbiota and vaccine-induced responses, advanced techniques to analyze the proteins, metabolites, and nucleic acid from microbiota should be applied. Furthermore, future analysis using system vaccinology approaches on diverse vaccine-induced immune responses in humans is promising
Furthermore, to determine the exact bacterial strain or species that have regulatory functions in vaccine-induced immune responses, metagenomic DNA extraction, massive next-generation sequencing, and bioinformatic analysis tools that have the potential to detect all microbiota in samples with increased sensitivity, should be applied (Fig. 2) (Lind & Pollard, 2021; Song et al., 2018; Wensel et al., 2022; Wu et al., 2022).
Intriguingly, vaccination also can lead to alterations in the microbiota (Guo et al., 2020; Uehara et al., 2022). Different vaccination methods were associated with different effects on gut microflora diversity in mice, and individuals who received the COVID-19 BNT162b2 mRNA vaccine showed higher oral bacterial diversity and a significantly reduced number of oral Bacteroides. Thus, research is needed to uncover the detailed bidirectional crosstalk between microbiota and the immune system.
The ideal goal in the field of immunology is to build a vaccination strategy that induces an effective protective immune response in infants, the elderly, and immunocompromised patients. Despite being the most vulnerable to infection, vaccines are less effective in these groups. Identifying the specific microbiota related to the effectiveness of each type of vaccine and determining the underlying mechanism could make it possible to design more effective personalized vaccination strategies.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
References
Badal, V. D., Vaccariello, E. D., Murray, E. R., Yu, K. E., Knight, R., Jeste, D. V., & Nguyen, T. T. (2020). The gut microbiome, aging, and longevity: A systematic review. Nutrients, 12, 3759.
Bianchini, S., Orabona, C., Camilloni, B., Berioli, M. G., Argentiero, A., Matino, D., Alunno, A., Albini, E., Vacca, C., Pallotta, M. T., et al. (2020). Effects of probiotic administration on immune responses of children and adolescents with type 1 diabetes to a quadrivalent inactivated influenza vaccine. Human Vaccines and Immunotherapeutics, 16, 86–94.
Bremel, R. D., & Homan, E. J. (2015). Extensive T-cell epitope repertoire sharing among human proteome, gastrointestinal microbiome, and pathogenic bacteria: Implications for the definition of self. Frontiers in Immunology, 6, 538.
Bunout, D., Barrera, G., Hirsch, S., Gattas, V., de la Maza, M. P., Haschke, F., Steenhout, P., Klassen, P., Hager, C., Avendano, M., et al. (2004). Effects of a nutritional supplement on the immune response and cytokine production in free-living Chilean elderly. Journal of Parenteral and Enteral Nutrition, 28, 348–354.
Carrasco Pro, S., Lindestam Arlehamn, C. S., Dhanda, S. K., Carpenter, C., Lindvall, M., Faruqi, A. A., Santee, C. A., Renz, H., Sidney, J., Peters, B., et al. (2018). Microbiota epitope similarity either dampens or enhances the immunogenicity of disease-associated antigenic epitopes. PLoS ONE, 13, e0196551.
Chac, D., Bhuiyan, T. R., Saha, A., Alam, M. M., Salma, U., Jahan, N., Chowdhury, F., Khan, A. I., Ryan, E. T., LaRocque, R., et al. (2021). Gut microbiota and development of Vibrio cholerae-specific long-term memory B cells in adults after whole-cell killed oral cholera vaccine. Infection and Immunity, 89, e00217-21.
Chapman, T. J., Pham, M., Bajorski, P., & Pichichero, M. E. (2022). Antibiotic use and vaccine antibody levels. Pediatrics, 149, e2021052061.
Chaudhary, N., Weissman, D., & Whitehead, K. A. (2021). mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nature Reviews Drug Discovery, 20, 817–838.
Cheung, K. S., Lam, L. K., Zhang, R., Ooi, P. H., Tan, J. T., To, W. P., Hui, C. H., Chan, K. H., Seto, W. K., Hung, I. F. N., et al. (2022). Association between recent usage of antibiotics and immunogenicity within six months after COVID-19 vaccination. Vaccines, 10, 1122.
