Abstract
Background
The N7-methylguanosine modification (m7G) of the 5′ cap structure in the mRNA plays a crucial role in gene expression. However, the relation between m7G and tumor immune remains unclear. Hence, we intended to perform a pan-cancer analysis of m7G which can help explore the underlying mechanism and contribute to predictive, preventive, and personalized medicine (PPPM / 3PM).
Methods
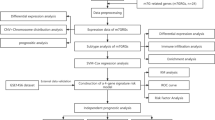
The gene expression, genetic variation, clinical information, methylation, and digital pathological section from 33 cancer types were downloaded from the TCGA database. Immunohistochemistry (IHC) was used to validate the expression of the m7G regulator genes (m7RGs) hub-gene. The m7G score was calculated by single-sample gene-set enrichment analysis. The association of m7RGs with copy number variation, clinical features, immune-related genes, TMB, MSI, and tumor immune dysfunction and exclusion (TIDE) was comprehensively assessed. CellProfiler was used to extract pathological section characteristics. XGBoost and random forest were used to construct the m7G score prediction model. Single-cell transcriptome sequencing (scRNA-seq) was used to assess the activation state of the m7G in the tumor microenvironment.
Results
The m7RGs were highly expressed in tumors and most of the m7RGs are risk factors for prognosis. Moreover, the cellular pathway enrichment analysis suggested that m7G score was closely associated with invasion, cell cycle, DNA damage, and repair. In several cancers, m7G score was significantly negatively correlated with MSI and TMB and positively correlated with TIDE, suggesting an ICB marker potential. XGBoost-based pathomics model accurately predicts m7G scores with an area under the ROC curve (AUC) of 0.97. Analysis of scRNA-seq suggests that m7G differs significantly among cells of the tumor microenvironment. IHC confirmed high expression of EIF4E in breast cancer. The m7G prognostic model can accurately assess the prognosis of tumor patients with an AUC of 0.81, which was publicly hosted at https://pan-cancer-m7g.shinyapps.io/Panca-m7g/.
Conclusion
The current study explored for the first time the m7G in pan-cancer and identified m7G as an innovative marker in predicting clinical outcomes and immunotherapeutic efficacy, with the potential for deeper integration with PPPM. Combining m7G within the framework of PPPM will provide a unique opportunity for clinical intelligence and new approaches.
















Similar content being viewed by others
Data availability
The data supporting the findings of this study are deposited in TCGA and GEO databases. The single cell sequencing datasets can be found in online repositories of GEO (GSE152938).
Code availability
The analyses methods and used packages are illustrated in the “Materials and methods” section. All other R and Python code and analyses are available from the corresponding author upon request.
References
Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer. 2021;127:3029–30. https://doi.org/10.1002/cncr.33587.
Cheng X, et al. Systematic pan-cancer analysis of KLRB1 with prognostic value and immunological activity across human tumors. J Immunol Res. 2022;2022:5254911. https://doi.org/10.1155/2022/5254911.
Grech G, et al. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6:9. https://doi.org/10.1186/s13167-015-0030-6.
Hamid O, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–44. https://doi.org/10.1056/NEJMoa1305133.
Doebele RC, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5:1049–57. https://doi.org/10.1158/2159-8290.Cd-15-0443.
Chen F, et al. Pan-cancer analysis of the prognostic and immunological role of HSF1: a potential target for survival and immunotherapy. Oxid Med Cell Longev. 2021;2021:5551036. https://doi.org/10.1155/2021/5551036.
Zhou X, et al. A pan-cancer analysis of CD161, a potential new immune checkpoint. Front Immunol. 2021;12:688215. https://doi.org/10.3389/fimmu.2021.688215.
Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. https://doi.org/10.1038/ng.2764.
Bodrova TA, et al. Introduction into PPPM as a new paradigm of public health service: an integrative view. EPMA J. 2012;3:16. https://doi.org/10.1186/1878-5085-3-16.
Malbec L, et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 2019;29:927–41. https://doi.org/10.1038/s41422-019-0230-z.
