Abstract
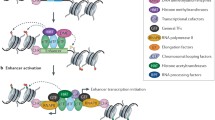
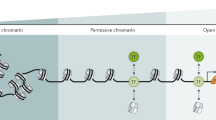
During organism development, a diversity of cell types emerges with disparate, yet stable profiles of gene expression with distinctive cellular functions. In addition to gene promoters, the genome contains enhancer regulatory sequences, which are implicated in cellular specialization by facilitating cell-type and tissue-specific gene expression. Enhancers are DNA binding elements characterized by highly sophisticated and various mechanisms of action allowing for the specific interaction of general and tissue-specific transcription factors (TFs). However, eukaryotic organisms package their genetic material into chromatin, generating a physical barrier for TFs to interact with their cognate sequences. The ability of TFs to bind DNA regulatory elements is also modulated by changes in the chromatin structure, including histone modifications, histone variants, ATP-dependent chromatin remodeling, and the methylation status of DNA. Furthermore, it has recently been revealed that enhancer sequences are also transcribed into a set of enhancer RNAs with regulatory potential. These interdependent processes act in the context of a complex network of chromatin interactions, which together contributes to a renewed vision of how gene activation is coordinated in a cell-type-dependent manner. In this review, we describe the interplay between genetic and epigenetic aspects associated with enhancers and discuss their possible roles on enhancer function.



Similar content being viewed by others
References
Khoury G, Gruss P (1983) Enhancer elements. Cell 33:313–314
Banerji J, Rusconi S, Schaffner W (1981) Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299–308
Banerji J, Olson L, Schaffner W (1983) A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33:729–740
Gillies SD, Morrison SL, Oi VT, Tonegawa S (1983) A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33:717–728
Walker MD, Edlund T, Boulet AM, Rutter WJ (1983) Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature 306:557–561
Edlund T, Walker MD, Barr PJ, Rutter WJ (1985) Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science 230:912–916
Goodbourn S, Zinn K, Maniatis T (1985) Human β-interferon gene expression is regulated by an inducible enhancer element. Cell 41:509–520
Schöler HR, Gruss P (1985) Cell type-specific transcriptional enhancement in vitro requires the presence of trans-acting factors. EMBO J 4:3305–3313
Ko MSH, Nakauchi H, Takahashi N (1990) The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J 9:2835–2842
Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DIK (1995) Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA 92:7125–7129
Sutherland HGE, Martin DIK, Whitelaw E (1997) A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol Cell Biol 3:1607–1614
Yie J, Senger K, Thanos D (1999) Mechanism by which the IFN-β enhanceosome activates transcription. Proc Natl Acad Sci USA 96:13108–13113
Sandaltzopoulos R, Becker PB (1998) Heat shock increases the reinitiation rate from potentiated chromatin templates. Mol Cell Biol 18:361–367
Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K (2012) Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22:490–503
Bulger M, Groudine M (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144:327–339
Ong CT, Corces VG (2011) Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12:283–293
De Laat W, Duboule D (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502:499–506
Calo E, Wysocka J (2013) Modification of enhancer chromatin: what, how and why? Mol Cell 49:825–837
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318
Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN et al (2009) ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28:1418–1428
Soufi A, Donahue G, Zaret KS (2012) Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151:994–1004
Spitz F, Furlong EE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13:613–626
Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161:555–568
Arnosti DN, Kulkarni MM (2005) Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94:890–898
Merika M, Williams AJ, Chen G, Collins T, Thanos D (1998) Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell 1:277–287
Panne D (2008) The enhaceosome. Curr Opin Struct Biol 18:236–242
Vega VB, Lin CY, Lai KS, Kong SL, Xie M, Su X et al (2006) Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol 7:R82
Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L et al (2010) Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol 6:456
Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241
Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A et al (2014) Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 7:1443–1455
Adams CC, Workman JL (1995) Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol 15:1405–1421
Bell O, Tiwari VK, Thomä NH, Schübeler D (2011) Determinants and dynamics of genome accessibility. Nat Rev Genet 12:554–564
Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Iwafuchi-Doi M, Zaret KS (2014) Pioneer transcription factors in cell reprogramming. Genes Dev 28:2679–2692
You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA (2011) OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci USA 108:14497–14502
Sammons MA, Zhu J, Drake AM, Berger SL (2014) TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res 25:179–188
Smith E, Shilatifard A (2014) Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21:210–219
Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB et al (2011) Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell 147:1283–1294
Malik S, Roeder RG (2005) Dynamic regulation of Pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30:256–263
Weake VM, Workman JL (2010) Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11:426–437
Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin remodeling complexes. Mol Cell Biol 5:1899–1910
Näär AM, Lemon BD, Tijan R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70:475–501
Becker PB, Hörz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71:247–273
ENCODE Project Consortium et al (2007) Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature 447:799–816
Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S et al (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903
Koch F, Andrau JC (2011) Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription 2:263–268
Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H et al (2011) H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30:4198–4210
Core LJ, Martins AL, Dank CG, Waters CT, Siepel A, Lis JT (2014) Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46:1311–1320
Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ et al (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107:21931–21936
Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21:1273–1283
Bogdanovic O, Fernandez-Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I et al (2012) Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22:2043–2053
Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N et al (2012) Tissue specific analysis of chromatin states identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44:148–156
Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT et al (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482:221–225
Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M et al (2010) NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem 285:15966–15977
Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR (2011) Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol 18:1358–1365
Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X et al (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 488:718–722
Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z et al (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717
Lyko F, Ramsahoye BA, Jaenisch R (2000) DNA methylation in Drosophila melanogaster. Nature 408:538–540
Cho H, Orphanides G, Sun X, Yang X-J, Ogryzko V, Lees E et al (1998) A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol 18:5355–5363
Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A et al (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858
Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D et al (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361–372
Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A et al (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136:3131–3141
Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE et al (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CB/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30:249–262
Holmqvist P-H, Mannervik M (2013) Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription 4:18–23
Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J (2014) Transcriptional coregulators: fine-tuning metabolism. Cell Metab 20:26–40
Cai L, Sutter BM, Li B, Tu BP (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42:426–437
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187
Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L (2011) SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell 44:410–423
Daitoku H, Sakamaki J, Fukamizu A (2001) Regulation of FoxO transcription factors by acetylation and protein–protein interactions. Biochem Biophys Acta 1813:1954–1960
Speranzini V, Pilotto S, Sixma TK, Mattevi A (2016) Touch, act and go: landing and operating on nucleosomes. EMBO J 35:376–388
Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ (2012) Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 32:2323–2334
Hödl M, Basler K (2012) Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol 22:2253–2257
Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J (2013) A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339:698–699
Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR (2012) Dynamics and memory of heterochromatin in living cells. Cell 149:1447–1460
Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z et al (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132:311–322
Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H et al (2006) Genome-scale mapping of DNase I sensitivity in vivo using DNA microarrays. Nat Methods 3:511–518
Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E et al (2012) The accessible chromatin landscape of human genome. Nature 489:75–82
He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q et al (2010) Nucleosome dynamics define transcriptional enhancers. Nat Genet 42:343–347
Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ et al (2008) Nucleosome organization in the Drosophila genome. Nature 453:358–364
Iyer V, Struhl K (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14:2570–2579
Anderson JD, Widom J (2001) Poly (dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol 21:3830–3839
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304
Shones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z et al (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887–898
Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641–645
Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X et al (2013) Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152:248–261
Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB et al (2004) Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36:955–957
Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG et al (2010) CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6:e1001023
Escamilla-Del-Arenal M, Recillas-Targa F (2008) GATA-1 modulates the chromatin structure and activity of the chicken α-globin 3′ enhancer. Mol Cell Biol 28:575–586
García-González E, Recillas-Targa F (2014) A regulatory element affects the activity and chromatin structure of the chicken α-globin 3′ enhancer. Biochim Biophys Acta 1839:1233–1241
Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ (2006) Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J 399:297–304
Kim SI, Bresnick EH, Bultman SJ (2009) BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res 37:6019–6027
Almer A, Rudolph H, Hinnen A, Hörz W (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction release additional upstream activating DNA elements. EMBO J 5:2689–2696
Richard-Foy H, Hager GL (1987) Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6:2321–2328
Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257
Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A et al (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511:611–615
Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A et al (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480:490–495
Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B et al (2012) An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489:83–90
Bock C, Beerman I, Lien W-H, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A (2012) DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 47:633–647
Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL et al (2013) Genome-wide analysis reveals TET-and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153:692–706
Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C et al (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152:1146–1159
Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C et al (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21:555–565
Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE (2011) 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12:R54
Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A et al (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149:1368–1380
Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M et al (2011) Genome wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473:394–397
Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K et al (2011) Genome wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25:679–684
Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I et al (2011) Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One 6:e21306
Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N (2012) MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151:1417–1430
Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J et al (2013) DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503:371–376
Arab K, Park YT, Lindroth AM, Schäfer A, Oakes C et al (2014) Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55:604–614
De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK et al (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8:e1000384
Natoli G, Andrau JC (2012) Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19
Mousavi K, Zare H, Koulnis M, Sartorelli V (2014) The emerging roles of eRNAs in transcriptional regulatory networks. RNA Biol 11:106–110
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106:11667–11672
Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G et al (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA et al (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494:497–501
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB et al (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18:1433–1445
Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT et al (2011) A rapid, extensive and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145:622–634
Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y et al (2013) Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498:511–515
Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY et al (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498:516–520
Mousavi K, Zare H, Dell’Orso S, Grontved L, Gutierrez-Cruz G, Derfoul A et al (2013) eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617
Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA et al (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49:524–535
Lam MTY, Li W, Rosenfeld MG, Glass CK (2014) Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39:170–182
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A et al (2012) Landscape of transcription in human cells. Nature 489:101–108
Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD et al (2013) Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51:310–325
Schaukowitch K, Joo JY, Liu X et al (2014) Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 56:29–41
Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J et al (2014) An atlas of active enhancers across human cell types and tissues. Nature 507:455–461
Jiang C, Pugh F (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10:161–172
Jansen A, Verstrepen KJ (2011) Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 75:301–320
Huang H, Liu H, Sun X (2013) Nucleosome distribution near the 3′ ends of genes in the human genome. Biosci Biotechnol Biochem 77:2051–2055
Duttke SH, Lacadie SA, Ibrahaim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U (2015) Human promoters are intrinsically directional. Mol Cell 57:674–684
Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY et al (2015) Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518:350–354
Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA et al (2012) Intragenic enhancers act as alternative promoters. Mol Cell 45:447–458
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL et al (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435
Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE (2010) The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol 342:146–156
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319
Malik S, Roeder RG (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11:761–772
Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M (2000) p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev 14:1899–1907
Pott S, Lieb JD (2015) What are super-enhancers? Nat Genet 47:8–12
Allen BL, Taatjes DJ (2015) The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16:155–166
Fiering S, Whitelaw E, Martin DI (2000) To be or not to be active: the stochastic nature of enhancer action. BioEssays 4:381–387
Plank JL, Dean A (2014) Enhancer function: mechanistic and genome-wide insights come together. Mol Cell 55:5–14
Heinz S, Romanoski CE, Benner C, Glass CK (2015) The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16:144–154
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N et al (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385
Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M et al (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472
Bouwman BA, de Laat W (2015) Getting the genome in shape: the formation of loops, domain and compartments. Genome Biol 16:154
Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K et al (2014) Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512:91–95
Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L et al (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47:598–606
Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY et al (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503:290–294
Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre BM et al (2015) The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res 25:582–597
Ghavi-Helm Y, Klein FA, Pakodzi T, Ciglar L, Noordermeer D, Huber W et al (2014) Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512:96–100
Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU et al (2015) CRISPR inversion of CTCF sites alters genome topology an enhancer/promoter function. Cell 162:900–910
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E et al (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025
Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489:109–113
Acknowledgments
We acknowledge Karin Meier for critical reading of the manuscript. This work was supported by the DGAPA-PAPIIT, UNAM (IN209403, IN203811 and IN201114), CONACyT (42653-Q, 128464 and 220503) and Fronteras de la Ciencia-2015 (Grant 290) to FR-T, and by a PhD fellowship from CONACyT and Programa de Apoyo a los Estudios del Posgrado (PAEP), UNAM to EG-G and RA-M. Additional support was provided by the Ph.D. Graduate Program, “Doctorado en Ciencias Bioquímicas y Biomédicas”, to the Instituto de Fisiología Celular, Universidad Nacional Autónoma de México.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
García-González, E., Escamilla-Del-Arenal, M., Arzate-Mejía, R. et al. Chromatin remodeling effects on enhancer activity. Cell. Mol. Life Sci. 73, 2897–2910 (2016). https://doi.org/10.1007/s00018-016-2184-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2184-3