Cho, I., & Blaser, M. J. (2012). The human microbiome: At the interface of health and disease. Nature Reviews Genetics, 13, 260–270.
Ciabattini, A., Nardini, C., Santoro, F., Garagnani, P., Franceschi, C., & Medaglini, D. (2018). Vaccination in the elderly: The challenge of immune changes with aging. Seminars in Immunology, 40, 83–94.
Ciabattini, A., Olivieri, R., Lazzeri, E., & Medaglini, D. (2019). Role of the microbiota in the modulation of vaccine immune responses. Frontiers in Microbiology, 10, 1305.
Clem, A. S. (2011). Fundamentals of vaccine immunology. Journal of Global Infectious Diseases, 3, 73–78.
Coelho, G. D. P., Ayres, L. F. A., Barreto, D. S., Henriques, B. D., Prado, M., & Passos, C. M. D. (2021). Acquisition of microbiota according to the type of birth: An integrative review. Revista Latino-Americana De Enfermagem, 29, e3446.
Coffman, R. L., Sher, A., & Seder, R. A. (2010). Vaccine adjuvants: Putting innate immunity to work. Immunity, 33, 492–503.
Collado, M. C., Meriluoto, J., & Salminen, S. (2007). Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Letters in Applied Microbiology, 45, 454–460.
Conlon, M. A., & Bird, A. R. (2014). The impact of diet and lifestyle on gut microbiota and human health. Nutrients, 7, 17–44.
Corrêa-Oliveira, R., Fachi, J. L., Vieira, A., Sato, F. T., & Vinolo, M. A. (2016). Regulation of immune cell function by short-chain fatty acids. Clinical and Translational Immunology, 5, e73.
Davidson, L. E., Fiorino, A. M., Snydman, D. R., & Hibberd, P. L. (2011). Lactobacillus GG as an immune adjuvant for live-attenuated influenza vaccine in healthy adults: A randomized double-blind placebo-controlled trial. European Journal of Clinical Nutrition, 65, 501–507.
DeJong, E. N., Surette, M. G., & Bowdish, D. M. E. (2020). The gut microbiota and unhealthy aging: Disentangling cause from consequence. Cell Host and Microbe, 28, 180–189.
Ding, R. X., Goh, W. R., Wu, R. N., Yue, X. Q., Luo, X., Khine, W. W. T., Wu, J. R., & Lee, Y. K. (2019). Revisit gut microbiota and its impact on human health and disease. Journal of Food and Drug Analysis, 27, 623–631.
Durack, J., & Lynch, S. V. (2019). The gut microbiome: Relationships with disease and opportunities for therapy. The Journal of Experimental Medicine, 216, 20–40.
Eslami, M., Bahar, A., Keikha, M., Karbalaei, M., Kobyliak, N. M., & Yousefi, B. (2020). Probiotics function and modulation of the immune system in allergic diseases. Allergologia Et Immunopathologia, 48, 771–788.
Fallani, M., Amarri, S., Uusijarvi, A., Adam, R., Khanna, S., Aguilera, M., Gil, A., Vieites, J. M., Norin, E., Young, D., et al. (2011). Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology, 157, 1385–1392.
Fang, H., Elina, T., Heikki, A., & Seppo, S. (2000). Modulation of humoral immune response through probiotic intake. FEMS Immunology and Medical Microbiology, 29, 47–52.
Fix, J., Chandrashekhar, K., Perez, J., Bucardo, F., Hudgens, M. G., Yuan, L., Twitchell, E., Azcarate-Peril, M. A., Vilchez, S., & Becker-Dreps, S. (2020). Association between gut microbiome composition and rotavirus vaccine response among Nicaraguan infants. The American Journal of Tropical Medicine and Hygiene, 102, 213–219.
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., Gordon, J. I., Relman, D. A., Fraser-Liggett, C. M., & Nelson, K. E. (2006). Metagenomic analysis of the human distal gut microbiome. Science, 312, 1355–1359.