Lin S, et al. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244-255.e245. https://doi.org/10.1016/j.molcel.2018.06.001.
Liu Y, Zhang Y, Chi Q, Wang Z, Sun B. Methyltransferase-like 1 (METTL1) served as a tumor suppressor in colon cancer by activating 7-methyguanosine (m7G) regulated let-7e miRNA/HMGA2 axis. Life Sci. 2020;249:117480. https://doi.org/10.1016/j.lfs.2020.117480.
Qin MM, et al. let-7i inhibits proliferation and migration of bladder cancer cells by targeting HMGA1. BMC Urol. 2019;19:53. https://doi.org/10.1186/s12894-019-0485-1.
Elghoroury EA, et al. Evaluation of miRNA-21 and miRNA Let-7 as prognostic markers in patients with breast cancer. Clin Breast Cancer. 2018;18:e721–6. https://doi.org/10.1016/j.clbc.2017.11.022.
Ma L, Zhao Q, Chen W, Zhang Y. Oncogene Lin28B increases chemosensitivity of colon cancer cells in a let-7-independent manner. Oncol Lett. 2018;15:6975–81. https://doi.org/10.3892/ol.2018.8250.
Huang Y, et al. METTL1 promotes neuroblastoma development through m(7)G tRNA modification and selective oncogenic gene translation. Biomarker research. 2022;10:68. https://doi.org/10.1186/s40364-022-00414-z.
Wang C, et al. Methyltransferase-like 1 regulates lung adenocarcinoma A549 cell proliferation and autophagy via the AKT/mTORC1 signaling pathway. Oncol Lett. 2021;21:330. https://doi.org/10.3892/ol.2021.12591.
Wang T, et al. Comprehensive analysis of nine m7G-related lncRNAs as prognosis factors in tumor immune microenvironment of hepatocellular carcinoma and experimental validation. Front Genet. 2022;13: 929035. https://doi.org/10.3389/fgene.2022.929035.
Chen M, et al. m7G regulator-mediated molecular subtypes and tumor microenvironment in kidney renal clear cell carcinoma. Front Pharmacol. 2022;13: 900006. https://doi.org/10.3389/fphar.2022.900006.
Su C, et al. Single-cell RNA sequencing in multiple pathologic types of renal cell carcinoma revealed novel potential tumor-specific markers. Front Oncol. 2021;11:719564. https://doi.org/10.3389/fonc.2021.719564.
Human genomics. The genotype-tissue expression (GTEx) pilot analysis. Science (New York, NY). 2015;348:648–60. https://doi.org/10.1126/science.1262110.
Linehan WM, Ricketts CJ. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16:539–52. https://doi.org/10.1038/s41585-019-0211-5.
Blum A, Wang P, Zenklusen J. C. SnapShot: TCGA-analyzed tumors. Cell. 2018;173:530. https://doi.org/10.1016/j.cell.2018.03.059.
Yang W, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955-961. https://doi.org/10.1093/nar/gks1111.
Fu Y, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nature cancer. 2020;1:800–10. https://doi.org/10.1038/s43018-020-0085-8.
Carpenter AE, et al. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. https://doi.org/10.1186/gb-2006-7-10-r100.
Rizvi AA, et al. gwasurvivr: an R package for genome-wide survival analysis. Bioinformatics (Oxford, England). 2019;35:1968–70. https://doi.org/10.1093/bioinformatics/bty920.
van Dyk E, Reinders MJ, Wessels LF. A scale-space method for detecting recurrent DNA copy number changes with analytical false discovery rate control. Nucleic Acids Res. 2013;41:e100. https://doi.org/10.1093/nar/gkt155.
Liu CJ, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics (Oxford, England). 2018;34:3771–2. https://doi.org/10.1093/bioinformatics/bty411.
Zhuang W, et al. An immunogenomic signature for molecular classification in hepatocellular carcinoma. Molecular therapy Nucleic acids. 2021;25:105–15. https://doi.org/10.1016/j.omtn.2021.06.024.
Wang G, et al. m7G-Associated subtypes, tumor microenvironment, and validation of prognostic signature in lung adenocarcinoma. Front Genet. 2022;13: 954840. https://doi.org/10.3389/fgene.2022.954840.
Xiao B, et al. Identification and verification of immune-related gene prognostic signature based on ssGSEA for osteosarcoma. Front Oncol. 2020;10: 607622. https://doi.org/10.3389/fonc.2020.607622.
Sherif S, et al. The immune landscape of solid pediatric tumors. J Exp Clin Cancer Res: CR. 2022;41:199. https://doi.org/10.1186/s13046-022-02397-z.
Misund K, et al. Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence. Leukemia. 2022;36:1887–97. https://doi.org/10.1038/s41375-022-01597-y.
Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can Res. 2017;77:e108–10. https://doi.org/10.1158/0008-5472.Can-17-0307.
Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-w102. https://doi.org/10.1093/nar/gkx247.
Yuan H, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900-d908. https://doi.org/10.1093/nar/gky939.
Patel SS, Lovko VJ, Lockey RF. Red Tide: overview and clinical manifestations. J Allergy Clin Immunol In practice. 2020;8:1219–23. https://doi.org/10.1016/j.jaip.2019.10.030.
Cao S, et al. Estimation of tumor cell total mRNA expression in 15 cancer types predicts disease progression. Nat Biotechnol. 2022;40(11):1624-1633. https://doi.org/10.1038/s41587-022-01342-x.
Mo X, et al. Immune infiltration and immune gene signature predict the response to fluoropyrimidine-based chemotherapy in colorectal cancer patients. Oncoimmunology. 2020;9:1832347. https://doi.org/10.1080/2162402x.2020.1832347.
Bishara AJ, Hittner JB. Testing the significance of a correlation with nonnormal data: comparison of Pearson, Spearman, transformation, and resampling approaches. Psychol Methods. 2012;17:399–417. https://doi.org/10.1037/a0028087.
Dantan E, et al. An original approach was used to better evaluate the capacity of a prognostic marker using published survival curves. J Clin Epidemiol. 2014;67:441–8. https://doi.org/10.1016/j.jclinepi.2013.10.022.
Fisher LD, Lin DY. Time-dependent covariates in the Cox proportional-hazards regression model. Annu Rev Public Health. 1999;20:145–57. https://doi.org/10.1146/annurev.publhealth.20.1.145.
Song M, Vogelstein B, Giovannucci EL, Willett WC, Tomasetti C. Cancer prevention: molecular and epidemiologic consensus. Science (New York, NY). 2018;361:1317–8. https://doi.org/10.1126/science.aau3830.
Cirillo D, Valencia A. Big data analytics for personalized medicine. Curr Opin Biotechnol. 2019;58:161–7. https://doi.org/10.1016/j.copbio.2019.03.004.
Carbine NE, Lostumbo L, Wallace J, Ko H. Risk-reducing mastectomy for the prevention of primary breast cancer. Cochrane Database Syst Rev. 2018;4:CD002748. https://doi.org/10.1002/14651858.CD002748.pub4.
He J, Huang Z, Han L, Gong Y, Xie C. Mechanisms and management of 3rd‑generation EGFR‑TKI resistance in advanced non‑small cell lung cancer (Review). Int J Oncol 2021; 59, https://doi.org/10.3892/ijo.2021.5270.
Argelaguet R, et al. Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 2018;14: e8124. https://doi.org/10.15252/msb.20178124.
Lakshminarasimhan R, Liang G. The role of DNA methylation in cancer. Adv Exp Med Biol. 2016;945:151–72. https://doi.org/10.1007/978-3-319-43624-1_7.
Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119–31. https://doi.org/10.1038/s41576-020-00295-8.
Xie S, et al. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int. 2020;20:585. https://doi.org/10.1186/s12935-020-01679-w.
Huang Y, et al. Exploration of potential roles of m5C-related regulators in colon adenocarcinoma prognosis. Front Genet. 2022;13: 816173. https://doi.org/10.3389/fgene.2022.816173.