Grassly, N. C., Praharaj, I., Babji, S., Kaliappan, S. P., Giri, S., Venugopal, S., Parker, E. P., Abraham, A., Muliyil, J., Doss, S., et al. (2016). The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: A double-blind randomised placebo-controlled trial in seronegative Indian infants. The Lancet Infectious Diseases, 16, 905–914.
Guo, C., Manjili, M. H., Subjeck, J. R., Sarkar, D., Fisher, P. B., & Wang, X. Y. (2013). Therapeutic cancer vaccines: Past, present, and future. Advances in Cancer Research, 119, 421–475.
Guo, J., Tang, J., Kang, T., Xiong, Y., Xiang, Z., & Qin, C. (2020). Different immunization methods lead to altered gut flora and varied responses to Mycobacterium tuberculosis infection in mice. The Journal of Infection in Developing Countries, 14, 1170–1177.
Hagan, T., Cortese, M., Rouphael, N., Boudreau, C., Linde, C., Maddur, M. S., Das, J., Wang, H., Guthmiller, J., Zheng, N. Y., et al. (2019). Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell, 178, 1313–1328.
Harris, V., Ali, A., Fuentes, S., Korpela, K., Kazi, M., Tate, J., Parashar, U., Wiersinga, W. J., Giaquinto, C., de Weerth, C., et al. (2018a). Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes, 9, 93–101.
Harris, V. C., Armah, G., Fuentes, S., Korpela, K. E., Parashar, U., Victor, J. C., Tate, J., de Weerth, C., Giaquinto, C., Wiersinga, W. J., et al. (2017). Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. The Journal of Infectious Diseases, 215, 34–41.
Harris, V. C., Haak, B. W., Handley, S. A., Jiang, B., Velasquez, D. E., Hykes, B. L., Jr., Droit, L., Berbers, G. A. M., Kemper, E. M., van Leeuwen, E. M. M., et al. (2018b). Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: A human, randomized-control proof-of-concept trial. Cell Host and Microbe, 24, 197–207.
Hegazy, A. N., West, N. R., Stubbington, M. J. T., Wendt, E., Suijker, K. I. M., Datsi, A., This, S., Danne, C., Campion, S., Duncan, S. H., et al. (2017). Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology, 153, 1320–1337.
Hogan, M. J., & Pardi, N. (2022). mRNA vaccines in the COVID-19 pandemic and beyond. Annual Review of Medicine, 73, 17–39.
Huda, M. N., Ahmad, S. M., Alam, M. J., Khanam, A., Kalanetra, K. M., Taft, D. H., Raqib, R., Underwood, M. A., Mills, D. A., & Stephensen, C. B. (2019). Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics, 143, e20181489.
Isolauri, E., Joensuu, J., Suomalainen, H., Luomala, M., & Vesikari, T. (1995). Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine, 13, 310–312.
Jespersen, L., Tarnow, I., Eskesen, D., Morberg, C. M., Michelsen, B., Bugel, S., Dragsted, L. O., Rijkers, G. T., & Calder, P. C. (2015). Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: A randomized, double-blind, placebo-controlled, parallel-group study. The American Journal of Clinical Nutrition, 101, 1188–1196.
Jia, G., Liu, X., Che, N., Xia, Y., Wang, G., Xiong, Z., Zhang, H., & Ai, L. (2020). Human-origin Lactobacillus salivarius AR809 protects against immunosuppression in S. aureus-induced pharyngitis via Akt-mediated NF-κB and autophagy signaling pathways. Food and Function, 11, 270–284.
Jia, L., Weng, S., Wu, J., Tian, X., Zhang, Y., Wang, X., Wang, J., Yan, D., Wang, W., Fang, F., et al. (2022). Preexisting antibodies targeting SARS-CoV-2 S2 cross-react with commensal gut bacteria and impact COVID-19 vaccine induced immunity. Gut Microbes, 14, 2117503.