Orellana EA, et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323-3338.e3314. https://doi.org/10.1016/j.molcel.2021.06.031.
Wang H, Franco F, Ho PC. Metabolic regulation of Tregs in cancer: opportunities for immunotherapy. Trends Cancer. 2017;3:583–92. https://doi.org/10.1016/j.trecan.2017.06.005.
Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–9. https://doi.org/10.1111/cas.14069.
Gao L, et al. Lung cancer deficient in the tumor suppressor GATA4 is sensitive to TGFBR1 inhibition. Nat Commun. 2019;10:1665. https://doi.org/10.1038/s41467-019-09295-7.
Grygielko ET, et al. Inhibition of gene markers of fibrosis with a novel inhibitor of transforming growth factor-beta type I receptor kinase in puromycin-induced nephritis. J Pharmacol Exp Ther. 2005;313:943–51. https://doi.org/10.1124/jpet.104.082099.
Saraiva M, Vieira P, O'Garra A. Biology and therapeutic potential of interleukin-10. J Exp Med 2020; 217, https://doi.org/10.1084/jem.20190418.
Itakura E, et al. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod Pathol. 2011;24:801–9. https://doi.org/10.1038/modpathol.2011.5.
Choueiry F et al. CD200 promotes immunosuppression in the pancreatic tumor microenvironment. J Immunother Cancer 2020; 8, https://doi.org/10.1136/jitc-2019-000189.
Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–70. https://doi.org/10.1016/j.molcel.2019.09.030.
Lin A, Zhang J, Luo P. Crosstalk between the MSI status and tumor microenvironment in colorectal cancer. Front Immunol. 2020;11:2039. https://doi.org/10.3389/fimmu.2020.02039.
Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39:154–73. https://doi.org/10.1016/j.ccell.2020.10.001.
Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–8. https://doi.org/10.1038/s41591-018-0136-1.
König IR, Fuchs O, Hansen G, von Mutius E & Kopp MV. What is precision medicine? Eur Respir J 2017; 50, https://doi.org/10.1183/13993003.00391-2017.
Han DS, et al. Nomogram predicting long-term survival after d2 gastrectomy for gastric cancer. J Clin Oncol: Off J Am Soc Clin Oncol. 2012;30:3834–40. https://doi.org/10.1200/jco.2012.41.8343.
Funding
This work received financial support from Guangxi Medical and Health Appropriate Technology Development and Promotion Application Project (S2021016); Guangxi Natural Science Foundation (2021JJA140081); and Basic Research Skills Enhancement Project for Young and Middle-aged Teachers in Universities in Guangxi (2021KY0087).
Author information
Authors and Affiliations
Contributions
Xiaoliang Huang, Zuyuan Chen, Xiaoyun Xiang, Yanling Liu, Xingqing Long, Xianwei Mo, Weizhong Tang Jungang Liu, Kezhen Li, and Mingjian Qin: conceived and designed the experiments; Xiaoliang Huang, Zuyuan Chen, Xiaoyun Xiang, Yanling Liu, Xingqing Long, Xianwei Mo, Chenyan Long, Weizhong Tang: analyzed the data; Xiaoliang Huang, Zuyuan Chen, Xiaoyun Xiang, Yanling Liu, Xingqing Long, Xianwei Mo, Weizhong Tang, Jungang Liu, Kezhen Li and Mingjian Qin: helped with reagents/materials/analysis tools; Xiaoliang Huang, Zuyuan Chen, Xiaoyun Xiang, Yanling Liu, Xingqing Long, Xianwei Mo, Chenyan Long, Weizhong Tang, Jungang Liu, Kezhen Li and Mingjian Qin: contributed to the writing of the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics and Human Subject Committee of Guangxi Medical University Cancer Hospital.
Consent for publication
Not applicable.
Conflict of interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, X., Chen, Z., Xiang, X. et al. Comprehensive multi-omics analysis of the m7G in pan-cancer from the perspective of predictive, preventive, and personalized medicine. EPMA Journal 13, 671–697 (2022). https://doi.org/10.1007/s13167-022-00305-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-022-00305-1