Kasturi, S. P., Skountzou, I., Albrecht, R. A., Koutsonanos, D., Hua, T., Nakaya, H. I., Ravindran, R., Stewart, S., Alam, M., Kwissa, M., et al. (2011). Programming the magnitude and persistence of antibody responses with innate immunity. Nature, 470, 543–547.
Khoshnood, S., Ghanavati, R., Shirani, M., Ghahramanpour, H., Sholeh, M., Shariati, A., Sadeghifard, N., & Heidary, M. (2022). Viral vector and nucleic acid vaccines against COVID-19: A narrative review. Frontiers in Microbiology, 13, 984536.
Kim, D., Kim, Y. G., Seo, S. U., Kim, D. J., Kamada, N., Prescott, D., Chamaillard, M., Philpott, D. J., Rosenstiel, P., Inohara, N., et al. (2016a). Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nature Medicine, 22, 524–530.
Kim, C. H., Park, J., & Kim, M. (2014). Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Network, 14, 277–288.
Kim, M., Qie, Y., Park, J., & Kim, C. H. (2016b). Gut microbial metabolites fuel host antibody responses. Cell Host and Microbe, 20, 202–214.
Konieczna, P., Schiavi, E., Ziegler, M., Groeger, D., Healy, S., Grant, R., & O’Mahony, L. (2015). Human dendritic cell DC-SIGN and TLR-2 mediate complementary immune regulatory activities in response to Lactobacillus rhamnosus JB-1. PLoS ONE, 10, e0120261.
Lépine, A. F. P., Konstanti, P., Borewicz, K., Resink, J. W., de Wit, N. J., de Vos, P., Smidt, H., & Mes, J. J. (2019). Combined dietary supplementation of long chain inulin and Lactobacillus acidophilus W37 supports oral vaccination efficacy against Salmonella Typhimurium in piglets. Scientific Reports, 9, 18017.
Lewnard, J. A., Lo, N. C., Arinaminpathy, N., Frost, I., & Laxminarayan, R. (2020). Childhood vaccines and antibiotic use in low- and middle-income countries. Nature, 581, 94–99.
Li, S., Plebanski, M., Smooker, P., & Gowans, E. J. (2015). Editorial: Why vaccines to HIV, HCV, and Malaria have so far failed-challenges to developing vaccines against immunoregulating pathogens. Frontiers in Microbiology, 6, 1318.
Lin, R., Zhang, Y., Chen, L., Qi, Y., He, J., Hu, M., Zhang, Y., Fan, L., Yang, T., Wang, L., et al. (2020). The effects of cigarettes and alcohol on intestinal microbiota in healthy men. Journal of Microbiology, 58, 926–937.
Lind, A. L., & Pollard, K. S. (2021). Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome, 9, 58.
Lloyd-Price, J., Abu-Ali, G., & Huttenhower, C. (2016). The healthy human microbiome. Genome Medicine, 8, 51.
Lloyd-Price, J., Mahurkar, A., Rahnavard, G., Crabtree, J., Orvis, J., Hall, A. B., Brady, A., Creasy, H. H., McCracken, C., Giglio, M. G., et al. (2017). Strains, functions and dynamics in the expanded human microbiome project. Nature, 550, 61–66.
Luu, M., Riester, Z., Baldrich, A., Reichardt, N., Yuille, S., Busetti, A., Klein, M., Wempe, A., Leister, H., Raifer, H., et al. (2021). Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nature Communications, 12, 4077.
Lynch, S. V., & Pedersen, O. (2016). The human intestinal microbiome in health and disease. The New England Journal of Medicine, 375, 2369–2379.
Matsubara, V. H., Ishikawa, K. H., Ando-Suguimoto, E. S., Bueno-Silva, B., Nakamae, A. E. M., & Mayer, M. P. A. (2017). Probiotic bacteria alter pattern-recognition receptor expression and cytokine profile in a human macrophage model challenged with Candida albicans and Lipopolysaccharide. Frontiers in Microbiology, 8, 2280.
Mu, Q., Kirby, J., Reilly, C. M., & Luo, X. M. (2017). Leaky gut as a danger signal for autoimmune diseases. Frontiers in Immunology, 8, 598.
Mullié, C., Yazourh, A., Thibault, H., Odou, M. F., Singer, E., Kalach, N., Kremp, O., & Romond, M. B. (2004). Increased poliovirus-specific intestinal antibody response coincides with promotion of Bifidobacterium longum-infantis and Bifidobacterium breve in infants: A randomized, double-blind, placebo-controlled trial. Pediatric Research, 56, 791–795.
Ng, S. C., Peng, Y., Zhang, L., Mok, C. K., Zhao, S., Li, A., Ching, J. Y., Liu, Y., Yan, S., Chan, D. L. S., et al. (2022). Gut microbiota composition is associated with SARS-CoV-2 vaccine immunogenicity and adverse events. Gut, 71, 1106–1116.
Oh, J. Z., Ravindran, R., Chassaing, B., Carvalho, F. A., Maddur, M. S., Bower, M., Hakimpour, P., Gill, K. P., Nakaya, H. I., Yarovinsky, F., et al. (2014). TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity, 41, 478–492.
Parada Venegas, D., De la Fuente, M. K., Landskron, G., Gonzalez, M. J., Quera, R., Dijkstra, G., Harmsen, H. J. M., Faber, K. N., & Hermoso, M. A. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Frontiers in Immunology, 10, 277.
Park, J., Kim, M., Kang, S. G., Jannasch, A. H., Cooper, B., Patterson, J., & Kim, C. H. (2015). Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunology, 8, 80–93.
Parker, E. P. K., Praharaj, I., Zekavati, A., Lazarus, R. P., Giri, S., Operario, D. J., Liu, J., Houpt, E., Iturriza-Gomara, M., Kampmann, B., et al. (2018). Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine, 36, 264–272.
Pérez, N., Iannicelli, J. C., Girard-Bosch, C., Gonzalez, S., Varea, A., Disalvo, L., Apezteguia, M., Pernas, J., Vicentin, D., & Cravero, R. (2010). Effect of probiotic supplementation on immunoglobulins, isoagglutinins and antibody response in children of low socio-economic status. European Journal of Nutrition, 49, 173–179.
Peroni, D. G., & Morelli, L. (2021). Probiotics as adjuvants in vaccine strategy: Is there more room for improvement? Vaccines, 9, 811.
Plotkin, S. (2014). History of vaccination. Proceedings of the National Academy of Sciences of the United States of America, 111, 12283–12287.
PrabhuDas, M., Adkins, B., Gans, H., King, C., Levy, O., Ramilo, O., & Siegrist, C. A. (2011). Challenges in infant immunity: Implications for responses to infection and vaccines. Nature Immunology, 12, 189–194.
Praharaj, I., Parker, E. P. K., Giri, S., Allen, D. J., Silas, S., Revathi, R., Kaliappan, S. P., John, J., Prasad, J. H., Kampmann, B., et al. (2019). Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: A study from south India. The Journal of Infectious Diseases, 219, 1178–1186.
Pulendran, B. (2009). Learning immunology from the yellow fever vaccine: Innate immunity to systems vaccinology. Nature Reviews Immunology, 9, 741–747.
Pulendran, B. (2019). Immunology taught by vaccines. Science, 366, 1074–1075.
Pulendran, B., Arunachalam, P. S., & O’Hagan, D. T. (2021). Emerging concepts in the science of vaccine adjuvants. Nature Reviews Drug Discovery, 20, 454–475.
Qiu, P., Ishimoto, T., Fu, L., Zhang, J., Zhang, Z., & Liu, Y. (2022). The gut microbiota in inflammatory bowel disease. Frontiers in Cellular and Infection Microbiology, 12, 733992.
Querec, T. D., Akondy, R. S., Lee, E. K., Cao, W., Nakaya, H. I., Teuwen, D., Pirani, A., Gernert, K., Deng, J., Marzolf, B., et al. (2009). Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature Immunology, 10, 116–125.
Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G. A. D., Gasbarrini, A., & Mele, M. C. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms, 7, 14.
Rizzardini, G., Eskesen, D., Calder, P. C., Capetti, A., Jespersen, L., & Clerici, M. (2012). Evaluation of the immune benefits of two probiotic strains Bifidobacterium animalis ssp. lactis, BB-12® and Lactobacillus paracasei ssp. paracasei, L. casei 431® in an influenza vaccination model: A randomised, double-blind, placebo-controlled study. The British Journal of Nutrition, 107, 876–884.
Robertson, R. C., Church, J. A., Edens, T. J., Mutasa, K., Min Geum, H., Baharmand, I., Gill, S. K., Ntozini, R., Chasekwa, B., Carr, L., et al. (2021). The fecal microbiome and rotavirus vaccine immunogenicity in rural Zimbabwean infants. Vaccine, 39, 5391–5400.
Rodrigues, C. M. C., & Plotkin, S. A. (2020). Impact of vaccines; health, economic and social perspectives. Frontiers in Microbiology, 11, 1526.
Rodriguez-Blanque, R., Sánchez-García, J. C., Cobos-Vargas, A., Aguilar Quesada, A., Maldonado-Lobon, J. A., Olivares, M., & Blanco-Rojo, R. (2022). Evaluation of the effect of Loigolactobacillus coryniformis K8 CECT 5711 consumption in health care workers exposed to COVID-19. Frontier in Nutrition, 9, 962566.
Roger, L. C., Costabile, A., Holland, D. T., Hoyles, L., & McCartney, A. L. (2010). Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology, 156, 3329–3341.
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., Costea, P. I., Godneva, A., Kalka, I. N., Bar, N., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature, 555, 210–215.
Rutayisire, E., Huang, K., Liu, Y., & Tao, F. (2016). The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review. BMC Gastroenterology, 16, 86.
Salazar, N., Valdés-Varela, L., González, S., Gueimonde, M., & de Los Reyes-Gavilan, C. G. (2017). Nutrition and the gut microbiome in the elderly. Gut Microbes, 8, 82–97.
Sanchez, H. N., Moroney, J. B., Gan, H., Shen, T., Im, J. L., Li, T., Taylor, J. R., Zan, H., & Casali, P. (2020). B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nature Communications, 11, 60.
Soh, S. E., Ong, D. Q., Gerez, I., Zhang, X., Chollate, P., Shek, L. P., Lee, B. W., & Aw, M. (2010). Effect of probiotic supplementation in the first 6 months of life on specific antibody responses to infant hepatitis B vaccination. Vaccine, 28, 2577–2579.
Song, E. J., Lee, E. S., & Nam, Y. D. (2018). Progress of analytical tools and techniques for human gut microbiome research. Journal of Microbiology, 56, 693–705.
Spinler, J. K., Taweechotipatr, M., Rognerud, C. L., Ou, C. N., Tumwasorn, S., & Versalovic, J. (2008). Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe, 14, 166–171.
Su, L. F., Kidd, B. A., Han, A., Kotzin, J. J., & Davis, M. M. (2013). Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity, 38, 373–383.
Swain, S. L., McKinstry, K. K., & Strutt, T. M. (2012). Expanding roles for CD4+ T cells in immunity to viruses. Nature Reviews Immunology, 12, 136–148.
Uehara, O., Abiko, Y., Nagasawa, T., Morikawa, T., Hiraki, D., Harada, F., Kawano, Y., Toraya, S., Matsuoka, H., Paudel, D., et al. (2022). Alterations in the oral microbiome of individuals with a healthy oral environment following COVID-19 vaccination. BMC Oral Health, 22, 50.
Ursell, L. K., Metcalf, J. L., Parfrey, L. W., & Knight, R. (2012). Defining the human microbiome. Nutrition Reviews, 70, S38–S44.
Van Puyenbroeck, K., Hens, N., Coenen, S., Michiels, B., Beunckens, C., Molenberghs, G., Van Royen, P., & Verhoeven, V. (2012). Efficacy of daily intake of Lactobacillus casei Shirota on respiratory symptoms and influenza vaccination immune response: A randomized, double-blind, placebo-controlled trial in healthy elderly nursing home residents. The American Journal of Clinical Nutrition, 95, 1165–1171.
Vatanen, T., Kostic, A. D., d’Hennezel, E., Siljander, H., Franzosa, E. A., Yassour, M., Kolde, R., Vlamakis, H., Arthur, T. D., Hamalainen, A. M., et al. (2016). Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell, 165, 842–853.
Wensel, C. R., Pluznick, J. L., Salzberg, S. L., & Sears, C. L. (2022). Next-generation sequencing: Insights to advance clinical investigations of the microbiome. The Journal of Clinical Investigation, 132, e154944.
Wilkinson, T. M., Li, C. K., Chui, C. S., Huang, A. K., Perkins, M., Liebner, J. C., Lambkin-Williams, R., Gilbert, A., Oxford, J., Nicholas, B., et al. (2012). Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nature Medicine, 18, 274–280.
Wimmers, F., & Pulendran, B. (2020). Emerging technologies for systems vaccinology-multi-omics integration and single-cell (epi)genomic profiling. Current Opinion in Immunology, 65, 57–64.
Wu, Z., Zhang, Q., Yang, J., Zhang, J., Fu, J., Dang, C., Liu, M., Wang, S., Lin, Y., Hao, J., et al. (2022). Significant alterations of intestinal symbiotic microbiota induced by intraperitoneal vaccination mediate changes in intestinal metabolism of NEW genetically improved farmed tilapia (NEW GIFT, Oreochromis niloticus). Microbiome, 10, 221.
Xu, J., Ren, Z., Cao, K., Li, X., Yang, J., Luo, X., Zhu, L., Wang, X., Ding, L., Liang, J., et al. (2021). Boosting vaccine-elicited respiratory mucosal and systemic COVID-19 immunity in mice with the oral Lactobacillus plantarum. Frontiers in Nutrition, 8, 789242.
Yang, W., Yu, T., Huang, X., Bilotta, A. J., Xu, L., Lu, Y., Sun, J., Pan, F., Zhou, J., Zhang, W., et al. (2020). Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nature Communications, 11, 4457.
Zeng, M. Y., Inohara, N., & Nunez, G. (2017). Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunology, 10, 18–26.
Zhang, N., & Bevan, M. J. (2011). CD8+ T cells: Foot soldiers of the immune system. Immunity, 35, 161–168.
Zhao, T., Li, J., Fui, Y., Ye, H., Liu, X., Li, G., Yang, X., & Yang, J. (2020). Influence of gut microbiota on mucosal IgA antibody response to the polio vaccine. npj Vaccines, 5, 47.
Zhao, H., Wang, D., Zhang, Z., Xian, J., & Bai, X. (2022). Effect of gut microbiota-derived metabolites on immune checkpoint inhibitor therapy: Enemy or friend? Molecules, 27, 4799.
Zheng, D., Liwinski, T., & Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Research, 30, 492–506.
Zimmermann, P., & Curtis, N. (2019). Factors that influence the immune response to vaccination. Clinical Microbiology Reviews, 32, e00084-18.
Zimmermann, P., Perrett, K. P., Ritz, N., Flanagan, K. L., Robins-Browne, R., van der Klis, F. R. M., Curtis, N., & the MIS BAIR group. (2020). Biological sex influences antibody responses to routine vaccinations in the first year of life. Acta Paediatrica, 109, 147–157.
Zitvogel, L., & Kroemer, G. (2021). Cross-reactivity between cancer and microbial antigens. Oncoimmunology, 10, 1877416.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01041639). Figures were created using BioRender.com and modified by Ji Young Bang and Yun Ji Kim.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no commercial or financial relationships that could be construed as potential conflicts of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, SH. Influence of Microbiota on Vaccine Effectiveness: “Is the Microbiota the Key to Vaccine-induced Responses?”. J Microbiol. 61, 483–494 (2023). https://doi.org/10.1007/s12275-023-00044-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00044-6